Theater in the Self-Cleaning Cell: Intrinsically Disordered Proteins or Protein Regions Acting with Membranes in Autophagy
Abstract
:1. Introduction
2. Extended Disordered Conformations Bind to Membranes via Coupling of Electrostatic and Hydrophobic Interactions
3. Disorder-To-Order Transitions at the Protein-Membrane Interface
4. Membrane Anchoring of Disordered Proteins and Regions by Posttranslational Modifications
5. Membrane Remodeling Orchestrated by Extended Disordered Conformations
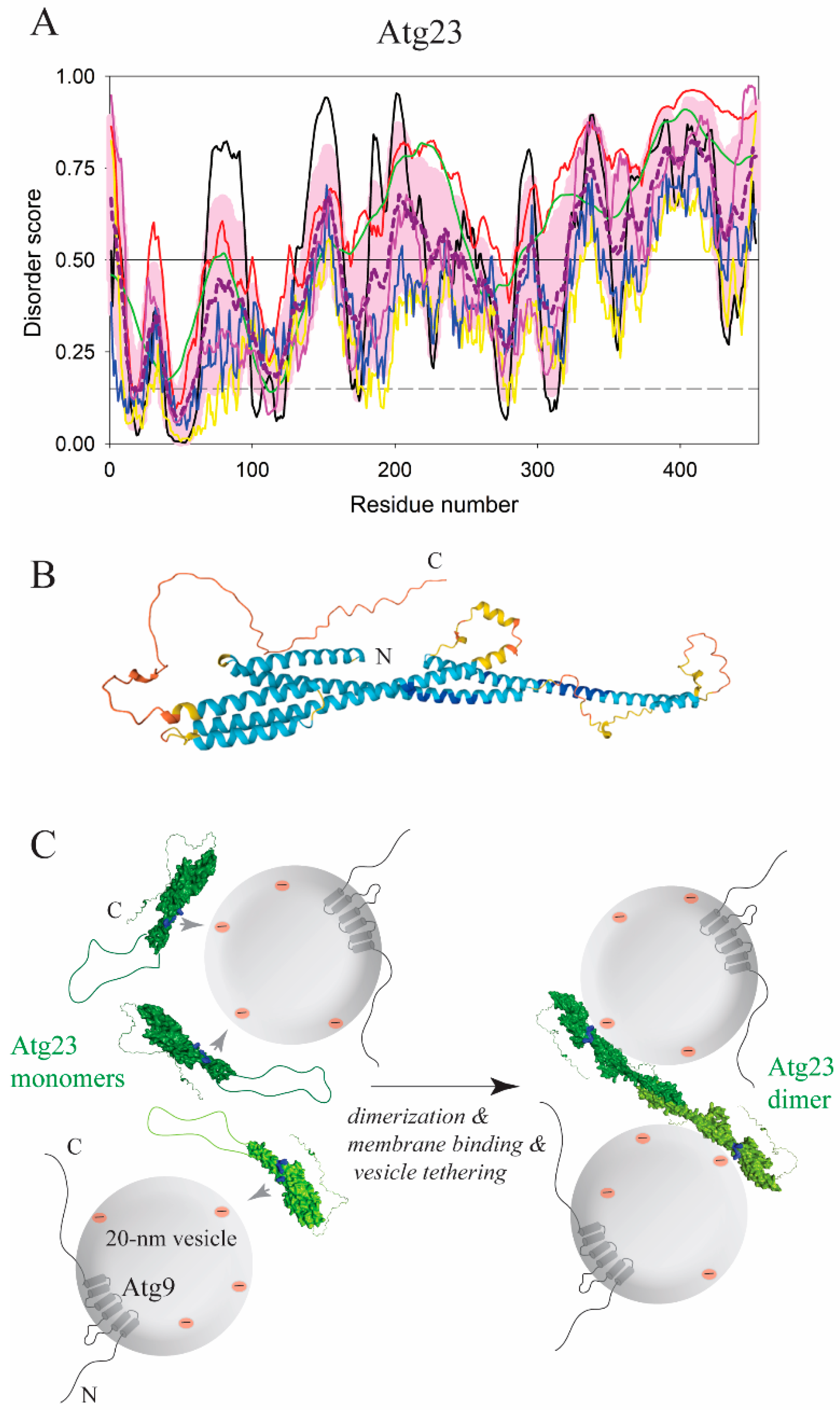
6. Unsolved Mechanisms of Membrane Association of Autophagy Proteins via a Disordered Region
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Dunker, A.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002, 11, 739–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Van Der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of Intrinsically Disordered Regions and Proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Uversky, V.N. The most important thing is the tail: Multitudinous functionalities of intrinsically disordered protein termini. FEBS Lett. 2013, 587, 1891–1901. [Google Scholar] [CrossRef] [Green Version]
- Fuxreiter, M.; Toth-Petroczy, A.; Kraut, D.A.; Matouschek, A.; Lim, R.Y.; Xue, B.; Kurgan, L.; Uversky, V.N. Disordered proteinaceous machines. Chem. Rev. 2014, 114, 6806–6843. [Google Scholar] [CrossRef] [Green Version]
- Habchi, J.; Tompa, P.; Longhi, S.; Uversky, V.N. Introducing Protein Intrinsic Disorder. Chem. Rev. 2014, 114, 6561–6588. [Google Scholar] [CrossRef] [Green Version]
- Jakob, U.; Kriwacki, R.; Uversky, V.N. Conditionally and Transiently Disordered Proteins: Awakening Cryptic Disorder To Regulate Protein Function. Chem. Rev. 2014, 114, 6779–6805. [Google Scholar] [CrossRef] [Green Version]
- Theillet, F.-X.; Binolfi, A.; Frembgen-Kesner, T.; Hingorani, K.; Sarkar, M.; Kyne, C.; Li, C.; Crowley, P.B.; Gierasch, L.; Pielak, G.J.; et al. Physicochemical Properties of Cells and Their Effects on Intrinsically Disordered Proteins (IDPs). Chem. Rev. 2014, 114, 6661–6714. [Google Scholar] [CrossRef]
- Tompa, P. Multisteric Regulation by Structural Disorder in Modular Signaling Proteins: An Extension of the Concept of Allostery. Chem. Rev. 2013, 114, 6715–6732. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N.; Davé, V.; Iakoucheva, L.M.; Malaney, P.; Metallo, S.J.; Pathak, R.R.; Joerger, A.C. Pathological Unfoldomics of Uncontrolled Chaos: Intrinsically Disordered Proteins and Human Diseases. Chem. Rev. 2014, 114, 6844–6879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Roey, K.; Uyar, B.; Weatheritt, R.J.; Dinkel, H.; Seiler, M.; Budd, A.; Gibson, T.J.; Davey, N.E. Short Linear Motifs: Ubiquitous and Functionally Diverse Protein Interaction Modules Directing Cell Regulation. Chem. Rev. 2014, 114, 6733–6778. [Google Scholar] [CrossRef] [PubMed]
- Fakhree, M.A.; Blum, C.; Claessens, M.M. Shaping membranes with disordered proteins. Arch. Biochem. Biophys. 2019, 677, 108163. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2013, 24, 24–41. [Google Scholar] [CrossRef] [Green Version]
- Wen, X.; Klionsky, D.J. An overview of macroautophagy in yeast. J. Mol. Biol. 2016, 428, 1681–1699. [Google Scholar] [CrossRef] [Green Version]
