In Silico Drug Design of Benzothiadiazine Derivatives Interacting with Phospholipid Cell Membranes
Abstract
1. Introduction
2. Methods
3. Results and Discussion
3.1. Characteristics of the Bilayer Systems
3.2. Local Structure of Benzothiadiazine Derivatives
3.2.1. Radial Distribution Functions
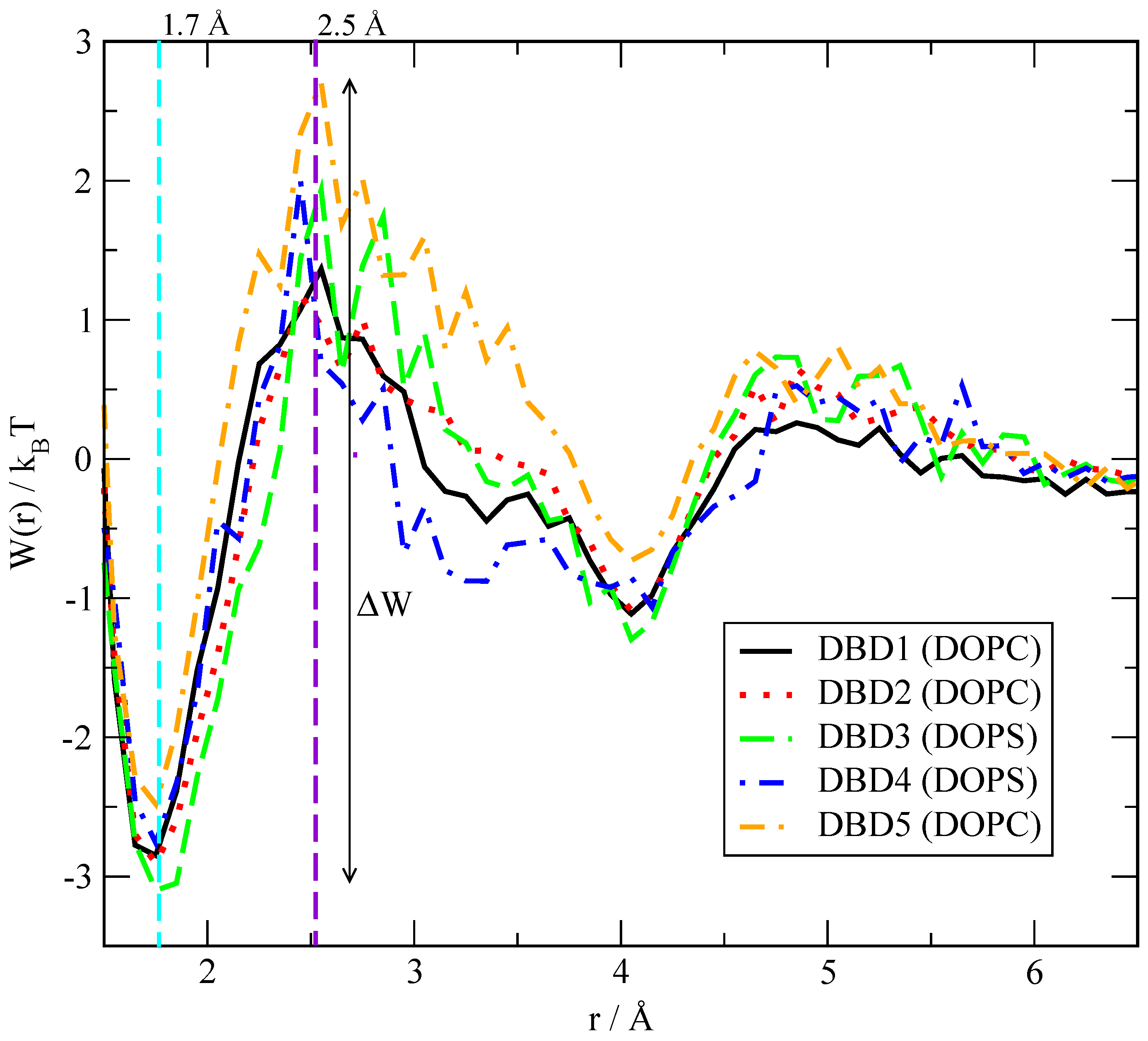
3.2.2. Potentials of Mean Force between Benzothiadiazine Derivatives and Lipids
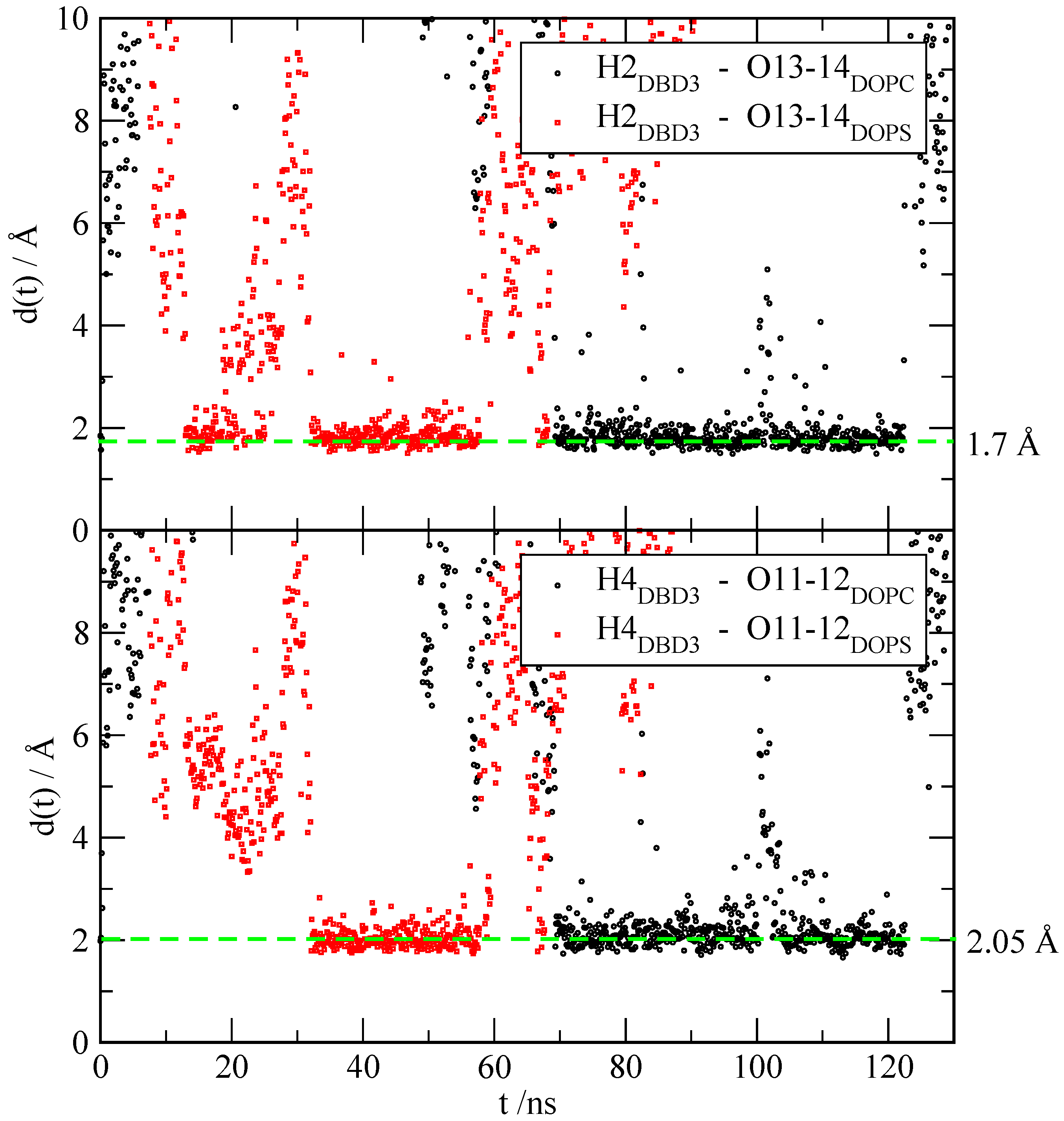
3.2.3. Time-Dependent Atomic Site–Site Distances
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Escribá, P.V.; Sastre, M.; García-Sevilla, J.A. Disruption of cellular signalling pathways by daunomycin through destabilization of nonlamellar membrane structures. Proc. Natl. Acad. Sci. USA 1995, 92, 7595. [Google Scholar] [CrossRef]
- Tong, S.; Lin, Y.; Lu, S.; Wang, M.; Bogdanov, M.; Zheng, L. Structural Insight into Substrate Selection and Catalysis of Lipid Phosphate Phosphatase PgpB in the Cell Membrane. J. Biol Chem. 2016, 291, 18342. [Google Scholar] [CrossRef]
- Doherty, G.J.; McMahon, H.T. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009, 78, 857. [Google Scholar] [CrossRef] [PubMed]
- Escribá, P.V.; González-Ros, J.M.; Goñi, F.M.; Kinnunen, P.K.; Vigh, L.; Sánchez-Magraner, L.; Fernández, A.M.; Busquets, X.; Horvath, I.; Barceló-Coblijn, G. Membranes: A meeting point for lipids, proteins and therapies. J. Cell Mol. Med. 2008, 12, 829. [Google Scholar] [CrossRef]
- Noutsi, P.; Gratton, E.; Chaieb, S. Assessment of Membrane Fluidity Fluctuations during Cellular Development Reveals Time and Cell Type Specificity. PLoS ONE. 2016, 11, e0158313. [Google Scholar] [CrossRef]
- Hakomori, S. Aberrant glycosylation in cancer cell membranes as focused on glycolipids: Overview and perspectives. Cancer Res. 1985, 45, 2405. [Google Scholar]
- Simons, K.; Ehehalt, R. Cholesterol, lipid rafts, and disease. J. Clin. Investig. 2002, 110, 597. [Google Scholar] [CrossRef] [PubMed]
- Vigh, L.; Escribá, P.V.; Sonnleitner, A.; Sonnleitner, M.; Piotto, S.; Maresca, B.; Horvath, I.; Harwood, J.L. The significance of lipid composition for membrane activity: New concepts and ways of assessing function. Prog. Lipid Res. 2005, 44, 303. [Google Scholar] [CrossRef]
