Prediction of the Long-Term Effect of Iron on Methane Yield in an Anaerobic Membrane Bioreactor Using Bayesian Network Meta-Analysis
Abstract
1. Introduction
2. Material and Methods
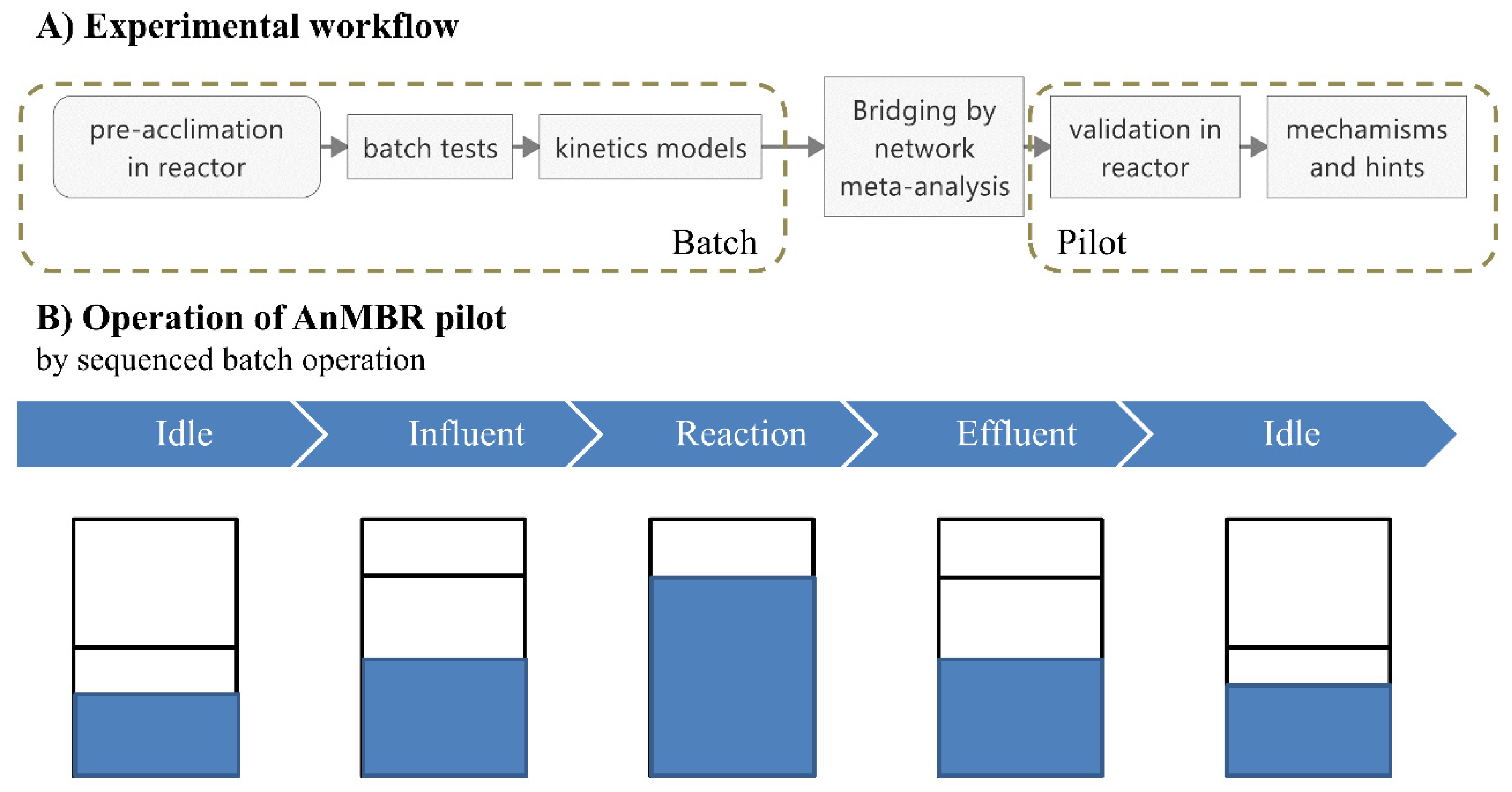
2.1. Scheme and Operation of the Anaerobic Membrane Bioreactor
2.2. Apparent Kinetics of Methanogenesis
2.3. Effects Evaluation by Bayesian Network Meta-Analysis
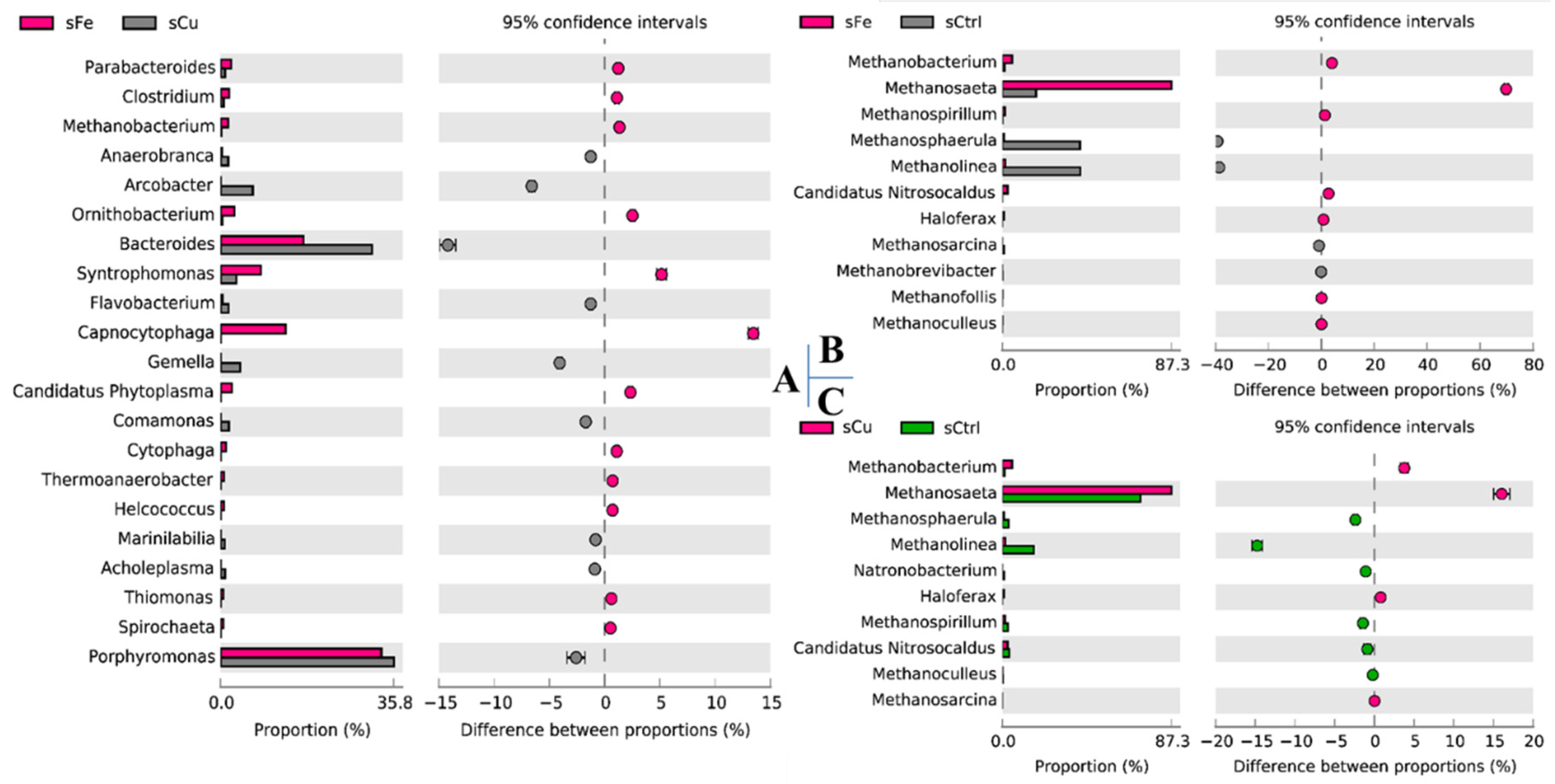
2.4. Microbial Community Analysis
2.5. Physicochemical Analysis
2.6. Statistical Analysis
3. Results
3.1. Enhancement of Cumulative Methane Production in the Batch Test
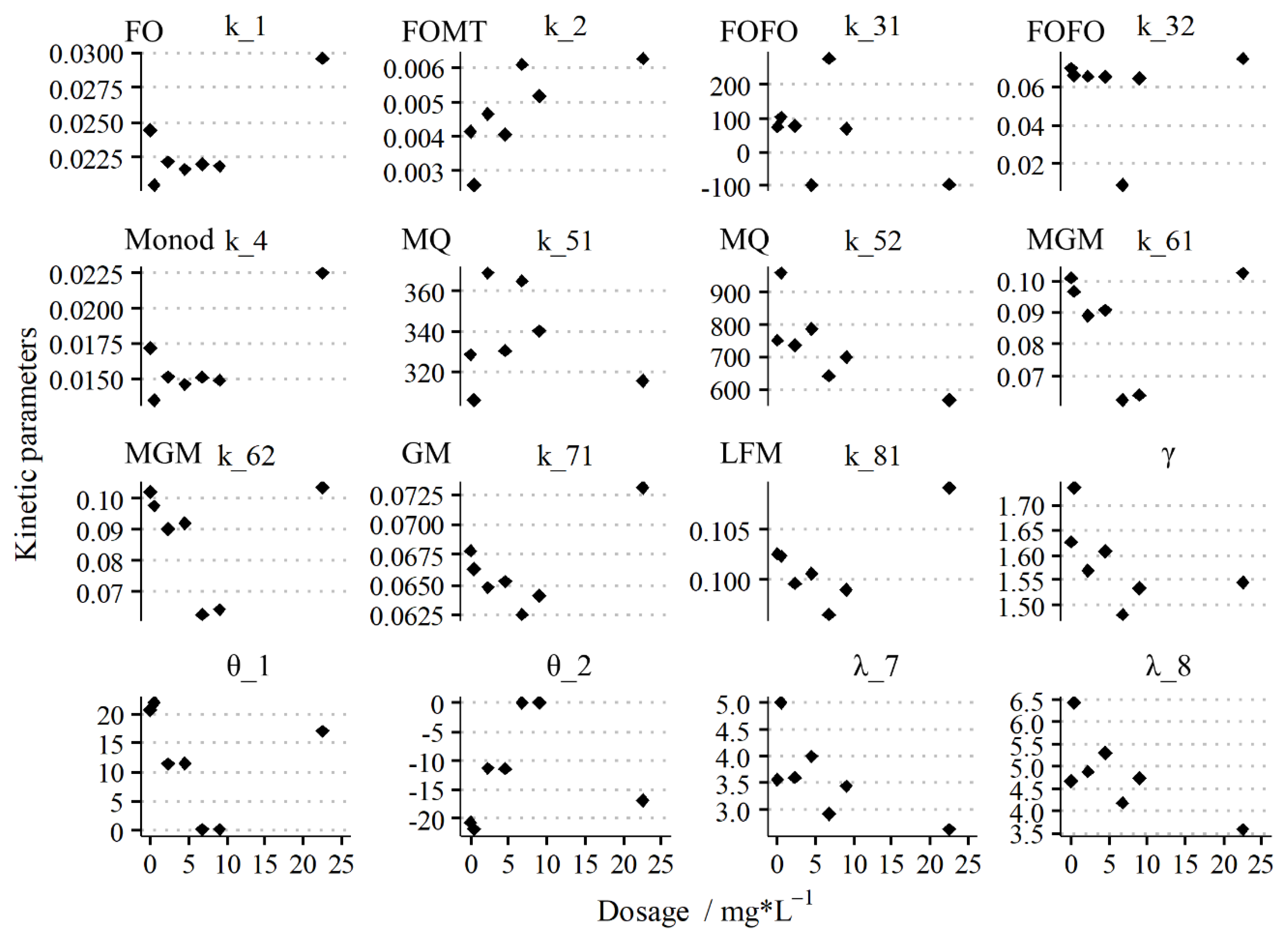
3.2. Effect of Dosages on the Kinetics Parameters in the Batch Test
3.3. Changing of Methanogenic Kinetic Patterns in the Batch Test
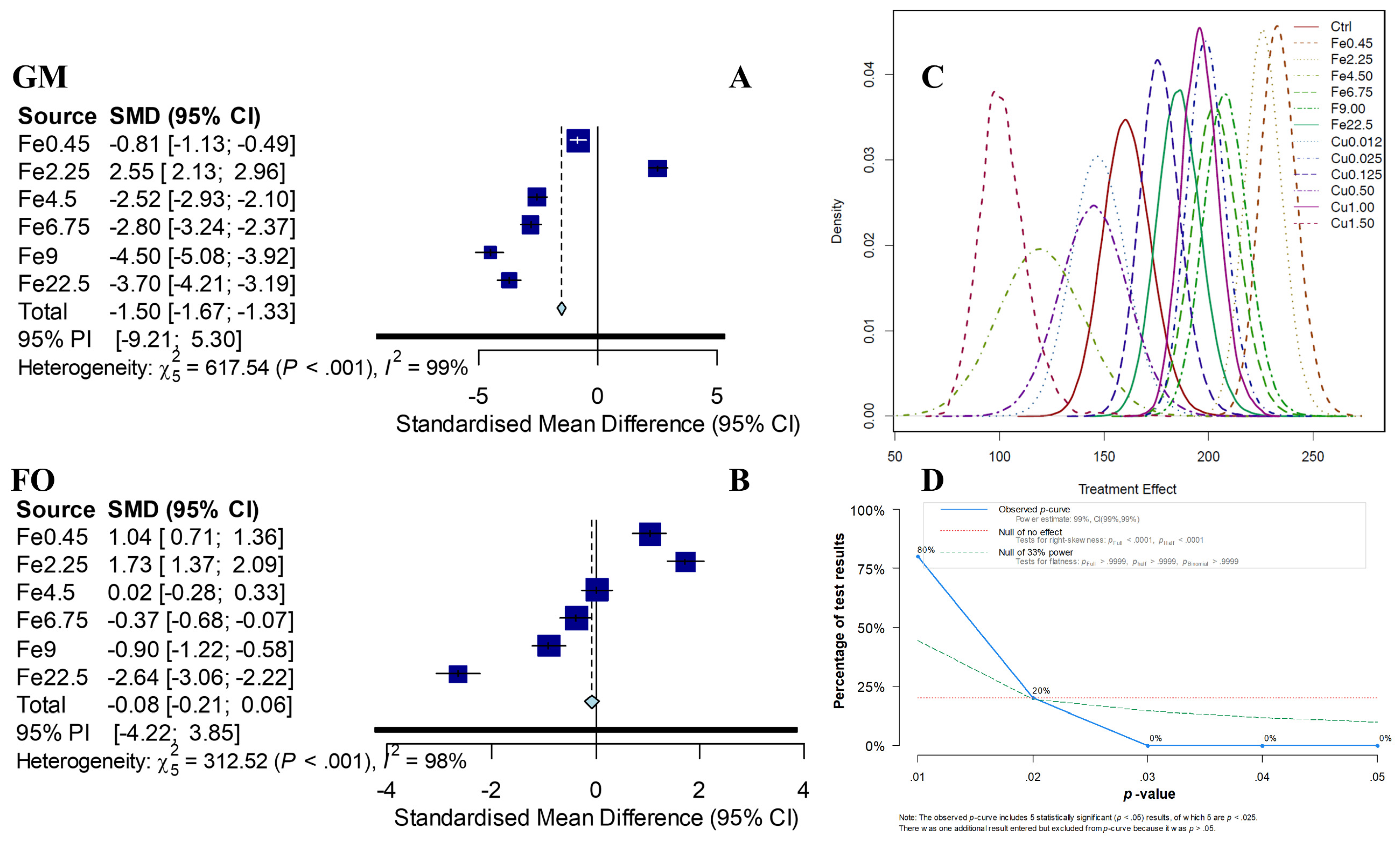
3.4. Effect of Dosage on Biochemical Methane Potential by GM-Based NMA
3.5. Changing of Methane Yield and Performance in Semicontinuous AnMBR
4. Discussion
4.1. Potential Mechanisms of Methanogenic Kinetic Response to Ferric
4.2. Deciphering the Methanogenic Kinetic Response to Ferric
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fletcher, S.E.M.; Schaefer, H. Rising methane: A new climate challenge. Science 2019, 364, 932–933. [Google Scholar] [CrossRef] [PubMed]
- Hafner, S.D.; Koch, K.; Carrere, H.; Astals, S.; Weinrich, S.; Rennuit, C. Software for biogas research: Tools for measurement and prediction of methane production. SoftwareX 2018, 7, 205–210. [Google Scholar] [CrossRef]