- Klionsky, D.J.; Petroni, G.; Amaravadi, R.K.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Pedro, J.M.B.; Cadwell, K.; Cecconi, F.; Choi, A.M.K.; et al. Autophagy in major human diseases. EMBO J. 2021, 40, e108863. [Google Scholar] [CrossRef]
- Popelka, H. Dancing while self-eating: Protein intrinsic disorder in autophagy. Prog. Mol. Biol. Transl. Sci. 2020, 263–305. [Google Scholar] [CrossRef]
- van der Lee, R.; Lang, B.; Kruse, K.; Gsponer, J.; de Groot, N.S.; Huynen, M.; Matouschek, A.; Fuxreiter, M.; Babu, M.M. Intrinsically Disordered Segments Affect Protein Half-Life in the Cell and during Evolution. Cell Rep. 2014, 8, 1832–1844. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Choi, W.; Hu, W.; Mi, N.; Guo, Q.; Ma, M.; Liu, M.; Tian, Y.; Lu, P.; Wang, F.-L.; et al. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 2012, 22, 473–489. [Google Scholar] [CrossRef] [Green Version]
- Rostislavleva, K.; Soler, N.; Ohashi, Y.; Zhang, L.; Pardon, E.; Burke, J.E.; Masson, G.R.; Johnson, C.; Steyaert, J.; Ktistakis, N.T.; et al. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 2015, 350, aac7365. [Google Scholar] [CrossRef] [Green Version]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, K.; Vucetic, S.; Radivojac, P.; Brown, C.J.; Dunker, A.K.; Obradovic, Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005, 3, 35–60. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Dunbrack, R.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. et Biophys. Acta (BBA) Proteins Proteom. 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [Green Version]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef] [Green Version]
- Dosztanyi, Z.; Csizmók, V.; Tompa, P.; Simon, I. The Pairwise Energy Content Estimated from Amino Acid Composition Discriminates between Folded and Intrinsically Unstructured Proteins. J. Mol. Biol. 2005, 347, 827–839. [Google Scholar] [CrossRef]
- Rajagopalan, K.; Mooney, S.M.; Parekh, N.; Getzenberg, R.H.; Kulkarni, P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J. Cell. Biochem. 2011, 112, 3256–3267. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Ruff, K.M.; Pappu, R.V. AlphaFold and Implications for Intrinsically Disordered Proteins. J. Mol. Biol. 2021, 433, 167208. [Google Scholar] [CrossRef]
- Azzaz, F.; Fantini, J. The epigenetic dimension of protein structure. Biomol. Concepts 2022, 13, 55–60. [Google Scholar] [CrossRef]
- Piovesan, D.; Monzon, A.M.; Tosatto, S.C. Intrinsic Protein Disorder, Conditional Folding and AlphaFold2. bioRxiv 2022. [Google Scholar]
- Nice, D.C.; Sato, T.K.; Stromhaug, P.E.; Emr, S.D.; Klionsky, D.J. Cooperative Binding of the Cytoplasm to Vacuole Targeting Pathway Proteins, Cvt13 and Cvt20, to Phosphatidylinositol 3-Phosphate at the Pre-autophagosomal Structure Is Required for Selective Autophagy. J. Biol. Chem. 2002, 277, 30198–30207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheever, M.L.; Sato, T.K.; de Beer, T.; Kutateladze, T.G.; Emr, S.D.; Overduin, M. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 2001, 3, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Theillet, F.-X.; Kalmar, L.; Tompa, P.; Han, K.-H.; Selenko, P.; Dunker, A.K.; Daughdrill, G.W.; Uversky, V.N. The alphabet of intrinsic disorder. Intrinsically Disord. Proteins 2013, 1, e24360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gatica, D.; Damasio, A.; Pascual, C.; Klionsky, D.J.; Ragusa, M.J.; Popelka, H. The carboxy terminus of yeast Atg13 binds phospholipid membrane via motifs that overlap with the Vac8-interacting domain. Autophagy 2019, 16, 1007–1020. [Google Scholar] [CrossRef] [PubMed]
