Reconstitution of Functional Integrin αIIbβ3 and Its Activation in Plasma Membrane-Mimetic Lipid Environments
Abstract
1. Introduction
2. Materials and Methods
3. Results
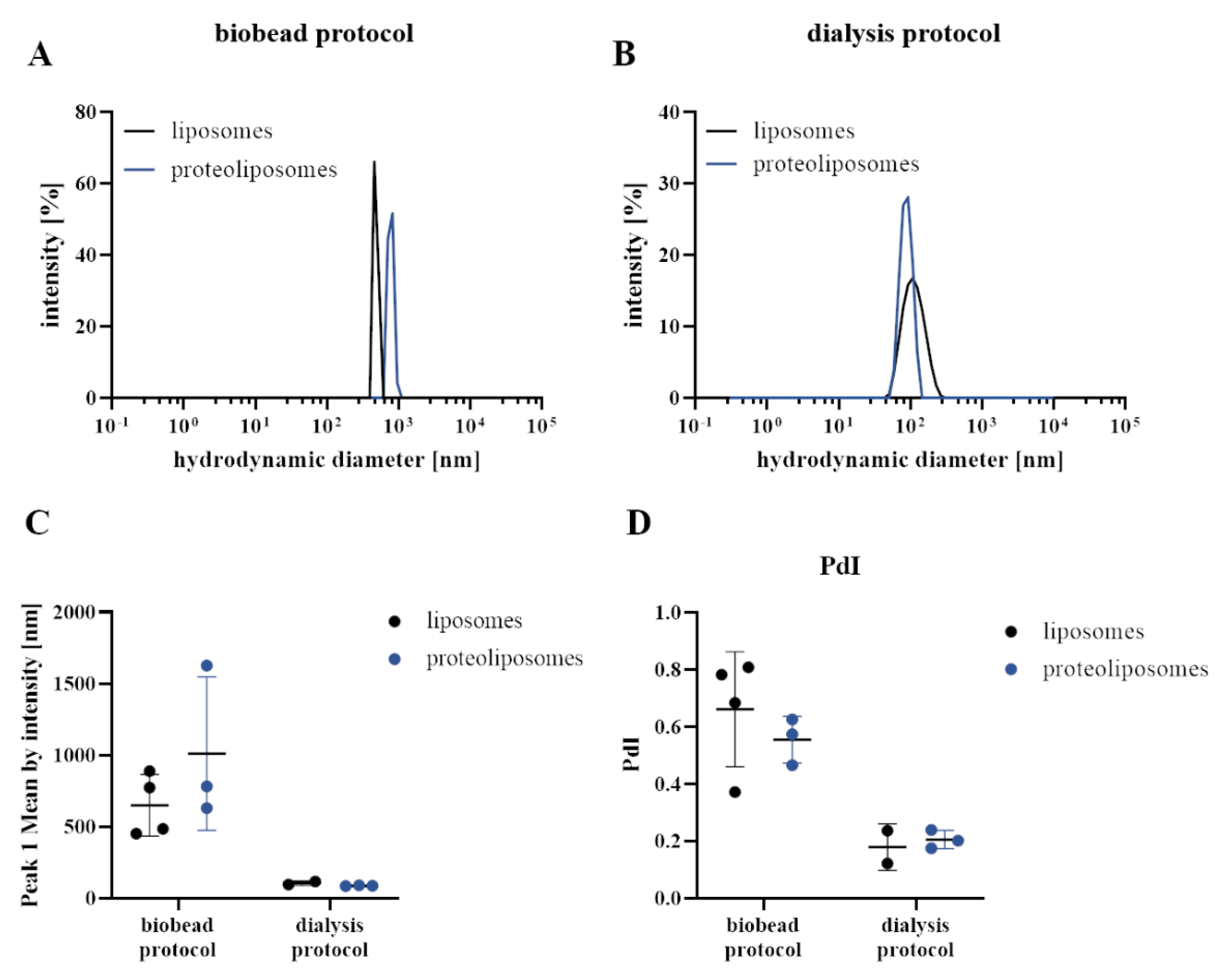
3.1. Different Reconstitution Procedures of Integrin into Cell Membrane Mimetic Liposome System
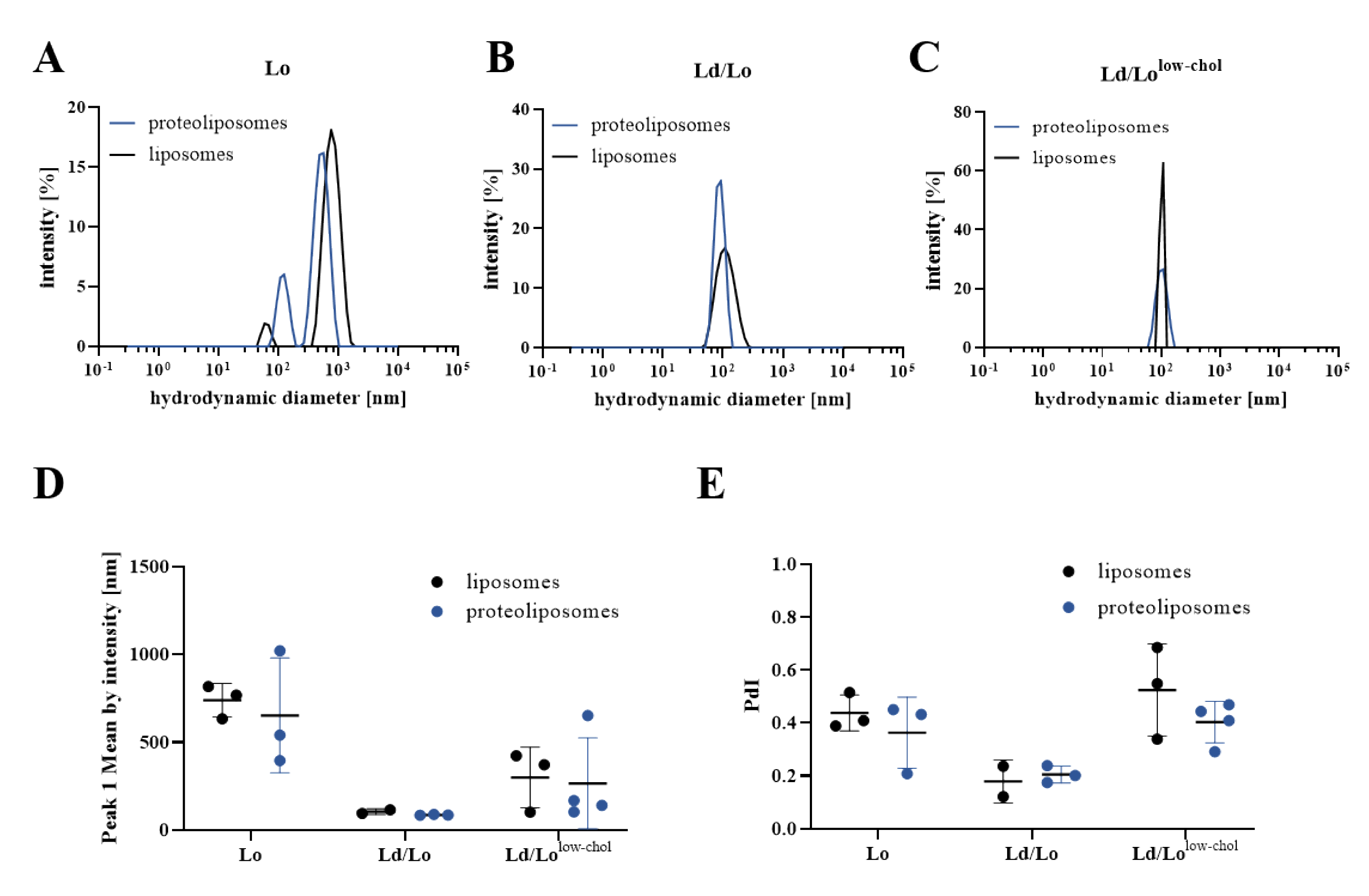
3.2. Effect of Cholesterol Content or Membrane Order on the αIIbβ3 Activation State
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Palacios-Acedo, A.L.; Mège, D.; Crescence, L.; Dignat-George, F.; Dubois, C.; Panicot-Dubois, L. Platelets, Thrombo-Inflammation, and Cancer: Collaborating With the Enemy. Front. Immunol. 2019, 10, 1805. [Google Scholar] [CrossRef]
- Bennett, J.S. Structure and function of the platelet integrin alphaIIbbeta3. J. Clin. Investig. 2005, 115, 3363–3369. [Google Scholar] [CrossRef]
- Delon, I.; Brown, N.H. Integrins and the actin cytoskeleton. Curr. Opin. Cell Biol. 2007, 19, 43–50. [Google Scholar] [CrossRef]
- Campbell, I.D.; Humphries, M.J. Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef]
- Zhu, J.; Zhu, J.; Negri, A.; Provasi, D.; Filizola, M.; Coller, B.S.; Springer, T.A. Closed headpiece of integrin αIIbβ3 and its complex with an αIIbβ3-specific antagonist that does not induce opening. Blood 2010, 116, 5050–5059. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.-H.; Carman, C.V.; Springer, T.A. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 2007, 25, 619–647. [Google Scholar] [CrossRef] [PubMed]
- Stalker, T.J.; Newman, D.K.; Ma, P.; Wannemacher, K.M.; Brass, L.F. Platelet signaling. Handb. Exp. Pharmacol. 2012, 59–85. [Google Scholar] [CrossRef]
- Erb, E.M.; Engel, J. Reconstitution of functional integrin into phospholipid vesicles and planar lipid bilayers. Methods Mol. Biol. 2000, 139, 71–82. [Google Scholar] [CrossRef]
- Gingras, A.R.; Ye, F.; Ginsberg, M.H. Reconstructing integrin activation in vitro. Methods Mol. Biol. 2013, 1046, 1–17. [Google Scholar] [CrossRef]
