Synthetic Cell as a Platform for Understanding Membrane-Membrane Interactions
Abstract
:1. Introduction
2. Model Bilayer Membranes
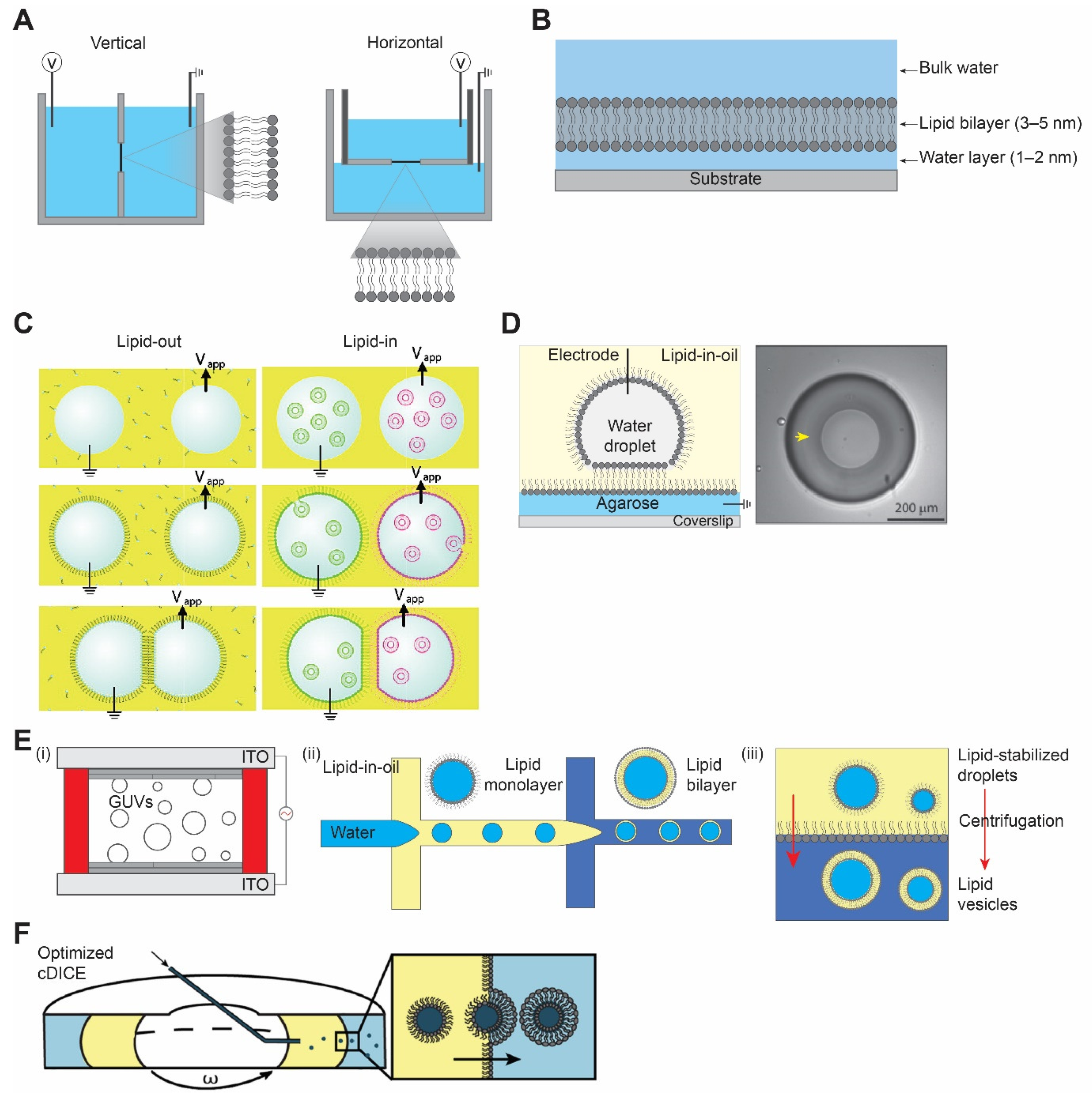
2.1. Planar Lipid Bilayer
2.1.1. Black Lipid Membrane
2.1.2. Droplet Interface Bilayer
2.2. Vesicle Preparation
2.2.1. Hydration Method
2.2.2. Droplet Microfluidics Method
2.2.3. Inverted Emulsion Method
2.2.4. cDICE Method
3. Membrane Protein Incorporation into Lipid Bilayer
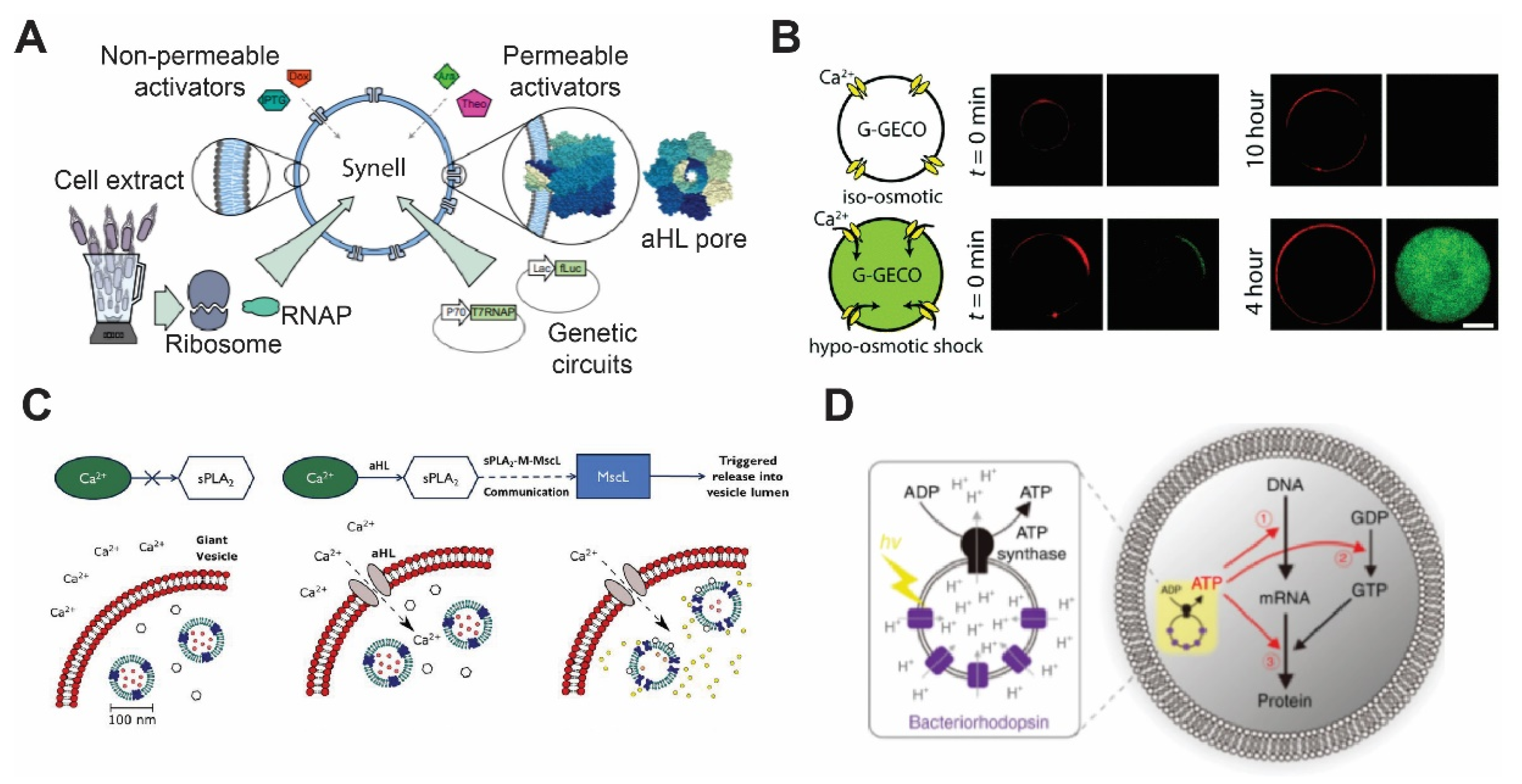
3.1. Alpha Hemolysin
3.2. Mechanosensitive Channel (MscL)
3.3. SUN Proteins
3.4. Bacteriorhodopsin
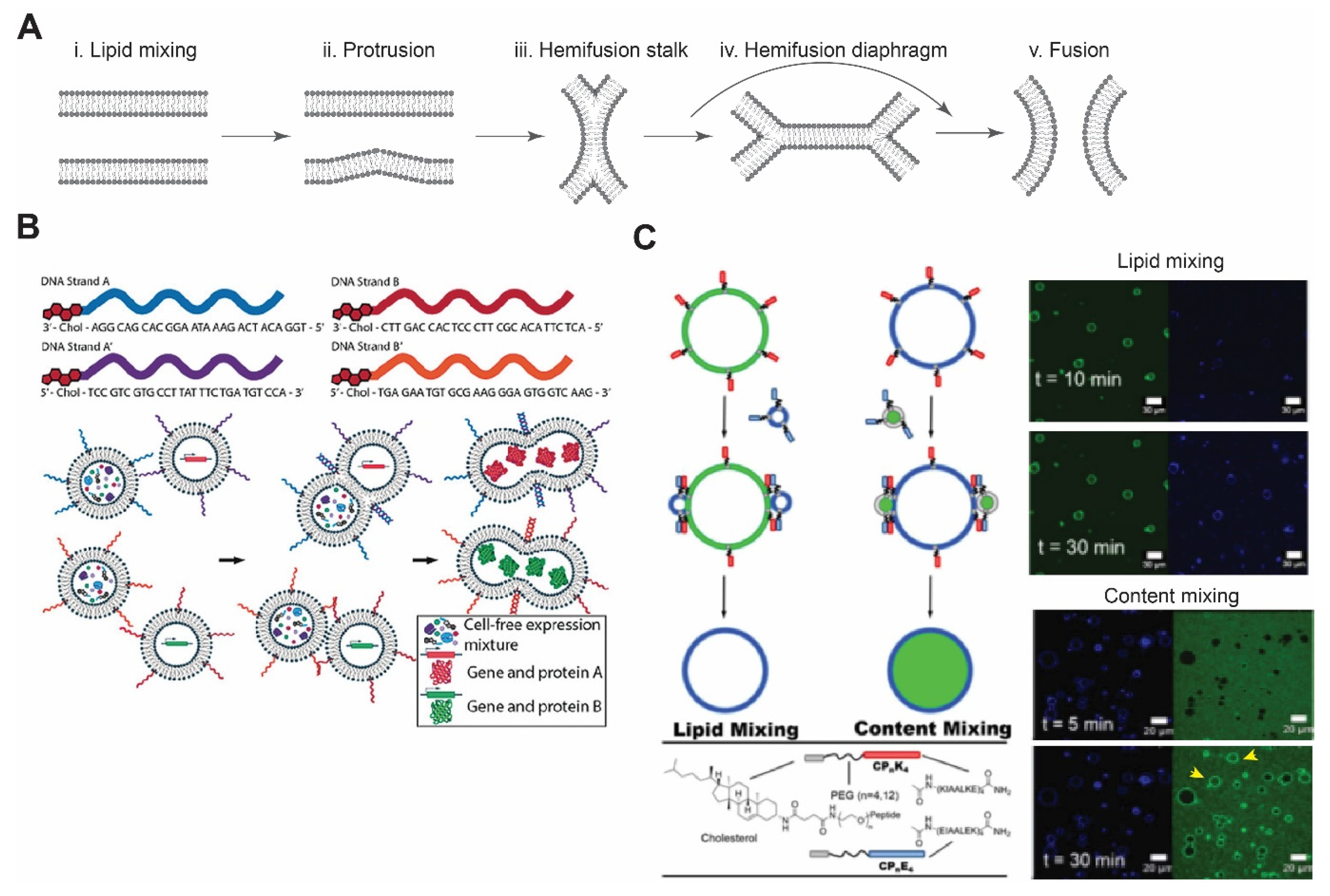
4. Membrane Fusion
4.1. DNA-Mediated Fusion
4.2. Peptide-Mediated Fusion
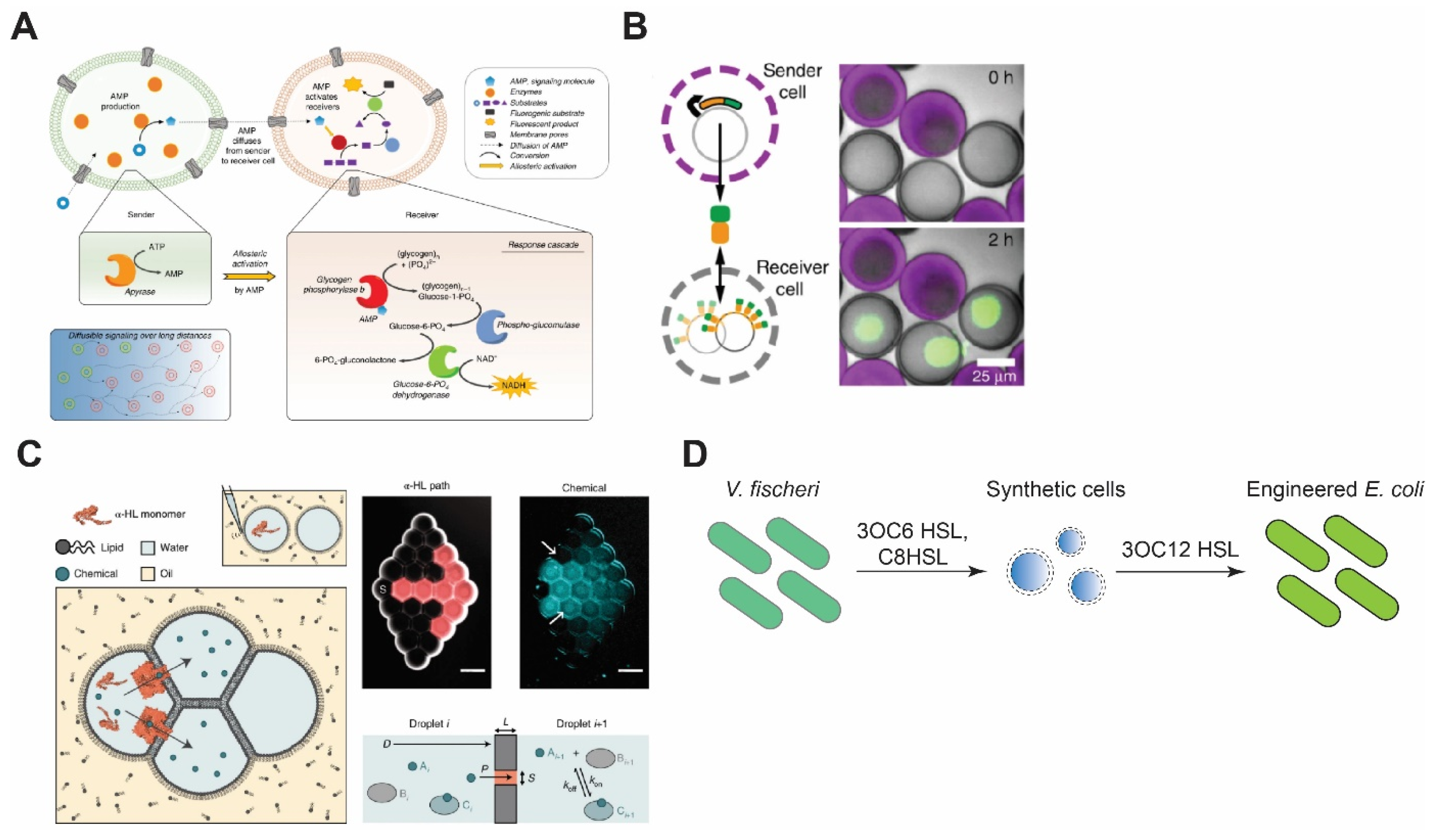
5. Intercellular Communication
5.1. Synthetic Cell–Synthetic Cell Communication
5.2. Synthetic Cell–Natural Cell Communication
6. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Nirenberg, M.W.; Matthaei, J.H. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc. Natl. Acad. Sci. USA 1961, 47, 1588–1602. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-M.; Swartz, J.R. Prolonging Cell-Free Protein Synthesis with a Novel ATP Regeneration System. Biotechnol. Bioeng. 1999, 66, 180–188. [Google Scholar] [CrossRef]
- Shimizu, Y.; Inoue, A.; Tomari, Y.; Suzuki, T.; Yokogawa, T.; Nishikawa, K.; Ueda, T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001, 19, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Damiano, L.; Stano, P. On the “Life-Likeness” of Synthetic Cells. Front. Bioeng. Biotechnol. 2020, 8, 953. [Google Scholar] [CrossRef]
- Ivanov, I.; Castellanos, S.L.; Balasbas, S.; Otrin, L.; Maruscaroniccaron, N.; Vidakovicacute-Koch, T.; Sundmacher, K. Bottom-Up Synthesis of Artificial Cells: Recent Highlights and Future Challenges. Annu. Rev. Chem. Biomol. Eng. 2021, 12, 287–308. [Google Scholar] [CrossRef]
- Noireaux, V.; Liu, A.P. The New Age of Cell-Free Biology. Annu. Rev. Biomed. Eng. 2020, 22, 51–77. [Google Scholar] [CrossRef] [Green Version]
- Groaz, A.; Moghimianavval, H.; Tavella, F.; Giessen, T.W.; Vecchiarelli, A.G.; Yang, Q.; Liu, A.P. Engineering spatiotemporal organization and dynamics in synthetic cells. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2021, 13. [Google Scholar] [CrossRef]
- McIlwain, B.C.; Ruprecht, M.T.; Stockbridge, R.B. Membrane Exporters of Fluoride Ion. Ann. Rev. Biochem. 2021, 90, 559–579. [Google Scholar] [CrossRef] [PubMed]
- Jackman, J.A.; Cho, N.-J. Supported Lipid Bilayer Formation: Beyond Vesicle Fusion. Langmuir 2020, 36, 1387–1400. [Google Scholar] [CrossRef] [PubMed]
- Robelek, R.; Lemker, E.S.; Wiltschi, B.; Kirste, V.; Naumann, R.; Oesterhelt, D.; Sinner, E.-K. Incorporation of In Vitro Synthesized GPCR into a Tethered Artificial Lipid Membrane System. Angew. Chemie Int. Ed. 2007, 46, 605–608. [Google Scholar] [CrossRef]
- Manzer, Z.A.; Ghosh, S.; Jacobs, M.L.; Krishnan, S.; Zipfel, W.R.; Piñeros, M.; Kamat, N.P.; Daniel, S. Cell-Free Synthesis of a Transmembrane Mechanosensitive Channel Protein into a Hybrid-Supported Lipid Bilayer. ACS Appl. Bio Mater. 2021, 4, 3101–3112. [Google Scholar] [CrossRef]
- Bashirzadeh, Y.; Liu, A.P. Encapsulation of the cytoskeleton: Towards mimicking the mechanics of a cell. Soft Matter 2019, 15, 8425–8436. [Google Scholar] [CrossRef] [PubMed]
- Mueller, P.; Rudin, D.O.; Ti Tien, H.; Wescott, W.C. Reconstitution of Cell Membrane Structure in vitro and its Transformation into an Excitable System. Nature 1962, 194, 979–980. [Google Scholar] [CrossRef]
- Mueller, P.; Rudin, D.O.; Ti Tien, H.; Wescott, W.C. Formation and Properties of Bimolecular Lipid Membranes; Elsevier: Amsterdam, The Netherlands, 1964; Volume 1, pp. 379–393. [Google Scholar]