- von Cossel, M.; Möhring, J.; Kiesel, A.; Lewandowski, I. Optimization of specific methane yield prediction models for biogas crops based on lignocellulosic components using non-linear and crop-specific configurations. Ind. Crops Prod. 2018, 120, 330–342. [Google Scholar] [CrossRef]
- Zang, Y.; Yang, Y.; Hu, Y.; Ngo, H.H.; Wang, X.C.; Li, Y.Y. Zero-valent iron enhanced anaerobic digestion of pre-concentrated domestic wastewater for bioenergy recovery: Characteristics and mechanisms. Bioresour. Technol. 2020, 310, 123441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lu, T.; Wang, Z.; Wang, Y.; Zhong, H.; Shen, P.; Wei, Y. Effects of magnetite on anaerobic digestion of swine manure: Attention to methane production and fate of antibiotic resistance genes. Bioresour. Technol. 2019, 291, 121847. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Z.; Lu, T.; Liu, J.; Wang, Y.; Shen, P.; Wei, Y. Response and mechanisms of the performance and fate of antibiotic resistance genes to nano-magnetite during anaerobic digestion of swine manure. J. Hazard. Mater. 2019, 366, 192–201. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Wu, J. Enhancement of methane production in anaerobic digestion process: A review. Appl. Energy 2019, 240, 120–137. [Google Scholar] [CrossRef]
- Baek, G.; Kim, J.; Lee, C. A long-term study on the effect of magnetite supplementation in continuous anaerobic digestion of dairy effluentEnhancement in process performance and stability. Bioresour. Technol. 2016, 222, 344–354. [Google Scholar] [CrossRef]