- Kim, Y.; Shanta, S.R.; Zhou, L.H.; Kim, K.P. Mass spectrometry based cellular phosphoinositides profiling and phospholipid analysis: A brief review. Exp. Mol. Med. 2010, 42, 1. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jolliet-Riant, P.; Tillement, J.P. Drug transfer across the blood-brain barrier and improvement of brain delivery. Fundam. Clin. Pharmacol. 1999, 13, 16. [Google Scholar] [CrossRef]
- Bodor, N.; Buchwald, P. Barriers to remember: Brain-targeting chemical delivery systems and Alzheimer’s disease. Drug Discov. Today 2002, 7, 766. [Google Scholar] [CrossRef]
- Waterhouse, R.N. Determination of lipophilicity and its use as a predictor of blood-brain barrier penetration of molecular imaging agents. Mol. Imaging Biol. 2003, 5, 376. [Google Scholar] [CrossRef] [PubMed]
- Rees, C.W. Polysulfur-Nitrogen Heterocyclic Chemistry. J. Heterocycl. Chem. 1992, 29, 639. [Google Scholar] [CrossRef]
- Vitaku, E.; Smith, D.T.; Njardarson, J.T. Analysis of the structural diversity, substitution patterns, and frequency of nitrogen heterocycles among U.S. FDA approved pharmaceuticals. J. Med. Chem. 2014, 57, 10257. [Google Scholar] [CrossRef] [PubMed]
- Horton, D.A.; Bourne, G.T.; Smythe, M.L. The combinatorial synthesis of bicyclic privileged structures or privileged substructures. Chem. Rev. 2003, 103, 893. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Kamal, R.; Kumar, V. Heterocyclic Analogues as Kinase Inhibitors: A Focus Review. Curr. Top. Med. Chem. 2017, 17, 2482. [Google Scholar] [CrossRef] [PubMed]
- Platts, M.M. Hydrochlorothiazide, a new oral diuretic. Br. Med. J. 1959, 1, 1565. [Google Scholar] [CrossRef][Green Version]
- Martínez, A.; Esteban, A.I.; Castro, A.; Gil, C.; Conde, S.; Andrei, G.; Snoeck, R.; Balzarini, J.; De Clercq, E. Novel potential agents for human cytomegalovirus infection: Synthesis and antiviral activity evaluation of benzothiadiazine dioxide acyclonucleosides. J. Med. Chem. 1999, 42, 1145. [Google Scholar] [CrossRef]
- Tait, A.; Luppi, A.; Hatzelmann, A.; Fossa, P.; Mosti, L. Synthesis, biological evaluation and molecular modelling studies on benzothiadiazine derivatives as PDE4 selective inhibitors. Bioorg. Med. Chem. 2005, 13, 1393. [Google Scholar] [CrossRef]
- Larsen, A.P.; Francotte, P.; Frydenvang, K.; Tapken, D.; Goffin, E.; Fraikin, P.; Caignard, D.H.; Lestage, P.; Danober, L.; Pirotte, B.; et al. Synthesis and Pharmacology of Mono-, Di-, and Trialkyl-Substituted 7-Chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides Combined with X-ray Structure Analysis to Understand the Unexpected Structure-Activity Relationship at AMPA Receptors. ACS Chem. Neurosci. 2016, 7, 378. [Google Scholar] [CrossRef]