- Heginbotham, L.; LeMasurier, M.; Kolmakova-Partensky, L.; Miller, C. Single Streptomyces lividans K+ Channels. J. Gen. Physiol. 1999, 114, 551–560. [Google Scholar] [CrossRef] [Green Version]
- Montal, M.; Mueller, P. Formation of Bimolecular Membranes from Lipid Monolayers and a Study of Their Electrical Properties. Proc. Natl. Acad. Sci. USA 1972, 69, 3561–3566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, W.L.; Chen, M.; Cronin, B.; Holden, M.A.; Bayley, H. Asymmetric Droplet Interface Bilayers. J. Am. Chem. Soc. 2008, 130, 5878–5879. [Google Scholar] [CrossRef]
- Heron, A.J.; Thompson, J.R.; Mason, A.E.; Wallace, M.I. Direct Detection of Membrane Channels from Gels Using Water-in-Oil Droplet Bilayers. J. Am. Chem. Soc. 2007, 129, 16042–16047. [Google Scholar] [CrossRef]
- Van de Cauter, L.; Fanalista, F.; van Buren, L.; De Franceschi, N.; Godino, E.; Bouw, S.; Danelon, C.; Dekker, C.; Koenderink, G.H.; Ganzinger, K.A. Optimized cDICE for Efficient Reconstitution of Biological Systems in Giant Unilamellar Vesicles. ACS Synth. Biol. 2021, 10, 1690–1702. [Google Scholar] [CrossRef]
- Tamm, L.K.; McConnell, H.M. Supported phospholipid bilayers. Biophys. J. 1985, 47, 105–113. [Google Scholar] [CrossRef] [Green Version]
- Sackmann, E. Supported Membranes: Scientific and Practical Applications. Science (80-.) 1996, 271, 43–48. [Google Scholar] [CrossRef]
- Johnson, S.J.; Bayerl, T.M.; McDermott, D.C.; Adam, G.W.; Rennie, A.R.; Thomas, R.K.; Sackmann, E. Structure of an adsorbed dimyristoylphosphatidylcholine bilayer measured with specular reflection of neutrons. Biophys. J. 1991, 59, 289–294. [Google Scholar] [CrossRef] [Green Version]
- Mou, J.; Yang, J.; Shao, Z. Accelerated Publications Tris(hydroxymethyl)aminomethane (C4H11NO3) Induced a Ripple Phase in Supported Unilamellar Phospholipid Bilayers1. Biochemistry 1975, 14, 35. [Google Scholar]
- Koenig, B.W.; Krueger, S.; Orts, W.J.; Majkrzak, C.F.; Berk, N.F.; Silverton, J.V.; Gawrisch, K. Neutron Reflectivity and Atomic Force Microscopy Studies of a Lipid Bilayer in Water Adsorbed to the Surface of a Silicon Single Crystal. Langmuir 1996, 12, 1343–1350. [Google Scholar] [CrossRef]
- Chen, Y.L.; Chen, S.; Frank, C.; Israelachvili, J. Molecular mechanisms and kinetics during the self-assembly of surfactant layers. J. Colloid Interface Sci. 1992, 153, 244–265. [Google Scholar] [CrossRef]
- Cremer, P.S.; Boxer, S.G. Formation and Spreading of Lipid Bilayers on Planar Glass Supports. J. Phys. Chem. B 1999, 103, 2554–2559. [Google Scholar] [CrossRef]
- Brian, A.A.; McConnell, H.M. Allogeneic stimulation of cytotoxic T cells by supported planar membranes. Proc. Natl. Acad. Sci. USA 1984, 81, 6159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, R.P.; Bérat, R.; Brisson, A.R. Formation of Solid-Supported Lipid Bilayers: An Integrated View. Langmuir 2006, 22, 3497–3505. [Google Scholar] [CrossRef]
- Kalb, E.; Frey, S.; Tamm, L.K. Formation of supported planar bilayers by fusion of vesicles to supported phospholipid monolayers. Biochim. Biophys. Acta-Biomembr. 1992, 1103, 307–316. [Google Scholar] [CrossRef]
- Crane, J.M.; Kiessling, V.; Tamm, L.K. Measuring lipid asymmetry in planar supported bilayers by fluorescence interference contrast microscopy. Langmuir 2005, 21, 1377–1388. [Google Scholar] [CrossRef]
- Zasadzinski, J.A.; Helm, C.A.; Longo, M.L.; Weisenhorn, A.L.; Gould, S.A.; Hansma, P.K. Atomic force microscopy of hydrated phosphatidylethanolamine bilayers. Biophys. J. 1991, 59, 755. [Google Scholar] [CrossRef] [Green Version]