- Martins, G.; Salvador, A.F.; Pereira, L.; Alves, M.M. Methane Production and Conductive Materials: A Critical Review. Environ. Sci. Technol. 2018, 52, 10241–10253. [Google Scholar] [CrossRef]
- Wu, X.; Tian, Z.; Lv, Z.; Chen, Z.; Liu, Y.; Yong, X.; Zhou, J.; Xie, X.; Jia, H.; Wei, P. Effects of copper salts on performance, antibiotic resistance genes, and microbial community during thermophilic anaerobic digestion of swine manure. Bioresour. Technol. 2020, 300, 122728. [Google Scholar] [CrossRef]
- Chan, P.C.; Lu, Q.; de Toledo, R.A.; Gu, J.D.; Shim, H. Improved anaerobic co-digestion of food waste and domestic wastewater by copper supplementation—Microbial community change and enhanced effluent quality. Sci. Total Environ. 2019, 670, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Howell, G.; Bennett, C.; Materić, D. A comparison of methods for early prediction of anaerobic biogas potential on biologically treated municipal solid waste. J. Environ. Manag. 2019, 232, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Strömberg, S.; Nistor, M.; Liu, J. Early prediction of Biochemical Methane Potential through statistical and kinetic modelling of initial gas production. Bioresour. Technol. 2015, 176, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; van Lier, J.B.; de Kreuk, M. Digestibility of waste aerobic granular sludge from a full-scale municipal wastewater treatment system. Water Res. 2020, 173, 115617. [Google Scholar] [CrossRef] [PubMed]