- Kastelowitz, N.; Tamura, R.; Onasoga, A.; Stalker, T.J.; White, O.R.; Brown, P.N.; Brodsky, G.L.; Brass, L.F.; Branchford, B.R.; Di Paola, J.; et al. Peptides derived from MARCKS block coagulation complex assembly on phosphatidylserine. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Fujioka, Y.; Alam, J.M.; Noshiro, D.; Mouri, K.; Ando, T.; Okada, Y.; May, A.I.; Knorr, R.L.; Suzuki, K.; Ohsumi, Y.; et al. Phase separation organizes the site of autophagosome formation. Nature 2020, 578, 301–305. [Google Scholar] [CrossRef]
- Pluhackova, K.; Wassenaar, T.A.; Kirsch, S.; Böckmann, R.A. Spontaneous Adsorption of Coiled-Coil Model Peptides K and E to a Mixed Lipid Bilayer. J. Phys. Chem. B 2015, 119, 4396–4408. [Google Scholar] [CrossRef]
- Cornell, R.B. Membrane lipid compositional sensing by the inducible amphipathic helix of CCT. Biochim. et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2015, 1861, 847–861. [Google Scholar] [CrossRef]
- Johnson, J.E.; Xie, M.; Singh, L.M.R.; Edge, R.; Cornell, R.B. Both Acidic and Basic Amino Acids in an Amphitropic Enzyme, CTP:Phosphocholine Cytidylyltransferase, Dictate Its Selectivity for Anionic Membranes. J. Biol. Chem. 2003, 278, 514–522. [Google Scholar] [CrossRef] [Green Version]
- Fan, W.; Nassiri, A.; Zhong, Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc. Natl. Acad. Sci. USA 2011, 108, 7769–7774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popelka, H.; Damasio, A.; Hinshaw, J.E.; Klionsky, D.J.; Ragusa, M.J. Structure and function of yeast Atg20, a sorting nexin that facilitates autophagy induction. Proc. Natl. Acad. Sci. USA 2017, 114, E10112–E10121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Weering, J.; Sessions, R.B.; Traer, C.J.; Kloer, D.P.; Bhatia, V.K.; Stamou, D.; Carlsson, S.R.; Hurley, J.H.; Cullen, P.J. Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules. EMBO J. 2012, 31, 4466–4480. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Otomo, C.; Leitner, A.; Ohashi, K.; Aebersold, R.; Lander, G.C.; Otomo, T. Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proc. Natl. Acad. Sci. USA 2018, 115, E9792–E9801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotani, T.; Kirisako, H.; Koizumi, M.; Ohsumi, Y.; Nakatogawa, H. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. USA 2018, 115, 10363–10368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osawa, T.; Kotani, T.; Kawaoka, T.; Hirata, E.; Suzuki, K.; Nakatogawa, H.; Ohsumi, Y.; Noda, N.N. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 2019, 26, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Busse, R.A.; Scacioc, A.; Krick, R.; Pérez-Lara, Á.; Thumm, M.; Kühnel, K. Characterization of PROPPIN-Phosphoinositide Binding and Role of Loop 6CD in PROPPIN-Membrane Binding. Biophys. J. 2015, 108, 2223–2234. [Google Scholar] [CrossRef] [Green Version]