- Egawa, H.; Furusawa, K. Liposome Adhesion on Mica Surface Studied by Atomic Force Microscopy. Langmuir 1999, 15, 1660–1666. [Google Scholar] [CrossRef]
- Castellana, E.T.; Cremer, P.S. Solid supported lipid bilayers: From biophysical studies to sensor design. Surf. Sci. Rep. 2006, 61, 429–444. [Google Scholar] [CrossRef] [PubMed]
- Tsofina, L.M.; Liberman, E.A.; Babakov, A.V. Production of Bimolecular Protein-Lipid Membranes in Aqueous Solution. Nature 1966, 212, 681–683. [Google Scholar] [CrossRef]
- Bayley, H.; Cronin, B.; Heron, A.; Holden, M.A.; Hwang, W.; Syeda, R.; Thompson, J.; Wallace, M. Droplet interface bilayers. Mol. Biosyst. 2008, 4, 1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funakoshi, K.; Suzuki, H.; Takeuchi, S. Lipid Bilayer Formation by Contacting Monolayers in a Microfluidic Device for Membrane Protein Analysis. Anal. Chem. 2006, 78, 8169–8174. [Google Scholar] [CrossRef]
- Leptihn, S.; Castell, O.K.; Cronin, B.; Lee, E.-H.; Gross, L.C.M.; Marshall, D.P.; Thompson, J.R.; Holden, M.; Wallace, M.I. Constructing droplet interface bilayers from the contact of aqueous droplets in oil. Nat. Protoc. 2013, 8, 1048–1057. [Google Scholar] [CrossRef] [PubMed]
- Holden, M.A.; Needham, D.; Bayley, H. Functional Bionetworks from Nanoliter Water Droplets. J. Am. Chem. Soc. 2007, 129, 8650–8655. [Google Scholar] [CrossRef]
- Kawano, R.; Tsuji, Y.; Kamiya, K.; Kodama, T.; Osaki, T.; Miki, N.; Takeuchi, S. A Portable Lipid Bilayer System for Environmental Sensing with a Transmembrane Protein. PLoS ONE 2014, 9, e102427. [Google Scholar] [CrossRef]
- Thompson, J.R.; Heron, A.J.; Santoso, Y.; Wallace, M.I. Enhanced Stability and Fluidity in Droplet on Hydrogel Bilayers for Measuring Membrane Protein Diffusion. Nano Lett. 2007, 7, 3875–3878. [Google Scholar] [CrossRef]
- Szostak, J.W.; Bartel, D.P.; Luisi, P.L. Synthesizing life. Nature 2001, 409, 387–390. [Google Scholar] [CrossRef]
- Discher, B.M.; Won, Y.Y.; Ege, D.S.; Lee, J.C.M.; Bates, F.S.; Discher, D.E.; Hammer, D.A. Polymersomes: Tough vesicles made from diblock copolymers. Science (80-.) 1999, 284, 1143–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reeves, J.P.; Dowben, R.M. Formation and properties of thin-walled phospholipid vesicles. J. Cell. Physiol. 1969, 73, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Tsumoto, K.; Matsuo, H.; Tomita, M.; Yoshimura, T. Efficient formation of giant liposomes through the gentle hydration of phosphatidylcholine films doped with sugar. Colloids Surfaces B Biointerfaces 2009, 68, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Hishida, M.; Seto, H.; Yamada, N.L.; Yoshikawa, K. Hydration process of multi-stacked phospholipid bilayers to form giant vesicles. Chem. Phys. Lett. 2008, 455, 297–302. [Google Scholar] [CrossRef] [Green Version]
- Litschel, T.; Schwille, P. Protein Reconstitution Inside Giant Unilamellar Vesicles. Annu. Rev. Biophys. 2021, 50, 525–548. [Google Scholar] [CrossRef]
- Angelova, M.I.; Dimitrov, D.S. Liposome electroformation. Faraday Discuss. Chem. Soc. 1986, 81, 303–311. [Google Scholar] [CrossRef]
- Shimanouchi, T.; Umakoshi, H.; Kuboi, R. Kinetic study on giant vesicle formation with electroformation method. Langmuir 2009, 25, 4835–4840. [Google Scholar] [CrossRef]
- Bucher, P.; Fischer, A.; Luisi, P.L.; Oberholzer, T.; Walde, P. Giant Vesicles as Biochemical Compartments: The Use of Microinjection Techniques. Langmuir 1998, 14, 2712–2721. [Google Scholar] [CrossRef]
- Majumder, S.; Wubshet, N.; Liu, A.P. Encapsulation of complex solutions using droplet microfluidics towards the synthesis of artificial cells. J. Micromech. Microeng. 2019, 29, 083001. [Google Scholar] [CrossRef]