- Ma, X.; Wei, J.; Wang, C.; Gu, D.; Hu, Y.; Sheng, R. Design, synthesis and biological evaluation of novel benzothiadiazine derivatives as potent PI3Kδ-selective inhibitors for treating B-cell-mediated malignancies. Eur. J. Med. Chem. 2019, 170, 112. [Google Scholar] [CrossRef] [PubMed]
- Shaik, T.B.; Malik, M.S.; Routhu, S.R.; Seddigi, Z.S.; Althagafi, I.I.; Kamal, A. Evaluation of anticancer and anti-mitotic properties of quinazoline and quinazolino-benzothiadiazine derivatives. Anti-Cancer Agents Med. Chem. 2020, 20, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Huwaimel, B.I.; Bhakta, M.; Kulkarni, C.A.; Milliken, A.S.; Wang, F.; Peng, A.; Brookes, P.S.; Trippier, P.C. Discovery of Halogenated Benzothiadiazine Derivatives with Anticancer Activity. Chem. Med. Chem. 2021, 16, 1143–1162. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, F.; Koshio, H.; Katayama, N.; Ishihara, T.; Kaizawa, H.; Taniuchi, Y.; Sato, K.; Sakai-Moritani, Y.; Kaku, S.; Kurihara, H.; et al. Design, synthesis and biological activity of YM-60828 derivatives. Part 2: Potent and orally-bioavailable factor Xa inhibitors based on benzothiadiazine-4-one template. Bioorg. Med. Chem. 2003, 11, 367–381. [Google Scholar] [CrossRef]
- Kamal, A.; Reddy, K.-S.; Ahmed, S.-K.; Khan, M.-N.; Sinha, R.-K.; Yadav, J.-S.; Arora, S.-K. Anti-tubercular agents. Part 3. Benzothiadiazine as a novel scaffold for anti-Mycobacterium activity. Bioorg. Med. Chem. 2006, 14, 650–658. [Google Scholar] [CrossRef]
- Kamal, A.; Shetti, R.-V.; Azeeza, S.; Ahmed, S.-K.; Swapna, P.; Reddy, A.-M.; Khan, I.-A.; Sharma, S.; Abdullah, S.-T. Anti-tubercular agents. Part 5: Synthesis and biological evaluation of benzothiadiazine 1,1-dioxide based congeners. Eur. J. Med. Chem. 2010, 45, 4545–4553. [Google Scholar] [CrossRef]
- Tait, A.; Luppi, A.; Franchini, S.; Preziosi, E.; Parenti, C.; Buccioni, M.; Marucci, G.; Leonardi, A.; Poggesi, E.; Brasili, L. 1,2,4-Benzothiadiazine derivatives as alpha1 and 5-HT1A receptor ligands. Bioorg. Med. Chem. Lett. 2005, 15, 1185–1188. [Google Scholar] [CrossRef]
- Hu, Z.; Martí, J.; Lu, H. Structure of benzothiadiazine at zwitterionic phospholipid cell membranes. J. Chem. Phys. 2021, 155, 154303. [Google Scholar] [CrossRef]
- Prior, I.A.; Lewis, P.D.; Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012, 72, 2457–2467. [Google Scholar] [CrossRef]