- Gopaldass, N.; Fauvet, B.; Lashuel, H.; Roux, A.; Mayer, A. Membrane scission driven by the PROPPIN Atg18. EMBO J. 2017, 36, 3274–3291. [Google Scholar] [CrossRef]
- Baskaran, S.; Ragusa, M.J.; Boura, E.; Hurley, J.H. Two-Site Recognition of Phosphatidylinositol 3-Phosphate by PROPPINs in Autophagy. Mol. Cell 2012, 47, 339–348. [Google Scholar] [CrossRef] [Green Version]
- Juris, L.; Montino, M.; Rube, P.; Schlotterhose, P.; Thumm, M.; Krick, R. PI 3P binding by Atg21 organises Atg8 lipidation. EMBO J. 2015, 34, 955–973. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, Y.; Kobayashi, T.; Yamamoto, H.; Hoshida, H.; Akada, R.; Inagaki, F.; Ohsumi, Y.; Noda, N.N. Structure-based Analyses Reveal Distinct Binding Sites for Atg2 and Phosphoinositides in Atg18. J. Biol. Chem. 2012, 287, 31681–31690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munzel, L.; Neumann, P.; Otto, F.B.; Krick, R.; Metje-Sprink, J.; Kroppen, B.; Karedla, N.; Enderlein, J.; Meinecke, M.; Ficner, R.; et al. Atg21 organizes Atg8 lipidation at the contact of the vacuole with the phagophore. Autophagy 2020, 17, 1458–1478. [Google Scholar] [CrossRef] [PubMed]
- Strong, L.M.; Chang, C.; Riley, J.F.; Boecker, C.A.; Flower, T.G.; Buffalo, C.Z.; Ren, X.; Stavoe, A.K.; Holzbaur, E.L.; Hurley, J.H. Structural basis for membrane recruitment of ATG16L1 by WIPI2 in autophagy. eLife 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Popelka, H.; Reinhart, E.F.; Metur, S.P.; Leary, K.A.; Ragusa, M.J.; Klionsky, D.J. Membrane Binding and Homodimerization of Atg16 Via Two Distinct Protein Regions is Essential for Autophagy in Yeast. J. Mol. Biol. 2021, 433, 166809. [Google Scholar] [CrossRef] [PubMed]
- Lystad, A.H.; Carlsson, S.R.; De La Ballina, L.R.; Kauffman, K.J.; Nag, S.; Yoshimori, T.; Melia, T.J.; Simonsen, A. Distinct functions of ATG16L1 isoforms in membrane binding and LC3B lipidation in autophagy-related processes. Nat. Cell Biol. 2019, 21, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Hong, S.B.; Lee, J.K.; Han, S.; Roh, K.-H.; Lee, K.-E.; Kim, Y.K.; Choi, E.-J.; Song, H.K. Insights into autophagosome maturation revealed by the structures of ATG5 with its interacting partners. Autophagy 2014, 11, 75–87. [Google Scholar] [CrossRef] [Green Version]
- Otomo, C.; Metlagel, Z.; Takaesu, G.; Otomo, T. Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy. Nat. Struct. Mol. Biol. 2013, 20, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Rogov, V.; Dotsch, V.; Johansen, T.; Kirkin, V. Interactions between Autophagy Receptors and Ubiquitin-like Proteins Form the Molecular Basis for Selective Autophagy. Mol. Cell 2014, 53, 167–178. [Google Scholar] [CrossRef] [Green Version]
- Sora, V.; Kumar, M.; Maiani, E.; Lambrughi, M.; Tiberti, M.; Papaleo, E. Structure and Dynamics in the ATG8 Family From Experimental to Computational Techniques. Front. Cell Dev. Biol. 2020, 8, 420. [Google Scholar] [CrossRef]
- Bhaskara, R.M.; Grumati, P.; Garcia-Pardo, J.; Kalayil, S.; Covarrubias-Pinto, A.; Chen, W.; Kudryashev, M.; Dikic, I.; Hummer, G. Curvature induction and membrane remodeling by FAM134B reticulon homology domain assist selective ER-phagy. Nat. Commun. 2019, 10, 2370. [Google Scholar] [CrossRef] [Green Version]