- Lorenceau, E.; Utada, A.S.; Link, D.R.; Cristobal, G.; Joanicot, M.; Weitz, D.A. Generation of polymerosomes from double-emulsions. Langmuir 2005, 21, 9183–9186. [Google Scholar] [CrossRef]
- Teh, S.Y.; Khnouf, R.; Fan, H.; Lee, A.P. Stable, biocompatible lipid vesicle generation by solvent extraction-based droplet microfluidics. Biomicrofluidics 2011, 5, 044113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karamdad, K.; Law, R.V.; Seddon, J.M.; Brooks, N.J.; Ces, O. Preparation and mechanical characterisation of giant unilamellar vesicles by a microfluidic method. Lab Chip 2015, 15, 557–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshpande, S.; Caspi, Y.; Meijering, A.E.C.; Dekker, C. Octanol-assisted liposome assembly on chip. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Stachowiak, J.C.; Richmond, D.L.; Li, T.H.; Liu, A.P.; Parekh, S.H.; Fletcher, D.A. Unilamellar vesicle formation and encapsulation by microfluidic jetting. Proc. Natl. Acad. Sci. USA 2008, 105, 4697–4702. [Google Scholar] [CrossRef] [Green Version]
- Funakoshi, K.; Suzuki, H.; Takeuchi, S. Formation of giant lipid vesiclelike compartments from a planar lipid membrane by a pulsed jet flow. J. Am. Chem. Soc. 2007, 129, 12608–12609. [Google Scholar] [CrossRef]
- Walde, P.; Cosentino, K.; Engel, H.; Stano, P. Giant Vesicles: Preparations and Applications. ChemBioChem 2010, 11, 848–865. [Google Scholar] [CrossRef]
- Pautot, S.; Frisken, B.J.; Weitz, D.A. Production of unilamellar vesicles using an inverted emulsion. Langmuir 2003, 19, 2870–2879. [Google Scholar] [CrossRef]
- Moga, A.; Yandrapalli, N.; Dimova, R.; Robinson, T. Optimization of the Inverted Emulsion Method for High-Yield Production of Biomimetic Giant Unilamellar Vesicles. ChemBioChem 2019, 20, 2674–2682. [Google Scholar] [CrossRef] [Green Version]
- Abkarian, M.; Loiseau, E.; Massiera, G. Continuous droplet interface crossing encapsulation (cDICE) for high throughput monodisperse vesicle design. Soft Matter 2011, 7, 4610–4614. [Google Scholar] [CrossRef]
- Bashirzadeh, Y.; Wubshet, N.H.; Liu, A.P. Confinement Geometry Tunes Fascin-Actin Bundle Structures and Consequently the Shape of a Lipid Bilayer Vesicle. Front. Mol. Biosci. 2020, 7. [Google Scholar] [CrossRef]
- Bashirzadeh, Y.; Redford, S.A.; Lorpaiboon, C.; Groaz, A.; Moghimianavval, H.; Litschel, T.; Schwille, P.; Hocky, G.M.; Dinner, A.R.; Liu, A.P. Actin crosslinker competition and sorting drive emergent GUV size-dependent actin network architecture. Commun. Biol. 2021, 4. [Google Scholar] [CrossRef]
- Bashirzadeh, Y.; Moghimianavval, H.; Liu, A.P. Encapsulated actomyosin patterns drive cell-like membrane shape changes. bioRxiv 2021. [Google Scholar] [CrossRef]
- Campillo, C.; Sens, P.; Köster, D.; Pontani, L.L.; Lévy, D.; Bassereau, P.; Nassoy, P.; Sykes, C. Unexpected membrane dynamics unveiled by membrane nanotube extrusion. Biophys. J. 2013, 104, 1248–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majeed, S.; Ahmad, A.B.; Sehar, U.; Georgieva, E.R. Lipid Membrane Mimetics in Functional and Structural Studies of Integral Membrane Proteins. Membranes 2021, 11, 685. [Google Scholar] [CrossRef] [PubMed]
- Bhakdi, S.; Tranum-Jensen, J. Alpha-toxin of Staphylococcus aureus. Microbiol. Rev. 1991, 55, 733. [Google Scholar] [CrossRef]
- Gouaux, J.E.; Braha, O.; Hobaugh, M.R.; Song, L.; Cheley, S.; Shustak, C.; Bayley, H. Subunit stoichiometry of staphylococcal alpha-hemolysin in crystals and on membranes: A heptameric transmembrane pore. Proc. Natl. Acad. Sci. USA 1994, 91, 12828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menestrina, G. Ionic channels formed byStaphylococcus aureus alpha-toxin: Voltage-dependent inhibition by divalent and trivalent cations. J. Membr. Biol. 1986, 90, 177–190. [Google Scholar] [CrossRef]