- Lu, H.; Martí, J. Long-lasting Salt Bridges Provide the Anchoring Mechanism of Oncogenic Kirsten Rat Sarcoma Proteins at Cell Membranes. J. Phys. Chem. Lett. 2020, 11, 9938–9945. [Google Scholar] [CrossRef]
- Jang, H.; Abraham, S.J.; Chavan, T.S.; Hitchinson, B.; Khavrutskii, L.; Tarasova, N.I.; Nussinov, R.; Gaponenko, V. Mechanisms of membrane binding of small GTPase K-Ras4B farnesylated hypervariable region. J. Biol. Chem. 2015, 290, 9465–9477. [Google Scholar] [CrossRef]
- Lu, S.; Jang, H.; Muratcioglu, S.; Gursoy, A.; Keskin, O.; Nussinov, R.; Zhang, J. Ras conformational ensembles, allostery, and signaling. Chem. Rev. 2016, 116, 6607–6665. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-L.; Sheng, R.; Jung, J.H.; Wang, L.; Stec, E.; O’Connor, M.J.; Song, S.; Bikkavilli, R.K.; Winn, R.A.; Lee, D.; et al. Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol. Nat. Chem. Biol. 2017, 13, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859. [Google Scholar] [CrossRef]
- Jo, S.; Lim, J.B.; Klauda, J.B.; Im, W. CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009, 97, 50. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926. [Google Scholar] [CrossRef]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D., Jr.; Pastor, R.W. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys. Chem. B 2010, 114, 7830. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.B.; Rogaski, B.; Klauda, J.B. Update of the cholesterol force field parameters in CHARMM. J. Phys. Chem. B 2012, 116, 203. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135. [Google Scholar] [CrossRef] [PubMed]
- Linse, B.; Linse, P. Tuning the smooth particle mesh Ewald sum: Application on ionic solutions and dipolar fluids. J. Chem. Phys. 2014, 141, 184114. [Google Scholar] [CrossRef]
- Heinzinger, K. Computer Modelling of Fluids Polymers and Solids; Springer: Berlin/Heidelberg, Germany, 1990; pp. 357–394. [Google Scholar]
- Brodholt, J.P. Molecular dynamics simulations of aqueous NaCl solutions at high pressures and temperatures. Chem. Geol. 1998, 151, 11. [Google Scholar] [CrossRef]
- Chowdhuri, S.; Chandra, A. Molecular dynamics simulations of aqueous NaCl and KCl solutions: Effects of ion concentration on the single-particle, pair, and collective dynamical properties of ions and water molecules. J. Chem. Phys. 2001, 115, 3732. [Google Scholar] [CrossRef]