- Popelka, H.; Klionsky, D.J. Molecular dynamics simulations reveal how the reticulon-homology domain of the autophagy receptor RETREG1/FAM134B remodels membranes for efficient selective reticulophagy. Autophagy 2020, 16, 585–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mardakheh, F.; Auciello, G.; Dafforn, T.; Rappoport, J.Z.; Heath, J.K. Nbr1 Is a Novel Inhibitor of Ligand-Mediated Receptor Tyrosine Kinase Degradation. Mol. Cell. Biol. 2010, 30, 5672–5685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popelka, H.; Uversky, V.N.; Klionsky, D.J. Identification of Atg3 as an intrinsically disordered polypeptide yields insights into the molecular dynamics of autophagy-related proteins in yeast. Autophagy 2014, 10, 1093–1104. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Qiu, Y.; Grace, C.R.R.; Liu, X.; Klionsky, D.J.; Schulman, B.A. A switch element in the autophagy E2 Atg3 mediates allosteric regulation across the lipidation cascade. Nat. Commun. 2019, 10, 3600. [Google Scholar] [CrossRef] [PubMed]
- Popelka, H.; Klionsky, D.J. Multiple structural rearrangements mediated by high-plasticity regions in Atg3 are key for efficient conjugation of Atg8 to PE during autophagy. Autophagy 2021, 17, 1805–1808. [Google Scholar] [CrossRef]
- Ye, Y.; Tyndall, E.R.; Bui, V.; Tang, Z.; Shen, Y.; Jiang, X.; Flanagan, J.M.; Wang, H.-G.; Tian, F. An N-terminal conserved region in human Atg3 couples membrane curvature sensitivity to conjugase activity during autophagy. Nat. Commun. 2021, 12, 374. [Google Scholar] [CrossRef]
- Iakoucheva, L.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [Green Version]
- Pejaver, V.; Hsu, W.-L.; Xin, F.; Dunker, A.K.; Uversky, V.N.; Radivojac, P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014, 23, 1077–1093. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N. Protein intrinsic disorder and structure-function continuum. Prog. Mol. Biol. Transl. Sci. 2019, 166, 1–17. [Google Scholar] [CrossRef]
- Darling, A.L.; Uversky, V.N. Intrinsic Disorder and Posttranslational Modifications: The Darker Side of the Biological Dark Matter. Front. Genet. 2018, 9, 158. [Google Scholar] [CrossRef]
- Bürgi, J.; Xue, B.; Uversky, V.N.; Van Der Goot, F.G. Intrinsic Disorder in Transmembrane Proteins: Roles in Signaling and Topology Prediction. PLoS ONE 2016, 11, e0158594. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Wrecked regulation of intrinsically disordered proteins in diseases: Pathogenicity of deregulated regulators. Front. Mol. Biosci. 2014, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Vucetic, S.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Functional Anthology of Intrinsic Disorder. 3. Ligands, Post-Translational Modifications, and Diseases Associated with Intrinsically Disordered Proteins. J. Proteome Res. 2007, 6, 1917–1932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Kim, H.-I.; Jeong, H.; Lee, M.; Jang, S.H.; Yoon, S.Y.; Park, Z.-Y.; Jun, Y.; Lee, C. Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways. Autophagy 2019, 16, 991–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gatica, D.; Wen, X.; Cheong, H.; Klionsky, D.J. Vac8 determines phagophore assembly site vacuolar localization during nitrogen starvation-induced autophagy. Autophagy 2020, 17, 1636–1648. [Google Scholar] [CrossRef]