- Song, L.; Hobaugh, M.R.; Shustak, C.; Cheley, S.; Bayley, H.; Gouaux, J.E. Structure of staphylococcal $α$-hemolysin, a heptameric transmembrane pore. Science (80-) 1996, 274, 1859–1865. [Google Scholar] [CrossRef]
- Winfree, E.; Liu, F.; Wenzler, L.A.; Seeman, N.C. Design and self-assembly of two-dimensional DNA crystals. Nature 1998, 394, 539–544. [Google Scholar] [CrossRef]
- Langecker, M.; Arnaut, V.; Martin, T.G.; List, J.; Renner, S.; Mayer, M.; Dietz, H.; Simmel, F.C. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 2012, 338, 932. [Google Scholar] [CrossRef] [Green Version]
- Go, K.; Li, C.-Y.; Ricci, M.; Prathyusha Bhamidimarri, S.; Yoo, J.; Gyenes, B.; Ohmann, A.; Winterhalter, M.; Aksimentiev, A.; Keyser, U.F. Large-Conductance Transmembrane Porin Made from DNA Origami. ACS Nano 2016, 10, 8207–8214. [Google Scholar] [CrossRef]
- Iwabuchi, S.; Kawamata, I.; Murata, S.; Nomura, S.M. A large, square-shaped, DNA origami nanopore with sealing function on a giant vesicle membrane. Chem. Commun. 2021, 57, 2990–2993. [Google Scholar] [CrossRef] [PubMed]
- Fragasso, A.; De Franceschi, N.; Stömmer, P.; Van Der Sluis, E.O.; Dietz, H.; Dekker, C. Reconstitution of Ultrawide DNA Origami Pores in Liposomes for Transmembrane Transport of Macromolecules. bioRxiv 2021. [Google Scholar] [CrossRef]
- Thomsen, R.P.; Malle, M.G.; Okholm, A.H.; Krishnan, S.; Bohr, S.S.-R.; Sørensen, R.S.; Ries, O.; Vogel, S.; Simmel, F.C.; Hatzakis, N.S.; et al. A large size-selective DNA nanopore with sensing applications. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Tomita, T.; Yasuda, T. Membrane-damaging action of staphylococcal alpha-toxin on phospholipid-cholesterol liposomes. Biochim. Biophys. Acta-Biomembr. 1987, 898, 257–265. [Google Scholar] [CrossRef]
- Tomita, T.; Watanabe, M.; Yasuda, T. Influence of membrane fluidity on the assembly of Staphylococcus aureus α-toxin, a channel-forming protein, in liposome membrane. J. Biol. Chem. 1992, 267, 13391–13397. [Google Scholar] [CrossRef]
- Wu, H.-C.; Bayley, H. Single-Molecule Detection of Nitrogen Mustards by Covalent Reaction within a Protein Nanopore. J. Am. Chem. Soc. 2008, 130, 6813–6819. [Google Scholar] [CrossRef]
- Chalmeau, J.; Monina, N.; Shin, J.; Vieu, C.; Noireaux, V. α-Hemolysin pore formation into a supported phospholipid bilayer using cell-free expression. Biochim. Biophys. Acta-Biomembr. 2011, 1808, 271–278. [Google Scholar] [CrossRef] [Green Version]
- Deshpande, S.; Brandenburg, F.; Lau, A.; Last, M.G.F.; Spoelstra, W.K.; Reese, L.; Wunnava, S.; Dogterom, M.; Dekker, C. Spatiotemporal control of coacervate formation within liposomes. Nat. Commun. 2019, 10, 1800. [Google Scholar] [CrossRef] [Green Version]
- Adamala, K.P.; Martin-Alarcon, D.A.; Guthrie-Honea, K.R.; Boyden, E.S. Engineering genetic circuit interactions within and between synthetic minimal cells. Nat. Chem. 2016, 9, 431–439. [Google Scholar] [CrossRef] [Green Version]
- Hilburger, C.E.; Jacobs, M.L.; Lewis, K.R.; Peruzzi, J.A.; Kamat, N.P. Controlling Secretion in Artificial Cells with a Membrane AND Gate. ACS Synth. Biol. 2019, 8, 1224–1230. [Google Scholar] [CrossRef]
- Majumder, S.; Garamella, J.; Wang, Y.L.; Denies, M.; Noireaux, V.; Liu, A.P. Cell-sized mechanosensitive and biosensing compartment programmed with DNA. Chem. Commun. 2017, 53, 7349–7352. [Google Scholar] [CrossRef]