- Nagy, G.; Gordillo, M.C.; Guàrdia, E.; Martí, J. Liquid water confined in carbon nanochannels at high temperatures. J. Phys. Chem. B 2007, 111, 12524. [Google Scholar] [CrossRef] [PubMed]
- Videla, P.; Sala, J.; Martí, J.; Guàrdia, E.; Laria, D. Aqueous electrolytes confined within functionalized silica nanopores. J. Chem. Phys. 2011, 135, 104503. [Google Scholar] [CrossRef]
- Sala, J.; Guàrdia, E.; Martí, J. Specific ion effects in aqueous eletrolyte solutions confined within graphene sheets at the nanometric scale. Phys. Chem. Chem. Phys. 2012, 14, 10799. [Google Scholar] [CrossRef]
- Calero, C.; Martí, J.; Guàrdia, E. 1H nuclear spin relaxation of liquid water from molecular dynamics simulations. J. Phys. Chem. B 2015, 119, 1966–1973. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.; Mashl, R.J.; Jakobsson, E.; Aluru, N.R. Electrolytic transport in modified carbon nanotubes. Nano Lett. 2003, 3, 1399. [Google Scholar] [CrossRef]
- Allen, T.W.; Andersen, O.S.; Roux, B. Molecular dynamics—Potential of mean force calculations as a tool for understanding ion permeation and selectivity in narrow channels. Biophys. Chem. 2006, 124, 251. [Google Scholar] [CrossRef]
- Yang, J.; Calero, C.; Martí, J. Diffusion and spectroscopy of water and lipids in fully hydrated dimyristoylphosphatidylcholine bilayer membranes. J. Chem. Phys. 2014, 140, 104901. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Martí, J. Binding and dynamics of melatonin at the interface of phosphatidylcholine-cholesterol membranes. PLoS ONE 2019, 14, e0224624. [Google Scholar] [CrossRef]
- Drew Bennett, W.F.; He, S.; Bilodeau, C.L.; Jones, D.; Sun, D.; Kim, H.; Allen, J.E.; Lightstone, F.C.; Ingólfsson, H.I. Predicting small molecule transfer free energies by combining molecular dynamics simulations and deep learning. J. Chem. Inf. Model. 2020, 60, 5375. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Martí, J. Cellular absorption of small molecules: Free energy landscapes of melatonin binding at phospholipid membranes. Sci. Rep. 2020, 10, 9235. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R. Smith, J.C.; Kasson, P.M. van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, E.; Hess, B.; Van Der Spoel, D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. Mol. Model. Annu. 2001, 7, 306. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Van der Spoel, D.; Van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43. [Google Scholar] [CrossRef]