- Janke, U.; Kulke, M.; Buchholz, I.; Geist, N.; Langel, W.; Delcea, M. Drug-induced activation of integrin alpha IIb beta 3 leads to minor localized structural changes. PLoS ONE 2019, 14, e0214969. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.-S.; Rice, W.J.; Stokes, D.L.; Coller, B.S. Three-dimensional reconstruction of intact human integrin αIIbβ3: New implications for activation-dependent ligand binding. Blood 2013, 122, 4165–4171. [Google Scholar] [CrossRef]
- Ye, F.; Hu, G.; Taylor, D.; Ratnikov, B.; Bobkov, A.A.; McLean, M.A.; Sligar, S.G.; Taylor, K.A.; Ginsberg, M.H. Recreation of the terminal events in physiological integrin activation. J. Cell Biol. 2010, 188, 157–173. [Google Scholar] [CrossRef] [PubMed]
- Jackman, J.A.; Cho, N.-J. Supported Lipid Bilayer Formation: Beyond Vesicle Fusion. Langmuir 2020, 36, 1387–1400. [Google Scholar] [CrossRef]
- Glazier, R.; Salaita, K. Supported lipid bilayer platforms to probe cell mechanobiology. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1465–1482. [Google Scholar] [CrossRef]
- Schmidt, T.; Suk, J.-E.; Ye, F.; Situ, A.J.; Mazumder, P.; Ginsberg, M.H.; Ulmer, T.S. Annular anionic lipids stabilize the integrin αIIbβ3 transmembrane complex. J. Biol. Chem. 2015, 290, 8283–8293. [Google Scholar] [CrossRef]
- Gaul, V.; Lopez, S.G.; Lentz, B.R.; Moran, N.; Forster, R.J.; Keyes, T.E. The lateral diffusion and fibrinogen induced clustering of platelet integrin αIIbβ3 reconstituted into physiologically mimetic GUVs. Integr. Biol. 2015, 7, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Siontorou, C.G.; Nikoleli, G.-P.; Nikolelis, D.P.; Karapetis, S.K. Artificial Lipid Membranes: Past, Present, and Future. Membranes 2017, 7, 38. [Google Scholar] [CrossRef]
- Rigaud, J.-L.; Lévy, D. Reconstitution of Membrane Proteins into Liposomes. In Liposomes, Part B; Elsevier: Amsterdam, The Netherlands, 2003; pp. 65–86. ISBN 9780121822750. [Google Scholar]
- Seddon, A.M.; Curnow, P.; Booth, P.J. Membrane proteins, lipids and detergents: Not just a soap opera. Biochim. Biophys. Acta 2004, 1666, 105–117. [Google Scholar] [CrossRef]