- Hindley, J.W.; Zheleva, D.G.; Elani, Y.; Charalambous, K.; Barter, L.M.C.; Booth, P.J.; Bevan, C.L.; Law, R.V.; Ces, O. Building a synthetic mechanosensitive signaling pathway in compartmentalized artificial cells. Proc. Natl. Acad. Sci. USA 2019, 116, 16711–16716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berhanu, S.; Ueda, T.; Kuruma, Y. Artificial photosynthetic cell producing energy for protein synthesis. Nat. Commun. 2019, 10, 1325. [Google Scholar] [CrossRef] [PubMed]
- Martinac, B.; Buechner, M.; Delcour, A.H.; Adler, J.; Kung, C. Pressure-sensitive ion channel in Escherichia coli. Proc. Natl. Acad. Sci. USA 1987, 84, 2297–2301. [Google Scholar] [CrossRef] [Green Version]
- Brohawn, S.G. How ion channels sense mechanical force: Insights from mechanosensitive K2P channels TRAAK, TREK1, and TREK2. Ann. N. Y. Acad. Sci. 2015, 1352, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Blount, P.; Iscla, I. Life with Bacterial Mechanosensitive Channels, from Discovery to Physiology to Pharmacological Target. Microbiol. Mol. Biol. Rev. 2020, 84. [Google Scholar] [CrossRef]
- Majumder, S.; Liu, A.P. Bottom-up synthetic biology: Modular design for making artificial platelets. Phys. Biol. 2018, 15, 013001. [Google Scholar] [CrossRef]
- Rosholm, K.R.; Baker, M.A.B.; Ridone, P.; Nakayama, Y.; Rohde, P.R.; Cuello, L.G.; Lee, L.K.; Martinac, B. Activation of the mechanosensitive ion channel MscL by mechanical stimulation of supported Droplet-Hydrogel bilayers. Sci. Rep. 2017, 7, 45180. [Google Scholar] [CrossRef] [Green Version]
- Haylock, S.; Friddin, M.S.; Hindley, J.W.; Rodriguez, E.; Charalambous, K.; Booth, P.J.; Barter, L.M.C.; Ces, O. Membrane protein mediated bilayer communication in networks of droplet interface bilayers. Commun. Chem. 2020, 3, 1–8. [Google Scholar] [CrossRef]
- Strutt, R.; Hindley, J.W.; Gregg, J.; Booth, P.J.; Harling, J.D.; Law, R.V.; Friddin, M.S.; Ces, O. Activating mechanosensitive channels embedded in droplet interface bilayers using membrane asymmetry. Chem. Sci. 2021, 12, 2138–2145. [Google Scholar] [CrossRef] [PubMed]
- Garamella, J.; Majumder, S.; Liu, A.P.; Noireaux, V. An Adaptive Synthetic Cell Based on Mechanosensing, Biosensing, and Inducible Gene Circuits. ACS Synth. Biol. 2019, 8, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- Koçer, A.; Walko, M.; Meijberg, W.; Feringa, B.L. Chemistry: A light-actuated nanovalve derived from a channel protein. Science (80-) 2005, 309, 755–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Tang, S.; Meng, L.; Li, X.; Wen, X.; Chen, S.; Niu, L.; Li, X.; Qiu, W.; Hu, H.; et al. Ultrasonic Control of Neural Activity through Activation of the Mechanosensitive Channel MscL. Nano Lett. 2018, 18, 4148–4155. [Google Scholar] [CrossRef]
- Koçer, A.; Walko, M.; Bulten, E.; Halza, E.; Feringa, B.L.; Meijberg, W. Rationally designed chemical modulators convert a bacterial channel protein into a pH-sensory valve. Angew. Chemie-Int. Ed. 2006, 45, 3126–3130. [Google Scholar] [CrossRef]
- Starr, D.A.; Fridolfsson, H.N. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 2010, 26, 421–444. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.L.; Burke, B. LINC complexes and nuclear positioning. Semin. Cell Dev. Biol. 2018, 82, 67–76. [Google Scholar] [CrossRef]
- Burke, B. LINC complexes as regulators of meiosis. Curr. Opin. Cell Biol. 2018, 52, 22–29. [Google Scholar] [CrossRef]
- Majumder, S.; Willey, P.T.; DeNies, M.S.; Liu, A.P.; Luxton, G. A synthetic biology platform for the reconstitution and mechanistic dissection of LINC complex assembly. J. Cell Sci. 2019, 132, jcs219451. [Google Scholar] [CrossRef] [Green Version]