- Reddy, K.D.; Malipeddi, J.; DeForte, S.; Pejaver, V.; Radivojac, P.; Uversky, V.N.; Deschenes, R.J. Physicochemical sequence characteristics that influence S-palmitoylation propensity. J. Biomol. Struct. Dyn. 2016, 35, 2337–2350. [Google Scholar] [CrossRef]
- Pantoom, S.; Konstantinidis, G.; Voss, S.; Han, H.; Hofnagel, O.; Li, Z.; Wu, Y.-W. RAB33B recruits the ATG16L1 complex to the phagophore via a noncanonical RAB binding protein. Autophagy 2020, 17, 2290–2304. [Google Scholar] [CrossRef]
- Matsui, T.; Jiang, P.; Nakano, S.; Sakamaki, Y.; Yamamoto, H.; Mizushima, N. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J. Cell Biol. 2018, 217, 2633–2645. [Google Scholar] [CrossRef]
- Mochida, K.; Yamasaki, A.; Matoba, K.; Kirisako, H.; Noda, N.N.; Nakatogawa, H. Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Ichimura, Y.; Ohsumi, Y. Atg8, a Ubiquitin-like Protein Required for Autophagosome Formation, Mediates Membrane Tethering and Hemifusion. Cell 2007, 130, 165–178. [Google Scholar] [CrossRef] [Green Version]
- Popelka, H.; Klionsky, D.J. Analysis of the native conformation of the LIR/AIM motif in the Atg8/LC3/GABARAP-binding proteins. Autophagy 2015, 11, 2153–2159. [Google Scholar] [CrossRef] [PubMed]
- Wirth, M.; Zhang, W.; Razi, M.; Nyoni, L.; Joshi, D.; O’Reilly, N.; Johansen, T.; Tooze, S.A.; Mouilleron, S. Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins. Nat. Commun. 2019, 10, 2055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chino, H.; Hatta, T.; Natsume, T.; Mizushima, N. Intrinsically Disordered Protein TEX264 Mediates ER-phagy. Mol. Cell 2019, 74, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Delorme-Axford, E.; Popelka, H.; Klionsky, D.J. TEX264 is a major receptor for mammalian reticulophagy. Autophagy 2019, 15, 1677–1681. [Google Scholar] [CrossRef] [Green Version]
- Ragusa, M.J.; Stanley, R.E.; Hurley, J.H. Architecture of the Atg17 Complex as a Scaffold for Autophagosome Biogenesis. Cell 2012, 151, 1501–1512. [Google Scholar] [CrossRef] [Green Version]
- Stjepanovic, G.; Davies, C.W.; Stanley, R.E.; Ragusa, M.J.; Kim, D.J.; Hurley, J.H. Assembly and dynamics of the autophagy-initiating Atg1 complex. Proc. Natl. Acad. Sci. USA 2014, 111, 12793–12798. [Google Scholar] [CrossRef] [Green Version]
- Ohashi, Y.; Soler, N.; Ortegón, M.G.; Zhang, L.; Kirsten, M.L.; Perisic, O.; Masson, G.; Burke, J.; Jakobi, A.; Apostolakis, A.A.; et al. Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex. Autophagy 2016, 12, 2129–2144. [Google Scholar] [CrossRef]
- Puri, C.; Renna, M.; Bento, C.F.; Moreau, K.; Rubinsztein, D.C. Diverse Autophagosome Membrane Sources Coalesce in Recycling Endosomes. Cell 2013, 154, 1285–1299. [Google Scholar] [CrossRef] [Green Version]
- Puri, C.; Renna, M.; Bento, C.F.; Moreau, K.; Rubinsztein, D.C. ATG16L1 meets ATG9 in recycling endosomes. Autophagy 2013, 10, 182–184. [Google Scholar] [CrossRef] [Green Version]