- Batstone, D.J.J.; Keller, J.; Angelidaki, I.; Kalyuzhnyi, S.V.V.; Pavlostathis, S.G.G.; Rozzi, A.; Sanders, W.T.M.T.; Siegrist, H.; Vavilin, V.A.A. The IWA Anaerobic Digestion Model No 1 (ADM1). Water Sci. Technol. 2002, 45, 65–73. [Google Scholar] [CrossRef]
- Lee, E.; Cumberbatch, J.; Wang, M.; Zhang, Q. Kinetic parameter estimation model for anaerobic co-digestion of waste activated sludge and microalgae. Bioresour. Technol. 2017, 228, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lang, Q.; Wu, S.; Li, W.; Bah, H.; Dong, R. Anaerobic digestion characteristics of pig manures depending on various growth stages and initial substrate concentrations in a scaled pig farm in Southern China. Bioresour. Technol. 2014, 156, 63–69. [Google Scholar] [CrossRef]
- Kainthola, J.; Kalamdhad, A.S.; Goud, V.V.; Goel, R. Fungal pretreatment and associated kinetics of rice straw hydrolysis to accelerate methane yield from anaerobic digestion. Bioresour. Technol. 2019, 286, 121368. [Google Scholar] [CrossRef]
- Yu, D.; Liu, J.; Sui, Q.; Wei, Y. Biogas-pH automation control strategy for optimizing organic loading rate of anaerobic membrane bioreactor treating high COD wastewater. Bioresour. Technol. 2016, 203, 62–70. [Google Scholar] [CrossRef]
- Tian, Z.; Zhang, Y.; Li, Y.; Chi, Y.; Yang, M. Rapid establishment of thermophilic anaerobic microbial community during the one-step startup of thermophilic anaerobic digestion from a mesophilic digester. Water Res. 2015, 69, 9–19. [Google Scholar] [CrossRef]
- Cheng, H.; Li, Y.; Kato, H.; Li, Y.Y. Enhancement of sustainable flux by optimizing filtration mode of a high-solid anaerobic membrane bioreactor during long-term continuous treatment of food waste. Water Res. 2020, 168, 115195. [Google Scholar] [CrossRef] [PubMed]