- Chen, W.; Dusa, F.; Witos, J.; Ruokonen, S.-K.; Wiedmer, S.K. Determination of the main phase transition temperature of phospholipids by nanoplasmonic sensing. Sci. Rep. 2018, 8, 14815. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.J.; Holian, B.L. The Nose–Hoover thermostat. J. Chem. Phys. 1985, 83, 4069–4074. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Crystal structure and pair potentials: A molecular-dynamics study. Phys. Rev. Lett. 1980, 45, 1196. [Google Scholar] [CrossRef]
- Pandey, P.R.; Roy, S. Headgroup mediated water insertion into the DPPC bilayer: A molecular dynamics study. J. Phys. Chem. B 2011, 115, 3155–3163. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Tristram-Nagle, S.; Nagle, J.F. Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation. Phys. Rev. E 2009, 80, 021931. [Google Scholar] [CrossRef] [PubMed]
- Martí, J.; Lu, H. Microscopic interactions of melatonin, serotonin and tryptophan with zwitterionic phospholipid membranes. Int. J. Mol. Sci. 2021, 22, 2842. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, H.; Jin, B. Fluorescence of tryptophan in aqueous solution. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 106, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Trzesniak, D.; Kunz, A.-P.E.; van Gunsteren, W.F. A comparison of methods to compute the potential of mean force. ChemPhysChem 2007, 8, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Guàrdia, E.; Rey, R.; Padró, J.A. Statistical errors in constrained molecular dynamics calculations of the mean force potential. Mol. Sim. 1992, 9, 201–211. [Google Scholar] [CrossRef]
- Kastner, J. Umbrella sampling. Wiley Interdiscip. Rev. Comp. Mol. Sci. 2011, 1, 932–942. [Google Scholar] [CrossRef]
- Geissler, P.L.; Dellago, C.; Chandler, D.; Hutter, J.; Parrinello, M. Autoionization in liquid water. Science 2001, 291, 2121–2124. [Google Scholar] [CrossRef]
- Martí, J.; Csajka, F.S.; Chandler, D. Stochastic transition pathways in the aqueous sodium chloride dissociation process. Chem. Phys. Lett. 2000, 328, 169–176. [Google Scholar] [CrossRef]
- Bolhuis, P.G.; Chandler, D.; Dellago, C.; Geissler, P.L. Transition path sampling: Throwing ropes over rough mountain passes, in the dark. Annu. Rev. Phys. Chem. 2002, 53, 291–318. [Google Scholar] [CrossRef]
- Martí, J.; Csajka, F.S. Transition path sampling study of flip-flop transitions in model lipid bilayer membranes. Phys. Rev. E 2004, 69, 061918. [Google Scholar] [CrossRef] [PubMed]