- Erb, E.M.; Tangemann, K.; Bohrmann, B.; Müller, B.; Engel, J. Integrin alphaIIb beta3 reconstituted into lipid bilayers is nonclustered in its activated state but clusters after fibrinogen binding. Biochemistry 1997, 36, 7395–7402. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, J.; Jang, J.; Ye, F.; Hong, S.J.; Petrich, B.G.; Ulmer, T.S.; Kim, C. Topological Adaptation of Transmembrane Domains to the Force-Modulated Lipid Bilayer Is a Basis of Sensing Mechanical Force. Curr. Biol. 2020, 30, 1614–1625.e5. [Google Scholar] [CrossRef]
- Martens, U.; Janke, U.; Möller, S.; Talbot, D.; Abou-Hassan, A.; Delcea, M. Interaction of fibrinogen-magnetic nanoparticle bioconjugates with integrin reconstituted into artificial membranes. Nanoscale 2020, 12, 19918–19930. [Google Scholar] [CrossRef]
- Richter, R.; Mukhopadhyay, A.; Brisson, A. Pathways of Lipid Vesicle Deposition on Solid Surfaces: A Combined QCM-D and AFM Study. Biophys. J. 2003, 85, 3035–3047. [Google Scholar] [CrossRef]
- van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, V.B.; Murphy, R.C.; Watson, S.P. Platelet lipidomics: Modern day perspective on lipid discovery and characterization in platelets. Circ. Res. 2014, 114, 1185–1203. [Google Scholar] [CrossRef] [PubMed]
- Sunshine, H.; Iruela-Arispe, M.L. Membrane lipids and cell signaling. Curr. Opin. Lipidol. 2017, 28, 408–413. [Google Scholar] [CrossRef]
- Lietha, D.; Izard, T. Roles of Membrane Domains in Integrin-Mediated Cell Adhesion. Int. J. Mol. Sci. 2020, 21, 5531. [Google Scholar] [CrossRef]
- Luchini, A.; Vitiello, G. Mimicking the Mammalian Plasma Membrane: An Overview of Lipid Membrane Models for Biophysical Studies. Biomimetics 2020, 6, 3. [Google Scholar] [CrossRef]
- Kaiser, H.-J.; Lingwood, D.; Levental, I.; Sampaio, J.L.; Kalvodova, L.; Rajendran, L.; Simons, K. Order of lipid phases in model and plasma membranes. Proc. Natl. Acad. Sci. USA 2009, 106, 16645–16650. [Google Scholar] [CrossRef]
- Veatch, S.L.; Keller, S.L. Separation of Liquid Phases in Giant Vesicles of Ternary Mixtures of Phospholipids and Cholesterol. Biophys. J. 2003, 85, 3074–3083. [Google Scholar] [CrossRef]
- Kahya, N.; Scherfeld, D.; Bacia, K.; Poolman, B.; Schwille, P. Probing lipid mobility of raft-exhibiting model membranes by fluorescence correlation spectroscopy. J. Biol. Chem. 2003, 278, 28109–28115. [Google Scholar] [CrossRef]