- Lay, J.J.; Lee, Y.J.; Noike, T. Feasibility of biological hydrogen production from organic fraction of municipal solid waste. Water Res. 1999, 33, 2579–2586. [Google Scholar] [CrossRef]
- Bratton, D.J.; Gaisl, T.; Schlatzer, C.; Kohler, M. Comparison of the effects of continuous positive airway pressure and mandibular advancement devices on sleepiness in patients with obstructive sleep apnoea: A network meta-analysis. Lancet Respir. Med. 2015, 3, 869–878. [Google Scholar] [CrossRef]
- Harrer, M.; Cuijpers, P.; Furukawa, T.A.; Ebert, D.D. Doing Meta-Analysis in R: A Hands-On Guide. 2019, pp. 1–265. Available online: https://bookdown.org/MathiasHarrer/Doing_Meta_Analysis_in_R/ (accessed on 31 January 2021).
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Yu, D.; Meng, X.; Liu, J.; Dian, L.; Sui, Q.; Zhang, J.; Zhong, H.; Wei, Y. Formation and characteristics of a ternary pH buffer system for in-situ biogas upgrading in two-phase anaerobic membrane bioreactor treating starch wastewater. Bioresour. Technol. 2018, 269, 57–66. [Google Scholar] [CrossRef]
- Zhang, J.J.; Cai, X.; Qi, L.; Shao, C.; Lin, Y.; Zhang, J.J.; Zhang, Y.; Shen, P.; Wei, Y. Effects of aeration strategy on the evolution of dissolved organic matter (DOM) and microbial community structure during sludge bio-drying. Appl. Microbiol. Biotechnol. 2015, 99, 7321–7331. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Batstone, D.J.; Lu, Y.; Jensen, P.D. Impact of dewatering technologies on specific methanogenic activity. Water Res. 2015, 82, 78–85. [Google Scholar] [CrossRef]