- Majumder, S.; Hsu, Y.-Y.; Liu, A.P. Direct reconstitution and study of SUN protein interactions in vitro using mammalian cell-free expression. bioRxiv 2021. [Google Scholar] [CrossRef]
- Oesterhelt, D.; Stoeckenius, W. Functions of a New Photoreceptor Membrane. Proc. Natl. Acad. Sci. USA 1973, 70, 2853–2857. [Google Scholar] [CrossRef] [Green Version]
- Stern, L.J.; Khorana, H.G. Structure-Function Studies on Bacteriorhodopsin. J. Biol. Chem. 1989, 264, 14202–14208. [Google Scholar] [CrossRef]
- Lanyi, J.K. Bacteriorhodopsin. Annu. Rev. Physiol. 2004, 66, 665–688. [Google Scholar] [CrossRef]
- Racker, E.; Stoeckenius, W. Reconstitution of Purple Membrane Vesicles Catalyzing Light-driven Proton Uptake and Adenosine Triphosphate Formation. J. Biol. Chem. 1974, 249, 662–663. [Google Scholar] [CrossRef]
- Dezi, M.; Di Cicco, A.; Bassereau, P.; Lévy, D. Detergent-mediated incorporation of transmembrane proteins in giant unilamellar vesicles with controlled physiological contents. Proc. Natl. Acad. Sci. USA 2013, 110, 7276–7281. [Google Scholar] [CrossRef] [Green Version]
- Kahya, N.; Pécheur, E.I.; De Boeij, W.P.; Wiersma, D.A.; Hoekstra, D. Reconstitution of Membrane Proteins into Giant Unilamellar Vesicles via Peptide-Induced Fusion. Biophys. J. 2001, 81, 1464–1474. [Google Scholar] [CrossRef] [Green Version]
- Kalmbach, R.; Chizhov, I.; Schumacher, M.C.; Friedrich, T.; Bamberg, E.; Engelhard, M. Functional Cell-free Synthesis of a Seven Helix Membrane Protein: In situ Insertion of Bacteriorhodopsin into Liposomes. J. Mol. Biol. 2007, 371, 639–648. [Google Scholar] [CrossRef]
- Shimono, K.; Goto, M.; Kikukawa, T.; Miyauchi, S.; Shirouzu, M.; Kamo, N.; Yokoyama, S. Production of functional bacteriorhodopsin by an Escherichia coli cell-free protein synthesis system supplemented with steroid detergent and lipid. Protein Sci. 2009, 18, 2160–2171. [Google Scholar] [CrossRef] [Green Version]
- Pitard, B.; Richard, P.; Duñarach, M.; Girault, G.; Rigaiud, J.-L. ATP Synthesis by the F0F1 ATP Synthase from Thermophilic Bacillus PS3 Reconstituted into Liposomes with Bacteriorhodopsin. Eur. J. Biochem. 1996, 235, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.-J.; Montemagno, C.D. Artificial Organelle: ATP Synthesis from Cellular Mimetic Polymersomes. Nano Lett. 2005, 5, 2538–2542. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; De Queiros Silveira, G.; Ma, X.; Xie, Y.; Wu, Y.A.; Barry, E.; Rajh, T.; Fry, H.C.; Laible, P.D.; Rozhkova, E.A. Light-Gated Synthetic Protocells for Plasmon-Enhanced Chemiosmotic Gradient Generation and ATP Synthesis. Angew. Chemie 2019, 131, 4950–4954. [Google Scholar] [CrossRef]
- Ahmad, R.; Kleineberg, C.; Nasirimarekani, V.; Su, Y.-J.; Pozveh, S.G.; Bae, A.; Sundmacher, K.; Bodenschatz, E.; Guido, I.; Vidaković-koch, T.; et al. Light-Powered Reactivation of Flagella and Contraction of Microtubule Networks: Toward Building an Artificial Cell. ACS Synth. Biol. 2021, 10, 1490–1504. [Google Scholar] [CrossRef]
- Lee, K.Y.; Park, S.-J.; Lee, K.A.; Kim, S.-H.; Kim, H.; Meroz, Y.; Mahadevan, L.; Jung, K.-H.; Ahn, T.K.; Parker, K.K.; et al. Photosynthetic artificial organelles sustain and control ATP-dependent reactions in a protocellular system. Nat. Biotechnol. 2018, 36, 530–535. [Google Scholar] [CrossRef] [PubMed]
- Chernomordik, L.V.; Kozlov, M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008, 15, 675–683. [Google Scholar] [CrossRef] [Green Version]
- Chernomordik, L.V.; Kozlov, M.M. Protein-Lipid Interplay in Fusion and Fission of Biological Membranes. Annu. Rev. Biochem. 2003, 72, 175–207. [Google Scholar] [CrossRef] [PubMed]
- Marsden, H.R.; Tomatsu, I.; Kros, A. Model systems for membrane fusion. Chem. Soc. Rev. 2011, 40, 1572–1585. [Google Scholar] [CrossRef] [PubMed]