- Rice, L.M.; Brennwald, P.; Brünger, A.T. Formation of a yeast SNARE complex is accompanied by significant structural changes. FEBS Lett. 1997, 415, 49–55. [Google Scholar] [CrossRef] [Green Version]
- Fiebig, K.M.; Rice, L.M.; Pollock, E.; Brunger, A. Folding intermediates of SNARE complex assembly. Nat. Genet. 1999, 6, 117–123. [Google Scholar] [CrossRef]
- Nair, U.; Jotwani, A.; Geng, J.; Gammoh, N.; Richerson, D.; Yen, W.-L.; Griffith, J.; Nag, S.; Wang, K.; Moss, T.; et al. SNARE Proteins Are Required for Macroautophagy. Cell 2011, 146, 290–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brennwald, P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell 1994, 79, 245–258. [Google Scholar] [CrossRef]
- Lane, S.R.; Liu, Y. Characterization of the palmitoylation domain of SNAP-25. J. Neurochem. 2002, 69, 1864–1869. [Google Scholar] [CrossRef]
- Veit, M.; Söllner, T.H.; Rothman, J.E. Multiple palmitoylation of synaptotagmin and the t-SNARE SNAP-25. FEBS Lett. 1996, 385, 119–123. [Google Scholar] [CrossRef] [Green Version]
- Diao, J.; Liu, R.; Rong, Y.; Zhao, M.; Zhang, J.; Lai, Y.; Zhou, Q.; Wilz, L.M.; Li, J.; Vivona, S.; et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 2015, 520, 563–566. [Google Scholar] [CrossRef] [Green Version]
- Cheong, H.; Yorimitsu, T.; Reggiori, F.; Legakis, J.E.; Wang, C.-W.; Klionsky, D.J. Atg17 Regulates the Magnitude of the Autophagic Response. Mol. Biol. Cell 2005, 16, 3438–3453. [Google Scholar] [CrossRef]
- Tucker, K.A.; Reggiori, F.; Dunn, W.A.; Klionsky, D.J. Atg23 Is Essential for the Cytoplasm to Vacuole Targeting Pathway and Efficient Autophagy but Not Pexophagy. J. Biol. Chem. 2003, 278, 48445–48452. [Google Scholar] [CrossRef] [Green Version]
- Hawkins, W.D.; Leary, K.A.; Andhare, D.; Popelka, H.; Klionsky, D.J.; Ragusa, M.J. Dimerization-dependent membrane tethering by Atg23 is essential for yeast autophagy. Cell Rep. 2022, 39, 110702. [Google Scholar] [CrossRef]
- Legakis, J.E.; Yen, W.-L.; Klionsky, D.J. A Cycling Protein Complex Required for Selective Autophagy. Autophagy 2007, 3, 422–432. [Google Scholar] [CrossRef] [Green Version]
- Rao, Y.; Perna, M.G.; Hofmann, B.; Beier, V.; Wollert, T. The Atg1–kinase complex tethers Atg9-vesicles to initiate autophagy. Nat. Commun. 2016, 7, 10338. [Google Scholar] [CrossRef] [PubMed]

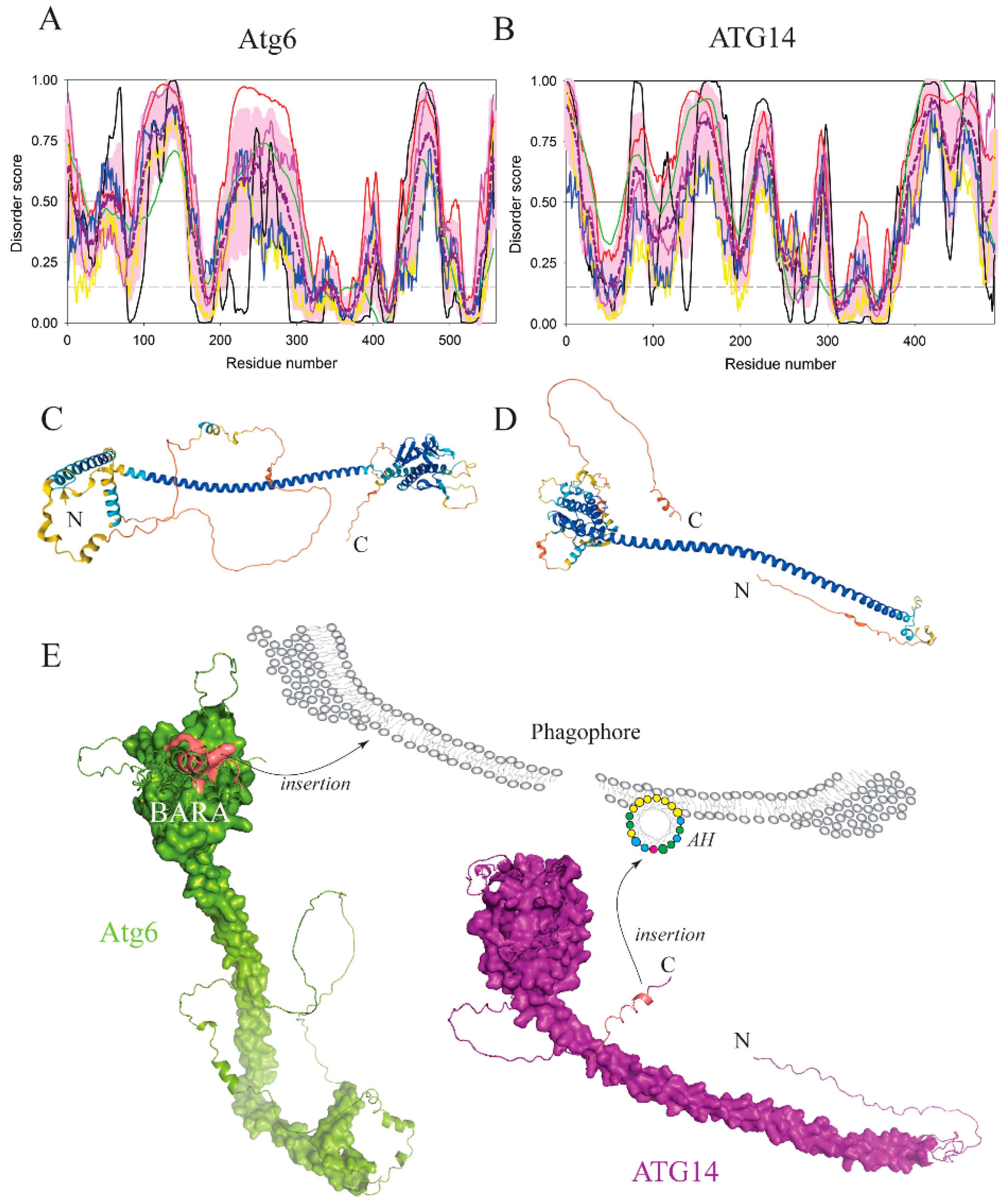
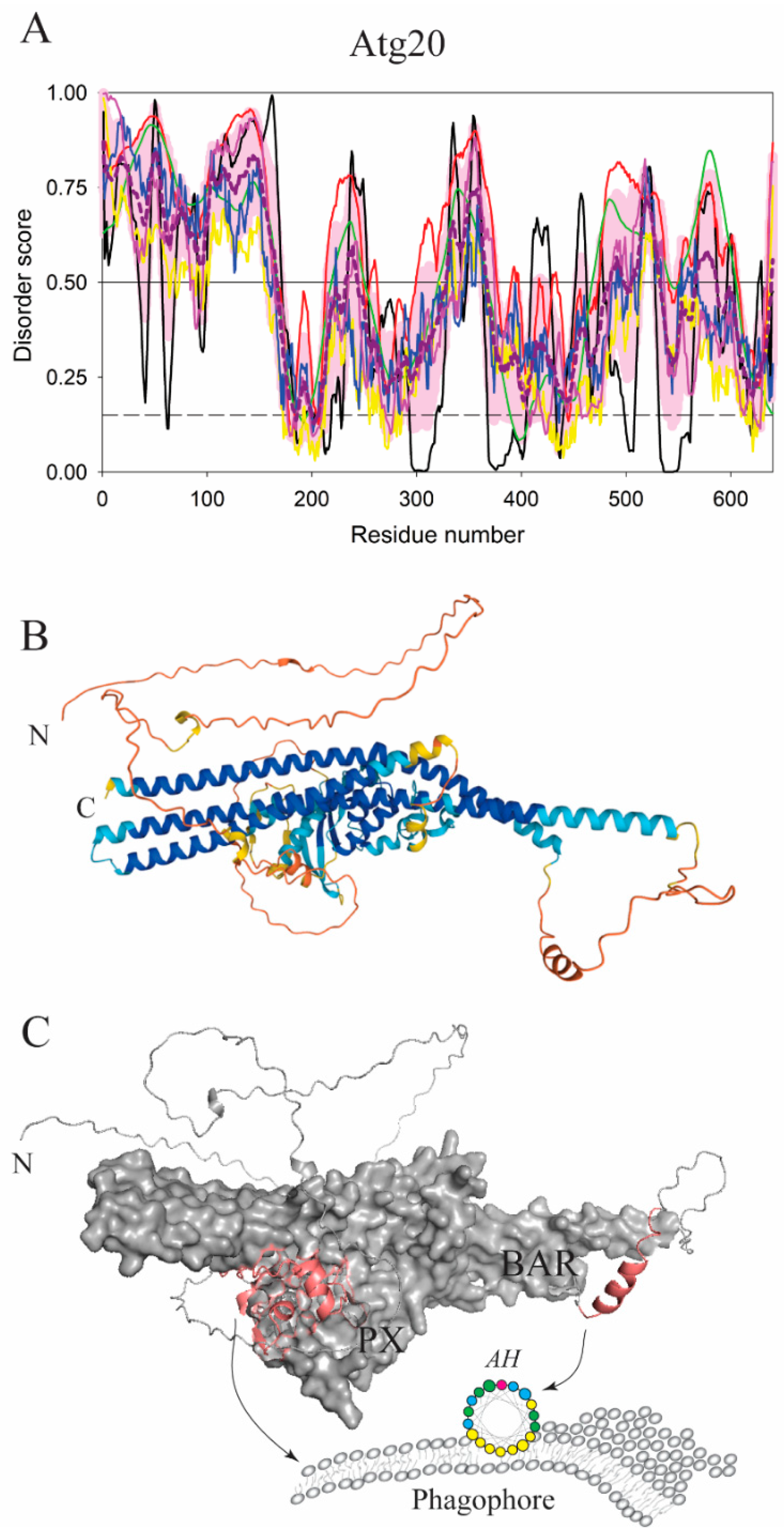
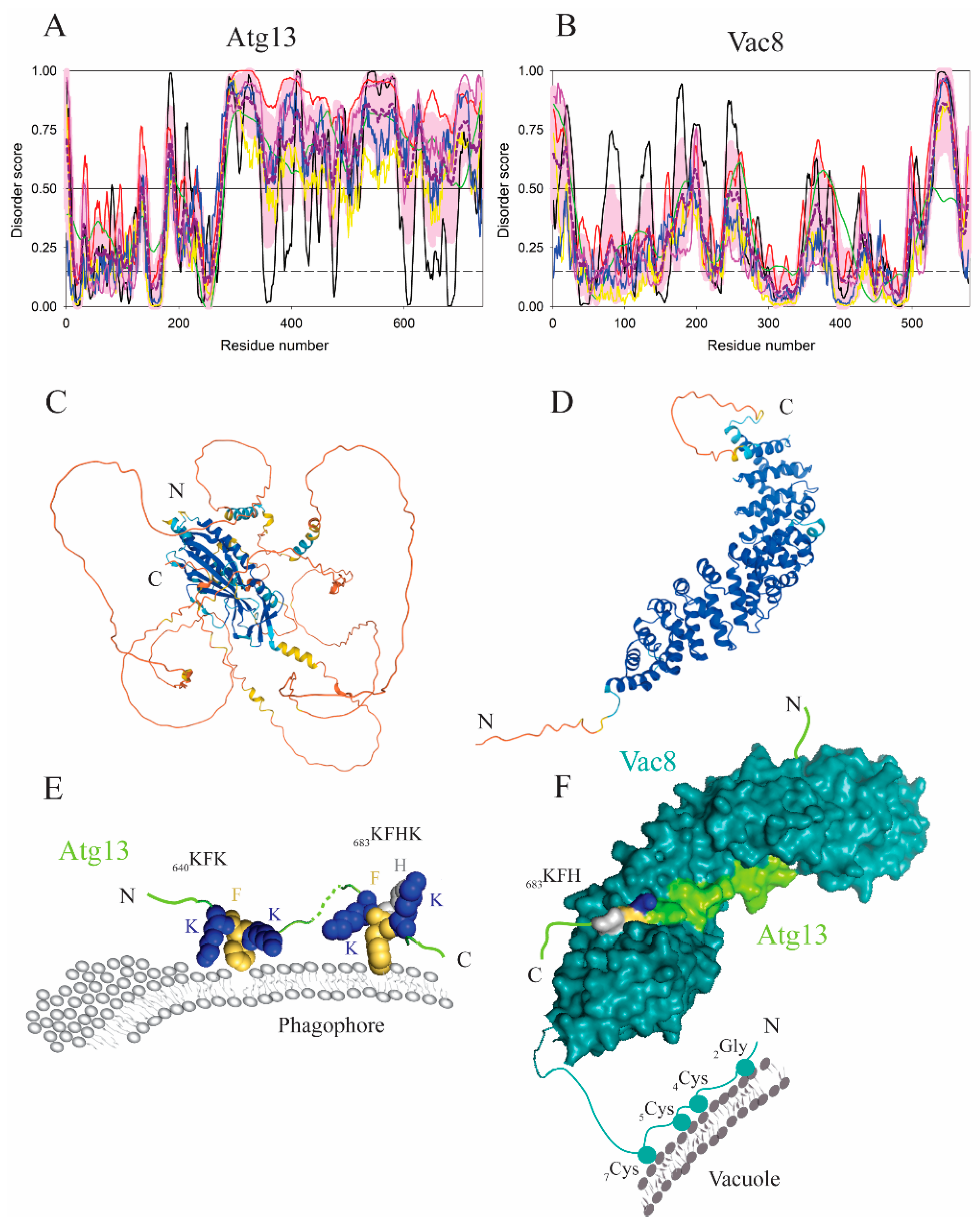
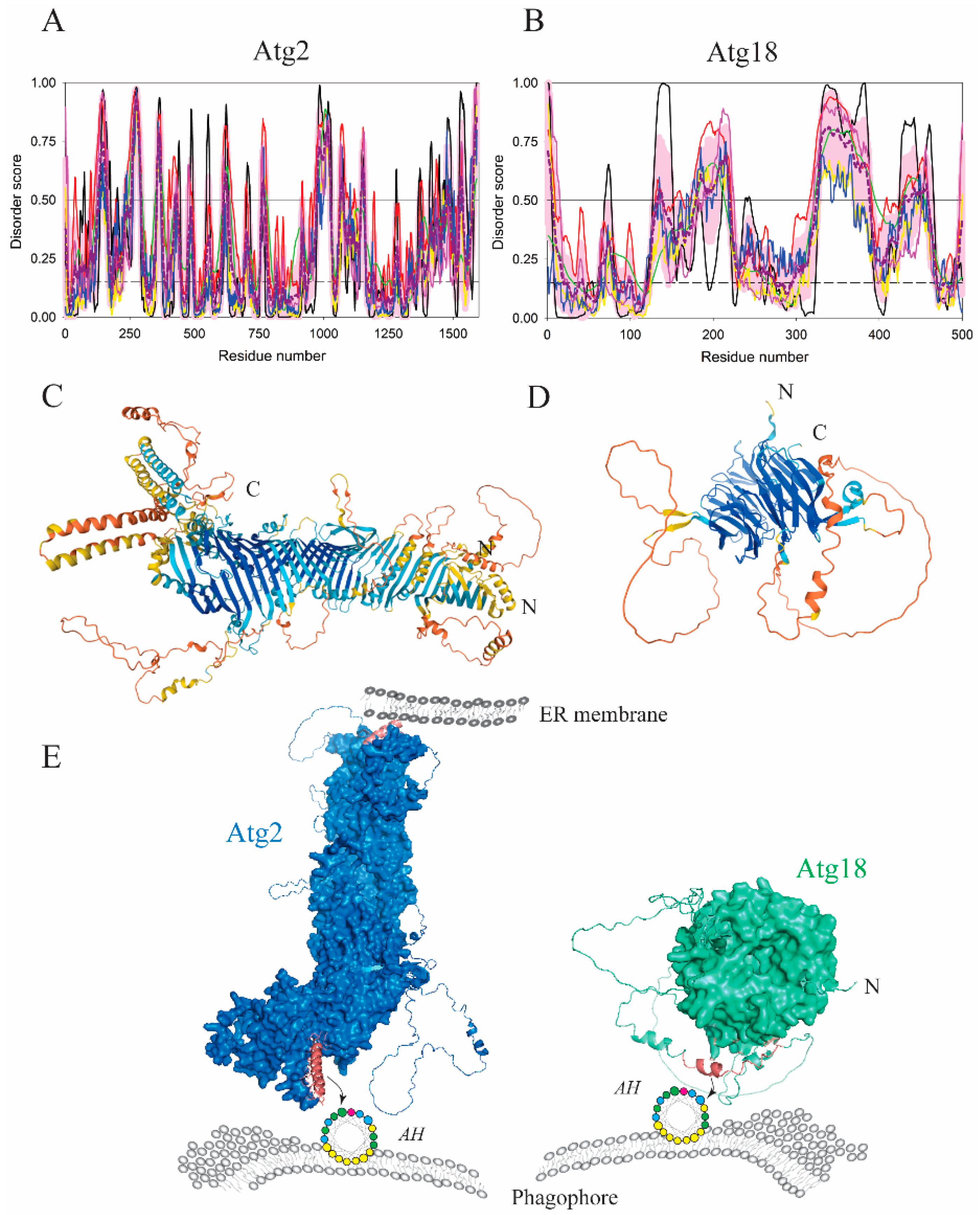
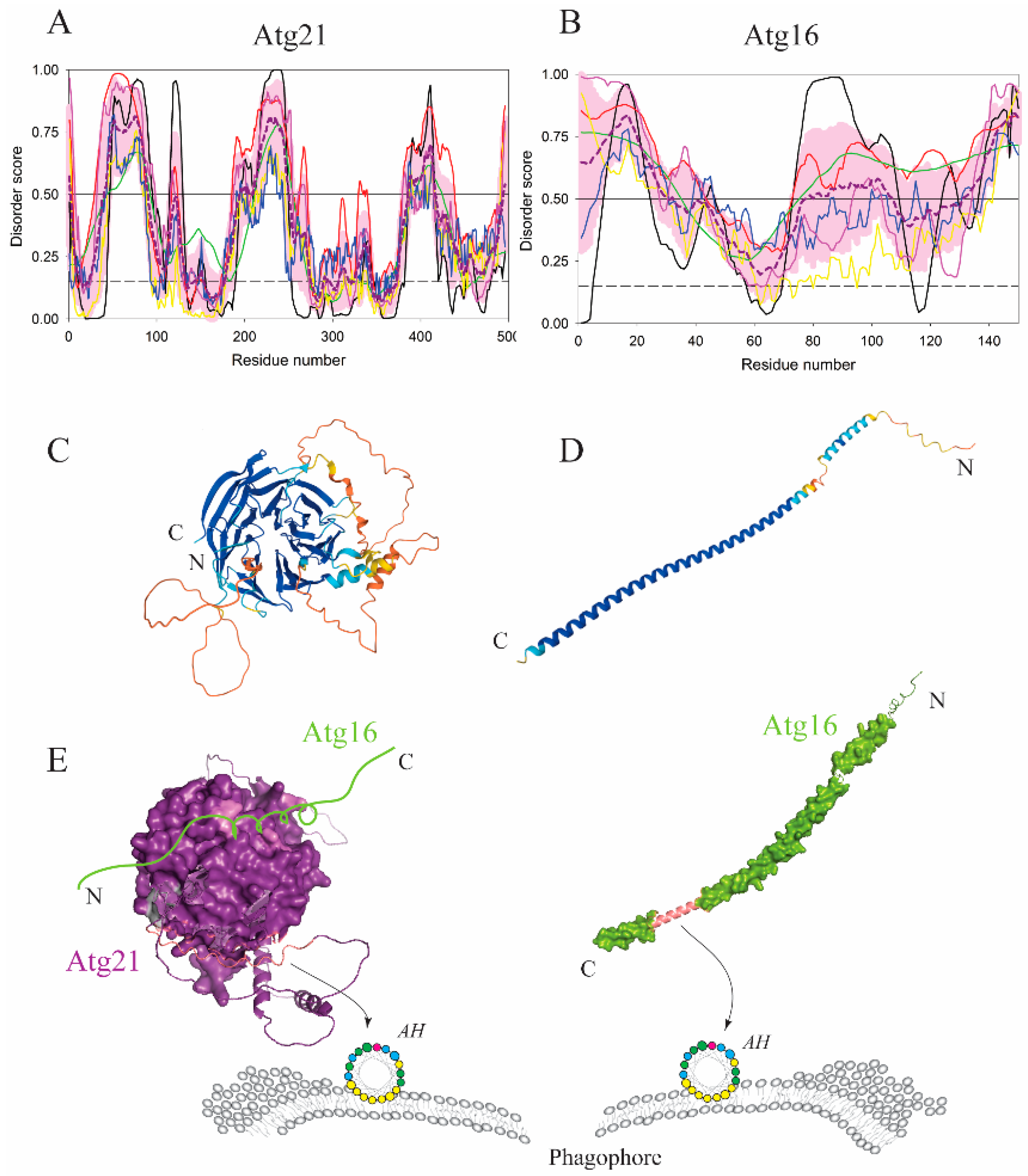

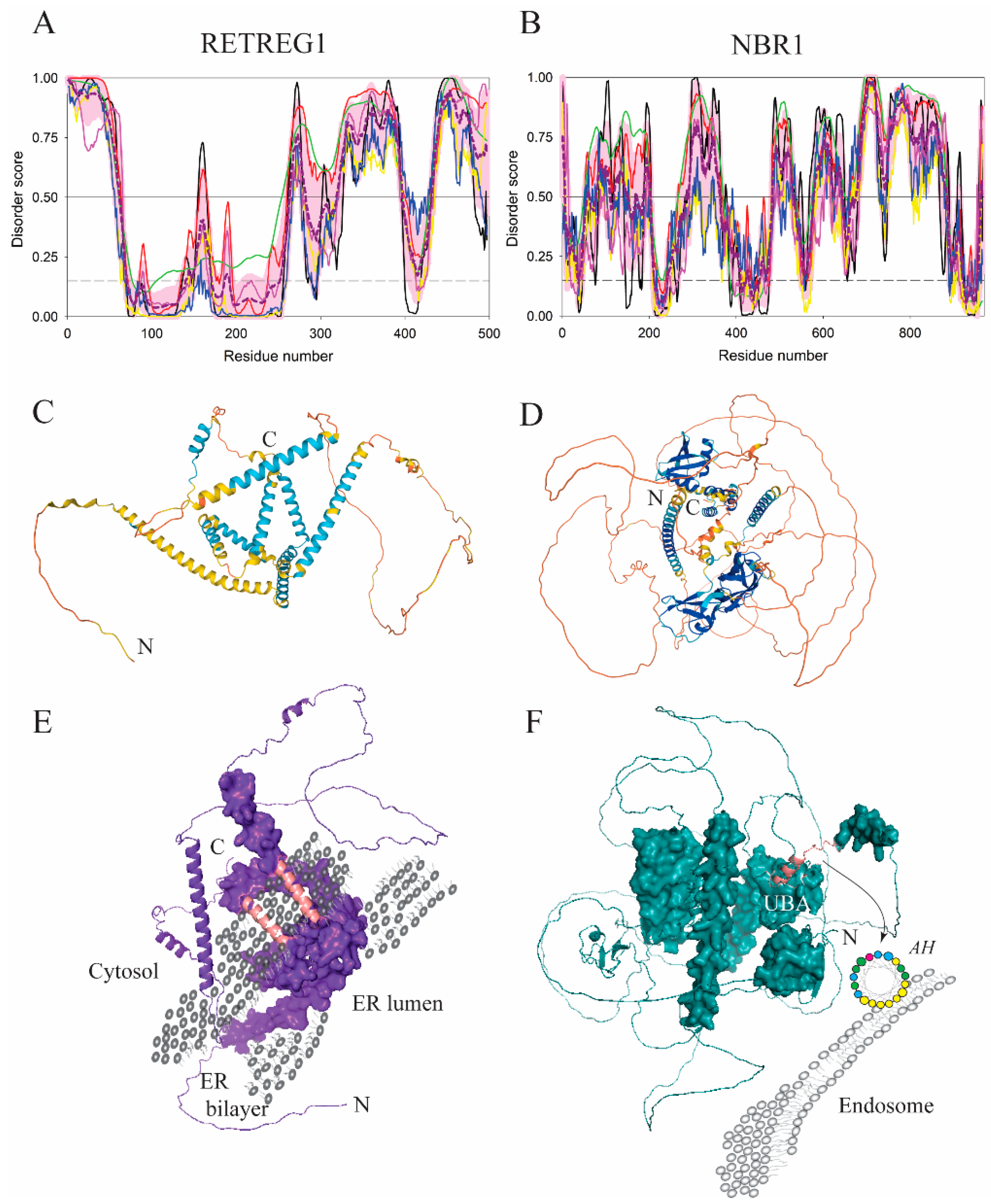
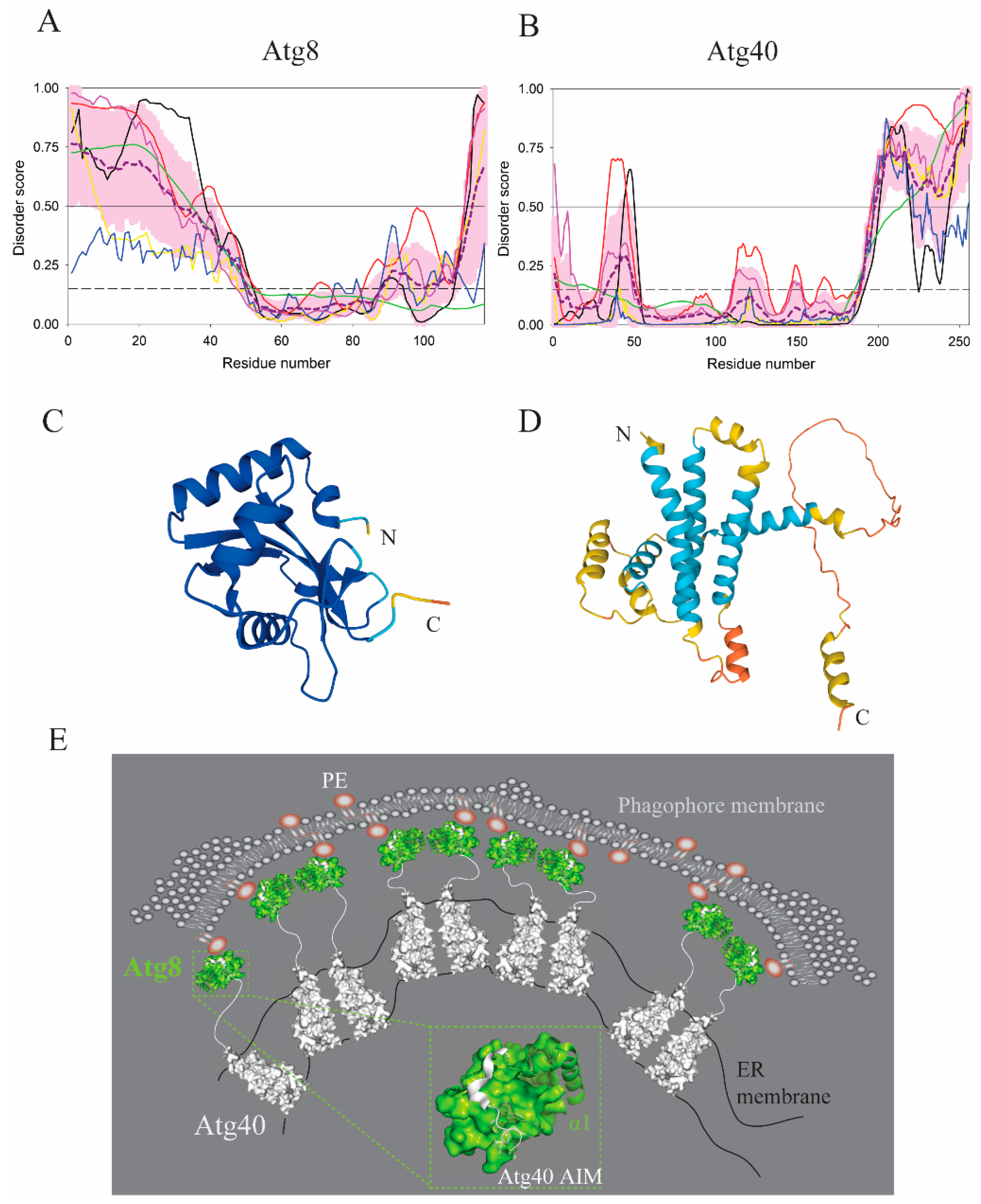
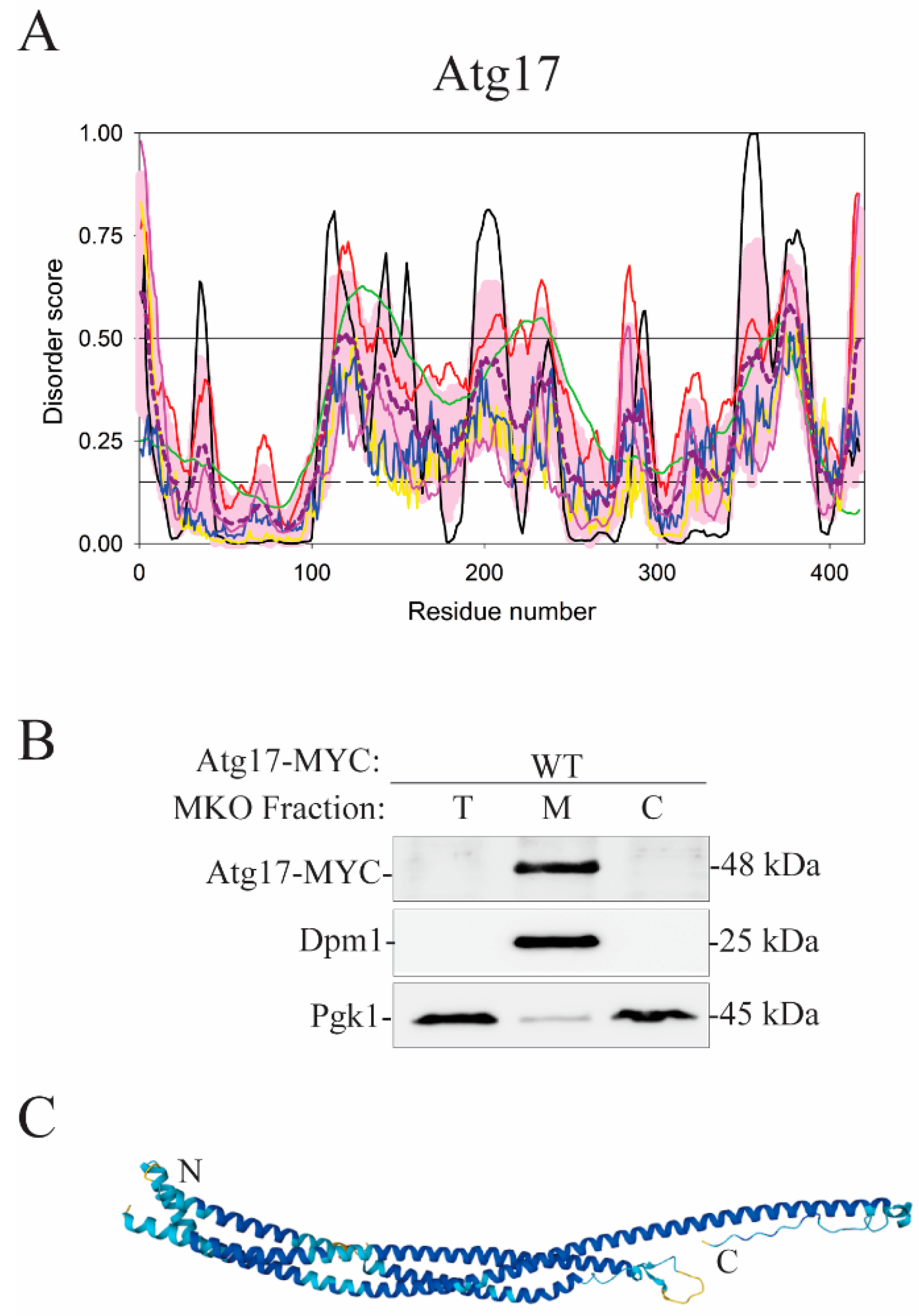
| Protein (length) | PPDR (%) | Positions of IDPRs | Act with Membranes | Molecular Mechanism MEDIATED by IDPR | PTM related to the Molecular Mechanism |
|---|---|---|---|---|---|
| Atg1 (897) | 42.8 | 1–17, 26–29, 45–53, 57–65, 119–121, 133–142, 328–563, 583–585, 628–683, 770–787, 854–864, 890–897 | Tethering | ? | ? |
| Atg2 (1592) | 29.9 | 35–41, 96–102, 112–161, 195–196, 230–296, 352–375, 399–405, 410–436, 474–491, 607–643, 697–704, 757–779, 914–919, 949–952, 964–1026, 1116–1121, 1145–1159, 1193–1196, 1429–1442, 1478–1485, 1527–1536, 1573–1592 | Binding | AH | Dephosphorylation? |