- Zhang, J.; Buhe, C.; Yu, D.; Zhong, H.; Wei, Y. Ammonia stress reduces antibiotic efflux but enriches horizontal gene transfer of antibiotic resistance genes in anaerobic digestion. Bioresour. Technol. 2020, 295, 122191. [Google Scholar] [CrossRef] [PubMed]
- Carbajosa, S.; Malki, M.; Caillard, R.; Lopez, M.F.; Palomares, F.J.; Martín-Gago, J.A.; Rodríguez, N.; Amils, R.; Fernández, V.M.; De Lacey, A.L. Electrochemical growth of Acidithiobacillus ferrooxidans on a graphite electrode for obtaining a biocathode for direct electrocatalytic reduction of oxygen. Biosens. Bioelectron. 2010, 26, 877–880. [Google Scholar] [CrossRef] [PubMed]
- Soussan, L.; Riess, J.; Erable, B.; Delia, M.L.; Bergel, A. Electrochemical reduction of CO2 catalysed by Geobacter sulfurreducens grown on polarized stainless steel cathodes. Electrochem. Commun. 2013, 28, 27–30. [Google Scholar] [CrossRef]
- Cruz Viggi, C.; Rossetti, S.; Fazi, S.; Paiano, P.; Majone, M.; Aulenta, F. Magnetite particles triggering a faster and more robust syntrophic pathway of methanogenic propionate degradation. Environ. Sci. Technol. 2014, 48, 7536–7543. [Google Scholar] [CrossRef] [PubMed]
- Hess, V.; Schuchmann, K.; Müller, V. The ferredoxin: NAD+ Oxidoreductase (Rnf) from the acetogen acetobacterium woodii requires na+ and is reversibly coupled to the membrane potential. J. Biol. Chem. 2013, 288, 31496–31502. [Google Scholar] [CrossRef] [PubMed]