- Nazari, M.; Kurdi, M.; Heerklotz, H. Classifying surfactants with respect to their effect on lipid membrane order. Biophys. J. 2012, 102, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Rodi, P.M.; Bocco Gianello, M.D.; Corregido, M.C.; Gennaro, A.M. Comparative study of the interaction of CHAPS and Triton X-100 with the erythrocyte membrane. Biochim. Biophys. Acta 2014, 1838, 859–866. [Google Scholar] [CrossRef]
- Rosen, M.J.; Kunjappu, J.T. Surfactants and Interfacial Phenomena; John Wiley & Sons, Inc: Hoboken, NJ, USA, 2012; ISBN 9781118228920. [Google Scholar]
- Coskun, Ü.; Grzybek, M.; Drechsel, D.; Simons, K. Regulation of human EGF receptor by lipids. Proc. Natl. Acad. Sci. USA 2011, 108, 9044–9048. [Google Scholar] [CrossRef]
- Ejsing, C.S.; Sampaio, J.L.; Surendranath, V.; Duchoslav, E.; Ekroos, K.; Klemm, R.W.; Simons, K.; Shevchenko, A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA 2009, 106, 2136–2141. [Google Scholar] [CrossRef]
- Hardy, G.J.; Nayak, R.; Alam, S.M.; Shapter, J.G.; Heinrich, F.; Zauscher, S. Biomimetic supported lipid bilayers with high cholesterol content formed by α-helical peptide-induced vesicle fusion. J. Mater. Chem. 2012, 22, 19506–19513. [Google Scholar] [CrossRef] [PubMed]
- Shattil, S.J.; Hoxie, J.A.; Cunningham, M.; Brass, L.F. Changes in the platelet membrane glycoprotein IIb.IIIa complex during platelet activation. J. Biol. Chem. 1985, 260, 11107–11114. [Google Scholar] [CrossRef]
- Ohi, M.; Li, Y.; Cheng, Y.; Walz, T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol. Proced. Online 2004, 6, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.-Y.; Zhu, J.; Eng, E.T.; Hudson, N.E.; Springer, T.A. β-Subunit Binding Is Sufficient for Ligands to Open the Integrin αIIbβ3 Headpiece. J. Biol. Chem. 2016, 291, 4537–4546. [Google Scholar] [CrossRef]
- Stetefeld, J.; McKenna, S.A.; Patel, T.R. Dynamic light scattering: A practical guide and applications in biomedical sciences. Biophys. Rev. 2016, 8, 409–427. [Google Scholar] [CrossRef]