| ATG3 (314) | 34.4 | 1–5, 25–30, 97–101, 129–180, 183–191, 235–250, 309–313 | Binding | AH | ? |
| Atg6 (557) | 55.8 | 1–6, 9–20, 38–40, 42–61, 73–80, 88–162, 199–307, 389–395, 399–406, 435–440, 446–490, 507–508, 539–-540, 550–557 | Binding | Disordered loop | ? |
| Atg8 (117) | 37.6 | 1–30, 35–42, 112–117 | Lipidation | PE conjugation | C-terminal cleavage |
| Atg13 (738) | 71.5 | 1–6, 30–37, 96–97, 129–143, 177–193, 204–212, 229–232, 272–738 | Binding | Lys/Phe-enriched motif | Dephosphoryation? |
| ATG14 (492) | 61.2 | 1–33,68–181, 212–242, 285–298, 384–492 | Binding | AH | Dephosphorylation? |
| Atg16 (150) | 80.7 | 1–43, 73–150 | Binding | AH | ? |
| ATG16L1 (607) | 51.1 | 1–30, 43–48, 53–165, 176–285, 289–307, 359–369, 407–408, 570–590 | Binding | AH | ? |
| Atg17 (417) | 23.7 | 1–6, 114–130, 137–141, 201–212, 218–223, 227–238, 280–288, 352–359, 365–382, 412–417 | Binding | ? | ? |
| Atg18 (500) | 38.8 | 1–9, 130–138, 153–163, 172–221, 316–391, 408–409, 427–459, 497–500 | Binding | AH | Dephosphorylation? |
| Atg20 (640) | 67.3 | 1–167, 213–248, 256–262, 300–376, 391–397, 417, 430–434, 474–540, 550–607, 635–640 | Binding/tubulation | Disordered loop/AH | Dephosphorylation? |
| Atg21 (496) | 44.4 | 1–4, 30–87, 117–123, 186–254, 264–271, 330–334, 336, 338–339, 380–438, 491–496 | Binding | AH | Dephosphorylation? |
| Atg23 (453) | 73.7 | 1–7, 28–35, 73–85, 100, 126–127, 139–169, 171–273, 284–453 | Tethering | Charged residues/? | ? |
| Atg40 (256) | 28.9 | 33–45, 196–256 | Remodeling | AIM for Atg8 | Phosphorylation? |
| RETREG1 (497) | 56.3 | 1–63, 157–165, 260–400, 431–497 | Binding/remodeling | AH/LIR for LC3 | Phosphorylation? |
| Vac8 (578) | 27.7 | 1–24, 158–161, 195–203, 242–268, 361–389, 430–435, 496–503, 518–566, 576–578 | Binding | PTM anchor | G2 myristoylation, C4, C5, C7 palmitoylation |
| NBR1 (966) | 62.1 | 1–5, 52–204, 267–366, 481–531, 581–643, 662–886, 961–966 | Binding | AH | Dephosphorylation? |
| RAB33B (229) | 35.8 | 1–26, 174–229 | Binding | PTM anchor | C227, C229 prenylation |
| YKT6 (198) | 27.8 | 1–4, 49–59, 130–143, 158–178, 187, 195–199 | Binding | PTM anchor | C194 palmitoylation, C195 prenylation |
| TEX264 (313) | 37.4 | 1–4, 40, 100–110, 213–313 | Space bridging | LIR for LC3 | Phosphorylation? |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popelka, H.; Uversky, V.N. Theater in the Self-Cleaning Cell: Intrinsically Disordered Proteins or Protein Regions Acting with Membranes in Autophagy. Membranes 2022, 12, 457. https://doi.org/10.3390/membranes12050457
Popelka H, Uversky VN. Theater in the Self-Cleaning Cell: Intrinsically Disordered Proteins or Protein Regions Acting with Membranes in Autophagy. Membranes. 2022; 12(5):457. https://doi.org/10.3390/membranes12050457
Chicago/Turabian StylePopelka, Hana, and Vladimir N. Uversky. 2022. "Theater in the Self-Cleaning Cell: Intrinsically Disordered Proteins or Protein Regions Acting with Membranes in Autophagy" Membranes 12, no. 5: 457. https://doi.org/10.3390/membranes12050457
APA StylePopelka, H., & Uversky, V. N. (2022). Theater in the Self-Cleaning Cell: Intrinsically Disordered Proteins or Protein Regions Acting with Membranes in Autophagy. Membranes, 12(5), 457. https://doi.org/10.3390/membranes12050457