- Meuer, J.; Kuettner, H.C.; Zhang, J.K.; Hedderich, R.; Metcalf, W.W. Genetic analysis of the archaeon Methanosarcina barkeri fusaro reveals a central role for ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc. Natl. Acad. Sci. USA 2002, 99, 5632–5637. [Google Scholar] [CrossRef]
- Buckel, W.; Thauer, R.K. Flavin-Based Electron Bifurcation, A New Mechanism of Biological Energy Coupling. Chem. Rev. 2018, 118, 3862–3886. [Google Scholar] [CrossRef] [PubMed]
- Shim, S.R.; Lee, J. Dose-response meta-analysis: Application and practice using the R software. Epidemiol. Health 2019, 41, e2019006. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, S.; Liang, D.; Li, N. Conductive materials in anaerobic digestion: From mechanism to application. Bioresour. Technol. 2020, 298, 122403. [Google Scholar] [CrossRef]
- Yu, D.; Li, C.; Wang, L.; Zhang, J.; Liu, J.; Wei, Y. Multiple effects of trace elements on methanogenesis in a two-phase anaerobic membrane bioreactor treating starch wastewater. Appl. Microbiol. Biotechnol. 2016, 100, 6631–6642. [Google Scholar] [CrossRef]
- Roussel, J.; Fermoso, F.G.; Collins, G.; Van Hullebusch, E.; Esposito, G.; Mucha, A.P. Trace Element Supplementation as a Management Tool for Anaerobic Digester Operation: Benefits and Risks; IWA Publishing: London, UK, 2018; Volume 17. [Google Scholar] [CrossRef][Green Version]

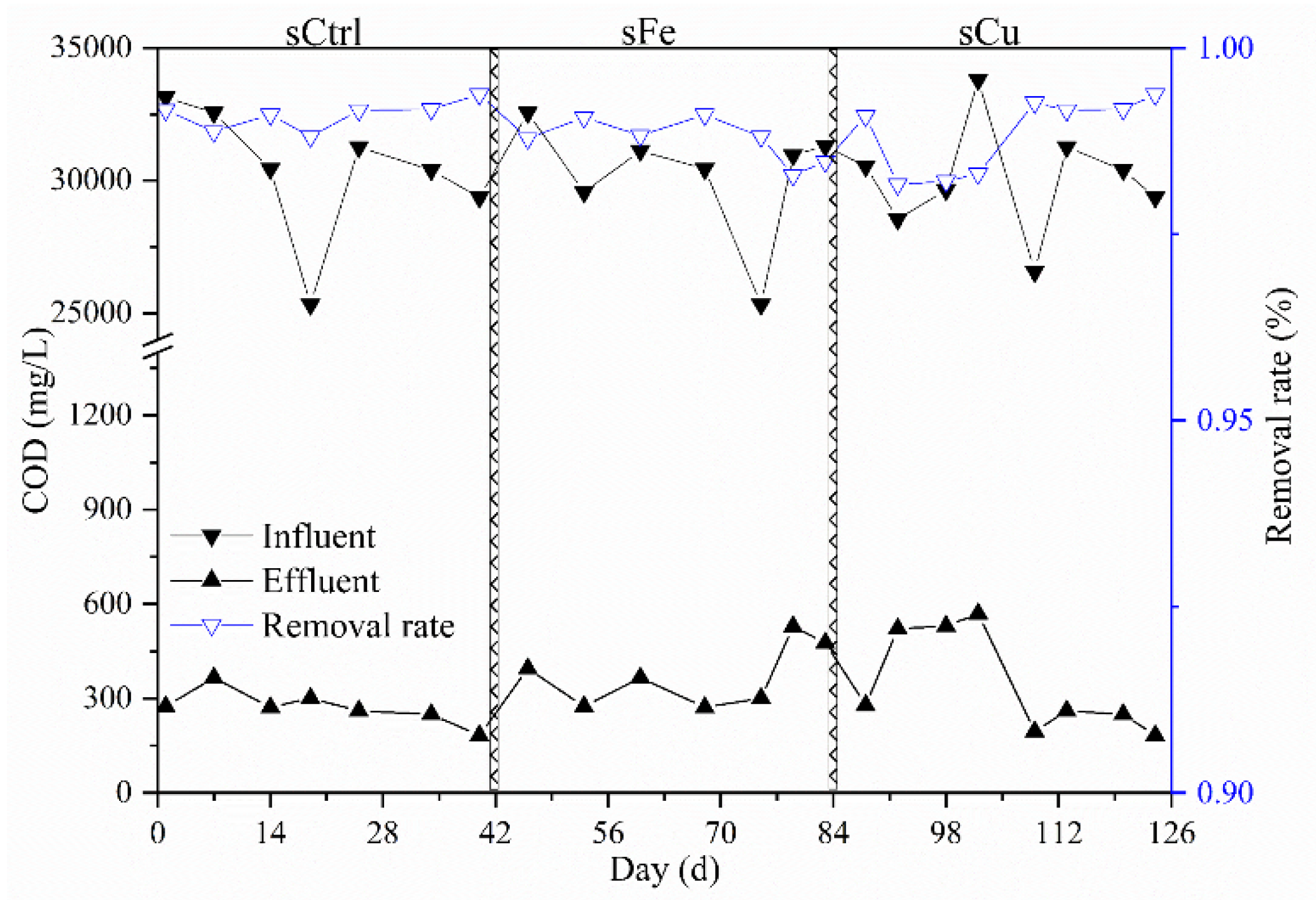
| Control Strategy (Biogas-pH strategy) | sCtrl | sFe | sFe Rate (%) | sCu | sCu Rate (%) |
|---|---|---|---|---|---|