- Zhang, K.; Chen, J. The regulation of integrin function by divalent cations. Cell Adhes. Migr. 2012, 6, 20–29. [Google Scholar] [CrossRef]
- Simons, K.; Sampaio, J.L. Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 2011, 3, a004697. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.-H.; Imanaka, M.Y.; Modahl, E.H.; Torres-Ocampo, A.P. Lipid Raft Phase Modulation by Membrane-Anchored Proteins with Inherent Phase Separation Properties. ACS Omega 2019, 4, 6551–6559. [Google Scholar] [CrossRef]
- Shahidi, F.; Zhong, Y. Lipid oxidation and improving the oxidative stability. Chem. Soc. Rev. 2010, 39, 4067–4079. [Google Scholar] [CrossRef]
- Zuidam, N.J.; Gouw, H.K.M.E.; Barenholz, Y.; Crommelin, D.J.A. Physical (in) stability of liposomes upon chemical hydrolysis: The role of lysophospholipids and fatty acids. Biochim. Biophys. Acta Biomembr. 1995, 1240, 101–110. [Google Scholar] [CrossRef]
- Bodin, S.; Soulet, C.; Tronchère, H.; Sié, P.; Gachet, C.; Plantavid, M.; Payrastre, B. Integrin-dependent interaction of lipid rafts with the actin cytoskeleton in activated human platelets. J. Cell Sci. 2005, 118, 759–769. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, L.; Tonggu, L. Membrane protein reconstitution for functional and structural studies. Sci. China Life Sci. 2015, 58, 66–74. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rebaud, S.; Maniti, O.; Girard-Egrot, A.P. Tethered bilayer lipid membranes (tBLMs): Interest and applications for biological membrane investigations. Biochimie 2014, 107 Pt A, 135–142. [Google Scholar] [CrossRef]
- Leitinger, B.; Hogg, N. The involvement of lipid rafts in the regulation of integrin function. J. Cell Sci. 2002, 115, 963–972. [Google Scholar] [CrossRef] [PubMed]
- López, J.A.; del Conde, I.; Shrimpton, C.N. Receptors, rafts, and microvesicles in thrombosis and inflammation. J. Thromb. Haemost. 2005, 3, 1737–1744. [Google Scholar] [CrossRef]
- Norambuena, A.; Schwartz, M.A. Effects of integrin-mediated cell adhesion on plasma membrane lipid raft components and signaling. Mol. Biol. Cell 2011, 22, 3456–3464. [Google Scholar] [CrossRef]