- Barducci, A.; Bussi, G.; Parrinello, M. Well-tempered metadynamics: A smoothly converging and tunable free-energy method. Phys. Rev. Lett. 2008, 100, 020603. [Google Scholar] [CrossRef] [PubMed]
- Chandler, D. Introduction to Modern Statistical Mechanics; Oxford University Press: Oxford, UK, 1987. [Google Scholar]
- Feyereisen, M.W.; Feller, D.; Dixon, D.A. Hydrogen bond energy of the water dimer. J. Phys. Chem. 1996, 100, 2993–2997. [Google Scholar] [CrossRef]
- Vega, C.; Abascal, J.L.F.; Conde, M.M.; Aragones, J.L. What ice can teach us about water interactions: A critical comparison of the performance of different water models. Faraday Discuss. 2009, 141, 251–276. [Google Scholar] [CrossRef]
- MacCallum, J.L.; Bennett, W.F.D.; Tieleman, D.P. Distribution of amino acids in a lipid bilayer from computer simulations. Biophys. J. 2008, 94, 3393–3404. [Google Scholar] [CrossRef]
- De Jesus, A.J.; Allen, T.W. The role of tryptophan side chains in membrane protein anchoring and hydrophobic mismatch. BBA Biomembr. 2013, 1828, 864–876. [Google Scholar] [CrossRef]
- Peters, G.H.; Werge, M.; Elf-Lind, M.N.; Madsen, J.J.; Velardez, G.F.; Westh, P. Interaction of neurotransmitters with a phospholipid bilayer: A molecular dynamics study. Chem. Phys. Lipids 2014, 184, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Martí, J. Effects of cholesterol on the binding of the precursor neurotransmitter tryptophan to zwitterionic membranes. J. Chem. Phys. 2018, 149, 164906. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Martí, J.; Padró, J.A.; Guàrdia, E. Computer simulation of molecular motions in liquids: Infrared spectra of water and heavy water. Mol. Sim. 1993, 11, 321–336. [Google Scholar] [CrossRef]
- Padró, J.A.; Martí, J. Response to “Comment on ‘An interpretation of the low-frequency spectrum of liquid water’” [J. Chem. Phys. 118, 452 (2003)]. J. Chem. Phys. 2004, 120, 1659–1660. [Google Scholar] [CrossRef]
- Padró, J.A.; Martí, J.; Guardia, E. Molecular dynamics simulation of liquid water at 523 K. J. Phys. Condens. Matter 1994, 6, 2283. [Google Scholar] [CrossRef]
- Martí, J.; Gordillo, M.C. Microscopic dynamics of confined supercritical water. Chem. Phys. Lett. 2002, 354, 227–232. [Google Scholar] [CrossRef]
- Martí, J. Dynamic properties of hydrogen-bonded networks in supercritical water. Phys. Rev. E 2000, 61, 449. [Google Scholar] [CrossRef] [PubMed]
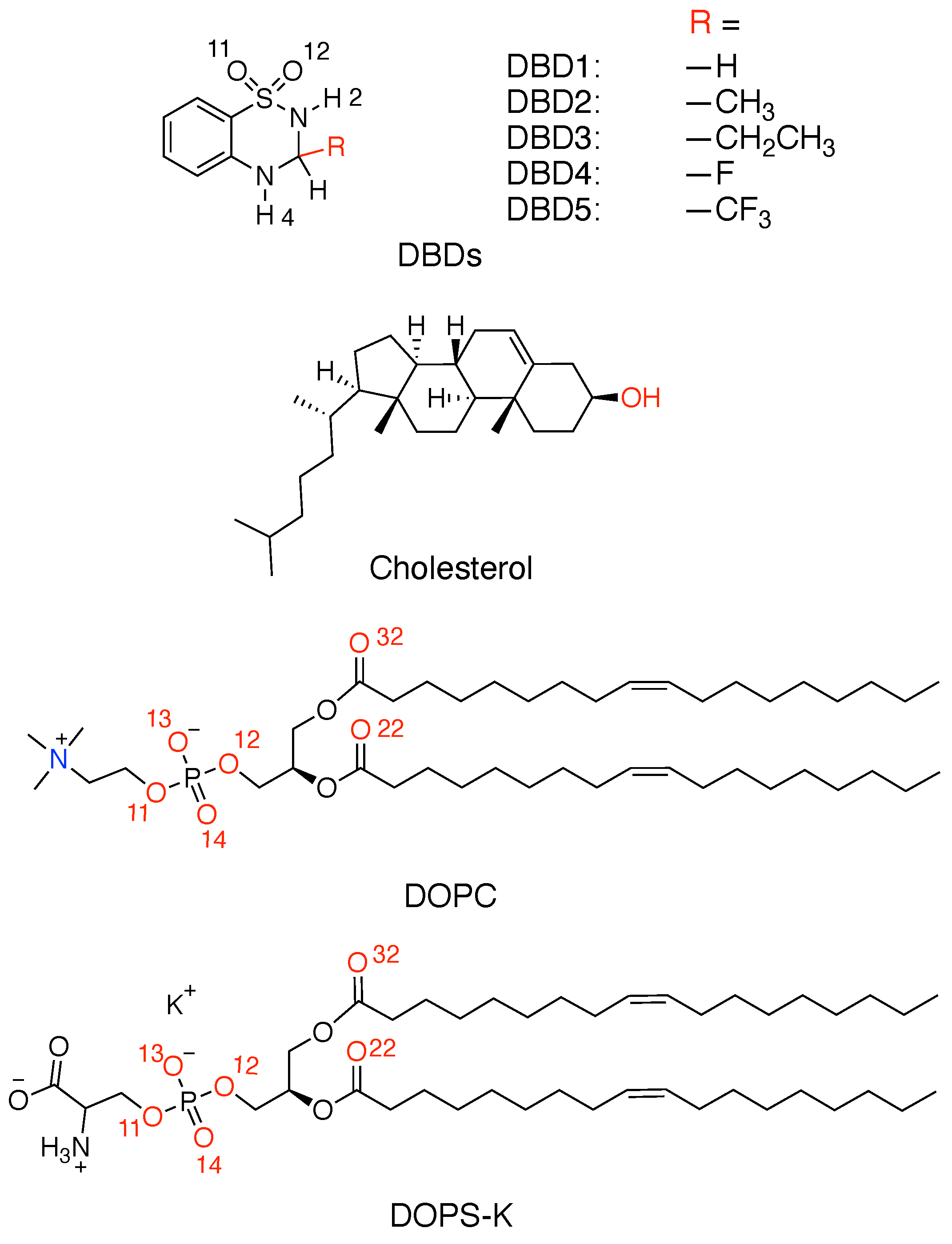

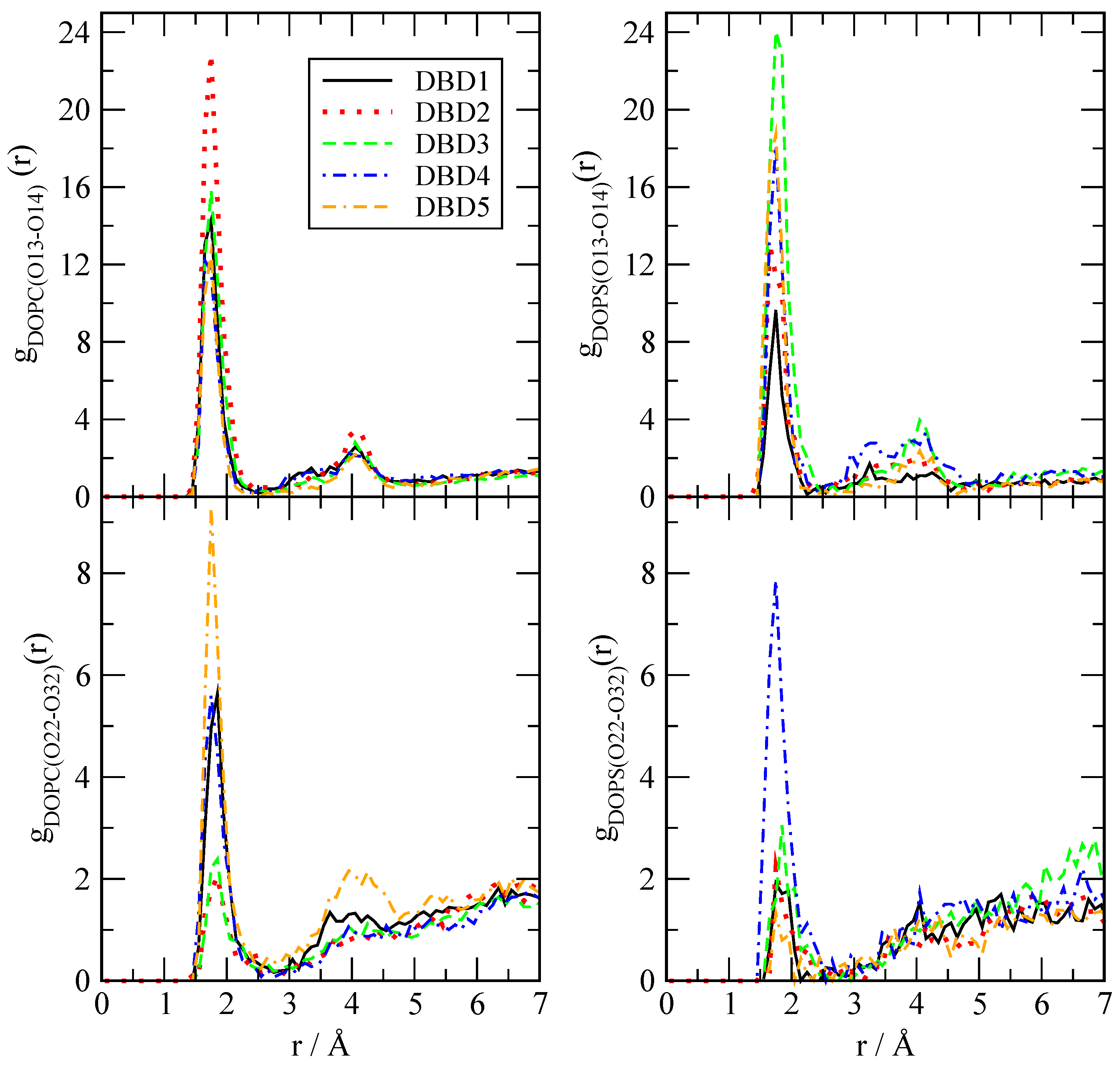