| Time (day) | 1–42 | 42–84 | 84–126 | ||
| Organic loading rate (kgCOD⋅kgVSS−1⋅d−1) | 11.8 ± 0.3 | 12.4 ± 0.4 | 4.6 | 12.1 ± 0.6 | 2.1 |
| Effluent COD (mg⋅L−1) | 273 ± 48 | 374 ± 93 | 37 | 349 ± 152 | 28 |
| Methane flow rate (NmL⋅h−1) | 593 ± 351 | 632 ± 397 | 6.6 | 619 ± 368 | 4.44 |
| Methane yield (NmL⋅gCODin−1) | 309 ± 51 | 316 ± 57 | 2.27 | 310 ± 62 | 0.32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, D.; Liang, Y.; Thejani Nilusha, R.; Ritigala, T.; Wei, Y. Prediction of the Long-Term Effect of Iron on Methane Yield in an Anaerobic Membrane Bioreactor Using Bayesian Network Meta-Analysis. Membranes 2021, 11, 100. https://doi.org/10.3390/membranes11020100
Yu D, Liang Y, Thejani Nilusha R, Ritigala T, Wei Y. Prediction of the Long-Term Effect of Iron on Methane Yield in an Anaerobic Membrane Bioreactor Using Bayesian Network Meta-Analysis. Membranes. 2021; 11(2):100. https://doi.org/10.3390/membranes11020100
Chicago/Turabian StyleYu, Dawei, Yushuai Liang, Rathmalgodagei Thejani Nilusha, Tharindu Ritigala, and Yuansong Wei. 2021. "Prediction of the Long-Term Effect of Iron on Methane Yield in an Anaerobic Membrane Bioreactor Using Bayesian Network Meta-Analysis" Membranes 11, no. 2: 100. https://doi.org/10.3390/membranes11020100
APA StyleYu, D., Liang, Y., Thejani Nilusha, R., Ritigala, T., & Wei, Y. (2021). Prediction of the Long-Term Effect of Iron on Methane Yield in an Anaerobic Membrane Bioreactor Using Bayesian Network Meta-Analysis. Membranes, 11(2), 100. https://doi.org/10.3390/membranes11020100