- Varshney, P.; Yadav, V.; Saini, N. Lipid rafts in immune signalling: Current progress and future perspective. Immunology 2016, 149, 13–24. [Google Scholar] [CrossRef]
- Hanein, D.; Volkmann, N. Conformational Equilibrium of Human Platelet Integrin Investigated by Three-Dimensional Electron Cryo-Microscopy. Subcell. Biochem. 2018, 87, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Gaus, K.; Le Lay, S.; Balasubramanian, N.; Schwartz, M.A. Integrin-mediated adhesion regulates membrane order. J. Cell Biol. 2006, 174, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Taub, R.; Gould, R.J.; Garsky, V.M.; Ciccarone, T.M.; Hoxie, J.; Friedman, P.A.; Shattil, S.J. A monoclonal antibody against the platelet fibrinogen receptor contains a sequence that mimics a receptor recognition domain in fibrinogen. J. Biol. Chem. 1989, 264, 259–265. [Google Scholar] [CrossRef]
- Head, B.P.; Patel, H.H.; Insel, P.A. Interaction of membrane/lipid rafts with the cytoskeleton: Impact on signaling and function: Membrane/lipid rafts, mediators of cytoskeletal arrangement and cell signaling. Biochim. Biophys. Acta 2014, 1838, 532–545. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janke, U.; Mitlehner, A.; Weide, A.; Gutmann, T.; Delcea, M. Reconstitution of Functional Integrin αIIbβ3 and Its Activation in Plasma Membrane-Mimetic Lipid Environments. Membranes 2021, 11, 499. https://doi.org/10.3390/membranes11070499
Janke U, Mitlehner A, Weide A, Gutmann T, Delcea M. Reconstitution of Functional Integrin αIIbβ3 and Its Activation in Plasma Membrane-Mimetic Lipid Environments. Membranes. 2021; 11(7):499. https://doi.org/10.3390/membranes11070499
Chicago/Turabian StyleJanke, Una, Alexandra Mitlehner, Aileen Weide, Theresia Gutmann, and Mihaela Delcea. 2021. "Reconstitution of Functional Integrin αIIbβ3 and Its Activation in Plasma Membrane-Mimetic Lipid Environments" Membranes 11, no. 7: 499. https://doi.org/10.3390/membranes11070499
APA StyleJanke, U., Mitlehner, A., Weide, A., Gutmann, T., & Delcea, M. (2021). Reconstitution of Functional Integrin αIIbβ3 and Its Activation in Plasma Membrane-Mimetic Lipid Environments. Membranes, 11(7), 499. https://doi.org/10.3390/membranes11070499