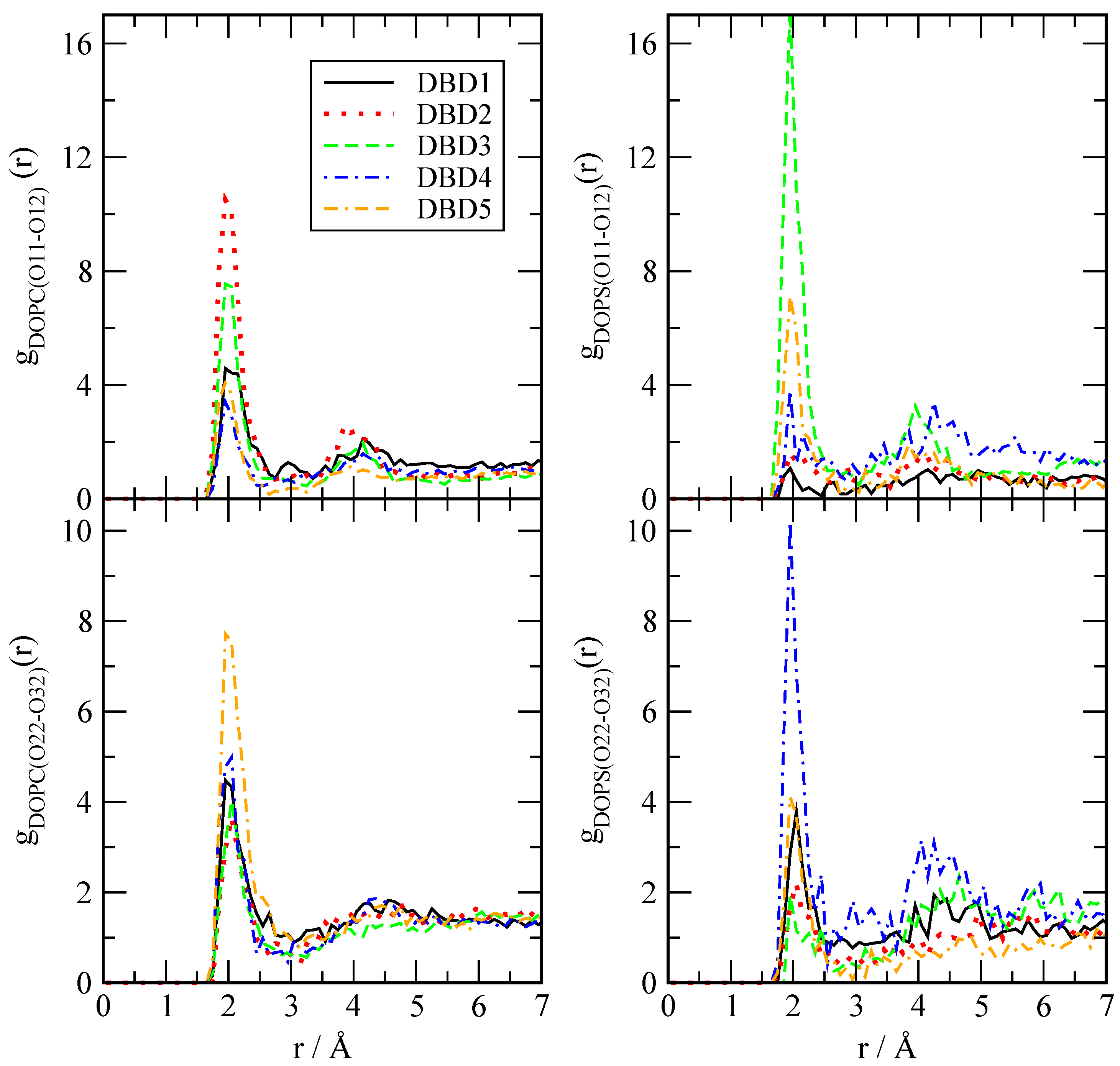
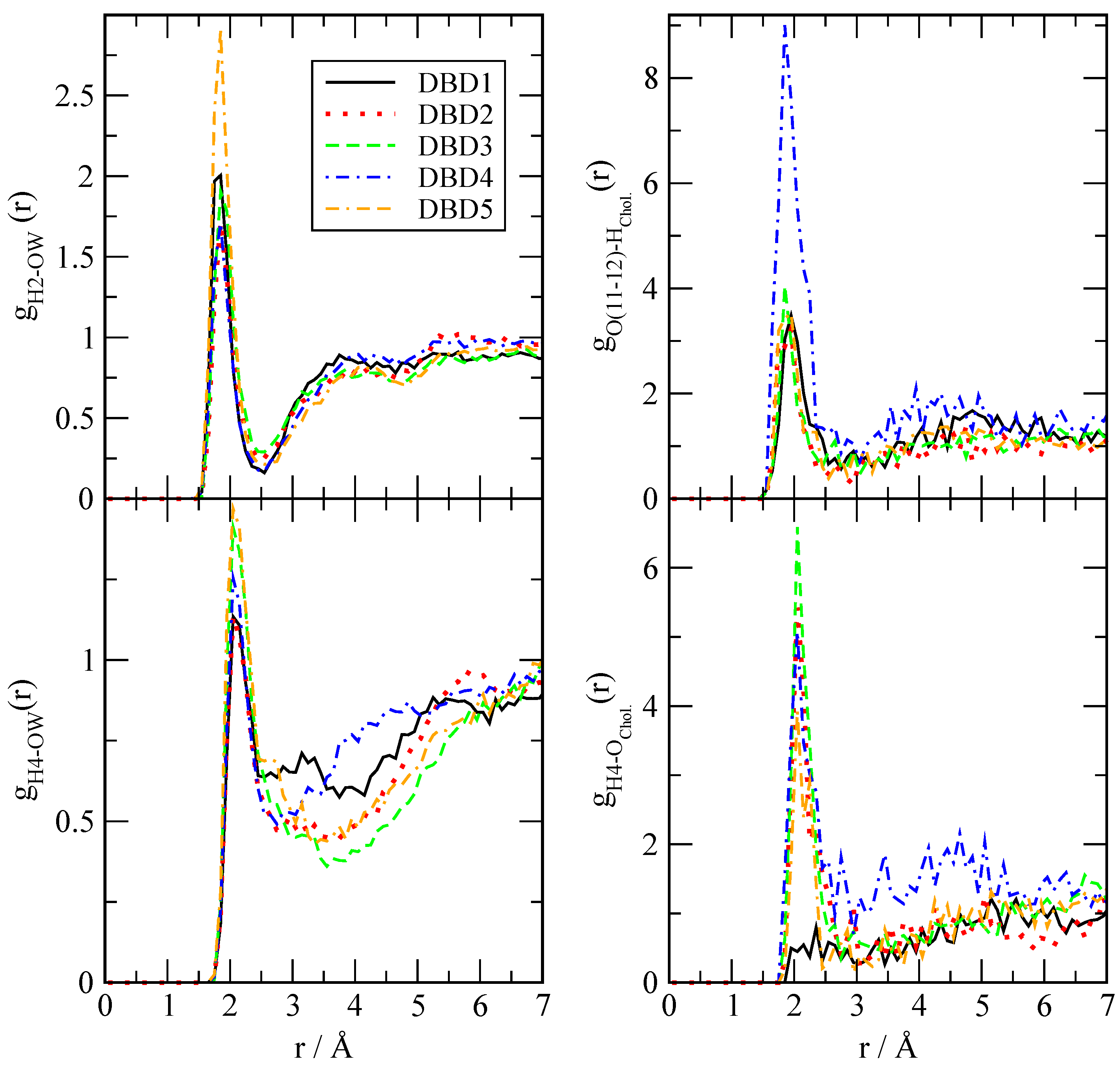
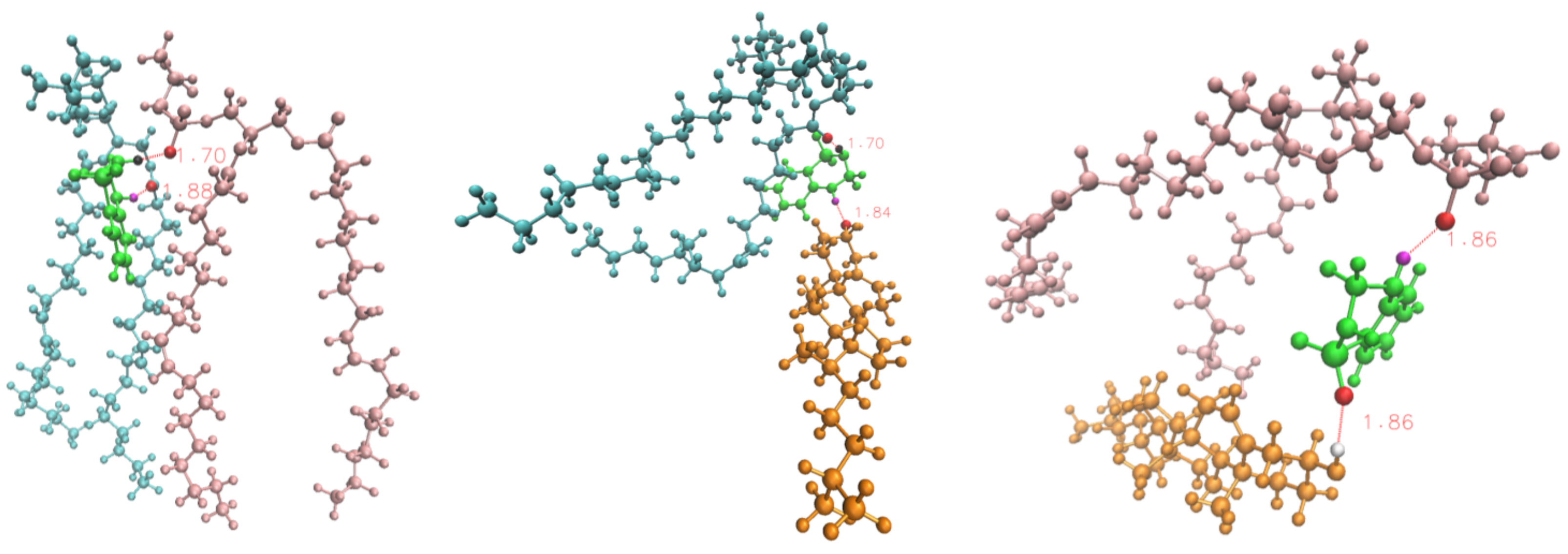
| DBD Derivative | A | |
|---|---|---|
| DBD1 | 52.18 | 43.04 |
| DBD2 | 52.20 | 43.02 |
| DBD3 | 52.23 | 42.98 |
| DBD4 | 52.19 | 43.02 |
| DBD5 | 52.23 | 42.97 |
| DBD Site | Lipid Site | |||||
|---|---|---|---|---|---|---|
| DBD1 | DBD2 | DBD3 | DBD4 | DBD5 | ||
| H2 | O13-14 DOPC | 4.2 | 4.0 | 4.0 | 4.2 | 5.2 |
| H2 | O22-32 DOPC | 3.4 | 3.2 | 2.5 | 4.4 | 3.7 |
| H2 | O13-14 DOPS | 4.2 | 3.8 | 5.0 | 4.7 | 4.7 |
| H2 | O22-32 DOPS | 2.8 | 3.5 | 3.3 | 3.0 | 2.2 |
| H4 | O11 DOPC | 2.0 | 2.5 | 2.4 | 1.9 | 3.1 |
| H4 | O22-32 DOPC | 1.6 | 2.2 | 1.9 | 2.3 | 2.3 |
| H4 | O11 DOPS | 2.3 | 1.2 | 3.5 | 1.8 | 3.8 |
| H4 | O22-32 DOPS | 1.8 | 1.6 | 2.0 | 4.0 | 3.9 |
| H4 | O Cholesterol | 1.2 | 3.2 | 2.9 | 2.4 | 3.4 |
| O11-12 | H Cholesterol | 1.8 | 2.4 | 2.2 | 2.6 | 2.3 |
| DBD Site | Lipid Site | Distance | |||||
|---|---|---|---|---|---|---|---|
| H2 | O13-14 DOPC | 1.7 | 67.4 | 73.7 | 63.2 | 28.4 | 38.5 |
| H2 | O22-32 DOPC | 1.8 | 20.4 | 11.8 | 12.3 | 19.4 | 24.6 |
| H2 | O13-14 DOPS | 1.7 | 6.9 | 8.8 | 27.0 | 9.6 | 9.7 |
| H2 | O22-32 DOPS | 1.8 | 1.4 | 1.9 | 2.3 | 3.1 | 0.9 |
| H4 | O11-12 DOPC | 1.9 | 42.5 | 64.4 | 61.5 | 15.6 | 35.9 |
| H4 | O22-32 DOPC | 2.0 | 16.5 | 14.0 | 13.0 | 16.6 | 28.4 |
| H4 | O11-12 DOPS | 1.9 | 1.3 | 1.9 | 24.8 | 1.9 | 6.4 |
| H4 | O22-32 DOPS | 2.0 | 1.8 | 1.4 | 0.9 | 2.6 | 1.9 |
| H4 | O Cholesterol | 2.0 | 0.5 | 3.4 | 4.4 | 3.7 | 3.6 |
| O11-12 | H Cholesterol | 1.9 | 4.6 | 4.5 | 6.6 | 10.3 | 6.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Z.; Marti, J. In Silico Drug Design of Benzothiadiazine Derivatives Interacting with Phospholipid Cell Membranes. Membranes 2022, 12, 331. https://doi.org/10.3390/membranes12030331
Hu Z, Marti J. In Silico Drug Design of Benzothiadiazine Derivatives Interacting with Phospholipid Cell Membranes. Membranes. 2022; 12(3):331. https://doi.org/10.3390/membranes12030331
Chicago/Turabian StyleHu, Zheyao, and Jordi Marti. 2022. "In Silico Drug Design of Benzothiadiazine Derivatives Interacting with Phospholipid Cell Membranes" Membranes 12, no. 3: 331. https://doi.org/10.3390/membranes12030331
APA StyleHu, Z., & Marti, J. (2022). In Silico Drug Design of Benzothiadiazine Derivatives Interacting with Phospholipid Cell Membranes. Membranes, 12(3), 331. https://doi.org/10.3390/membranes12030331