- Stengel, G.; Zahn, R.; Höök, F. DNA-Induced Programmable Fusion of Phospholipid Vesicles. J. Am. Chem. Soc. 2007, 129, 9584–9585. [Google Scholar] [CrossRef]
- Chan, Y.-H.M.; van Lengerich, B.; Boxer, S.G. Lipid-anchored DNA mediates vesicle fusion as observed by lipid and content mixing. Biointerphases 2008, 3, FA17–FA21. [Google Scholar] [CrossRef] [Green Version]
- Stengel, G.; Simonsson, L.; Campbell, R.A.; Höök, F. Determinants for Membrane Fusion Induced by Cholesterol-Modified DNA Zippers. J. Phys. Chem. B 2008, 112, 8264–8274. [Google Scholar] [CrossRef]
- Peruzzi, J.A.; Jacobs, M.L.; Vu, T.Q.; Wang, K.S.; Kamat, N.P. Barcoding Biological Reactions with DNA-Functionalized Vesicles. Angew. Chemie 2019, 131, 18856–18863. [Google Scholar] [CrossRef]
- Mora, N.L.; Boyle, A.L.; van Kolck, B.J.; Rossen, A.; Pokorná, Š.; Koukalová, A.; Šachl, R.; Hof, M.; Kros, A. Controlled Peptide-Mediated Vesicle Fusion Assessed by Simultaneous Dual-Colour Time-Lapsed Fluorescence Microscopy. Sci. Reports 2020 101 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.; Guha, S.; Diederichsen, U. SNARE protein analog-mediated membrane fusion. J. Pept. Sci. 2015, 21, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Mazur, F.; Chandrawati, R. Membrane Fusion Models for Bioapplications. ChemNanoMat 2021, 7, 223–237. [Google Scholar] [CrossRef]
- Bennett, M.K.; Calakos, N.; Scheller, R.H. Syntaxin: A synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science (80-) 1992, 257, 255–259. [Google Scholar] [CrossRef]
- Söllner, T.; Whiteheart, S.W.; Brunner, M.; Erdjument-Bromage, H.; Geromanos, S.; Tempst, P.; Rothman, J.E. SNAP receptors implicated in vesicle targeting and fusion. Nat. 1993 3626418 1993, 362, 318–324. [Google Scholar] [CrossRef]
- Jahn, R.; Scheller, R.H. SNAREs—Engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006, 7, 631–643. [Google Scholar] [CrossRef]
- Robson Marsden, H.; Elbers, N.A.; Bomans, P.H.H.; Sommerdijk, N.A.J.M.; Kros, A. A Reduced SNARE Model for Membrane Fusion. Angew. Chemie Int. Ed. 2009, 48, 2330–2333. [Google Scholar] [CrossRef]
- Litowski, J.R.; Hodges, R.S. Designing Heterodimeric Two-stranded α-Helical Coiled-coils. J. Biol. Chem. 2002, 277, 37272–37279. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.B.; Bassler, B.L. Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.; Bassler, B.L. Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 2019, 17, 371–382. [Google Scholar] [CrossRef]
- Thüroff, F.; Goychuk, A.; Reiter, M.; Frey, E. Bridging the gap between single-cell migration and collective dynamics. Elife 2019, 8. [Google Scholar] [CrossRef]
- Ladoux, B.; Mège, R.-M. Mechanobiology of collective cell behaviours. Nat. Rev. Mol. Cell Biol. 2017, 18, 743–757. [Google Scholar] [CrossRef] [PubMed]
- George, M.; Bullo, F.; Campàs, O. Connecting individual to collective cell migration. Sci. Rep. 2017, 7, 9720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buddingh, B.C.; Elzinga, J.; van Hest, J.C.M. Intercellular communication between artificial cells by allosteric amplification of a molecular signal. Nat. Commun. 2020, 11, 1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niederholtmeyer, H.; Chaggan, C.; Devaraj, N.K. Communication and quorum sensing in non-living mimics of eukaryotic cells. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Dupin, A.; Simmel, F.C. Signalling and differentiation in emulsion-based multi-compartmentalized in vitro gene circuits. Nat. Chem. 2019, 11, 32–39. [Google Scholar] [CrossRef]
- Lentini, R.; Martín, N.Y.; Forlin, M.; Belmonte, L.; Fontana, J.; Cornella, M.; Martini, L.; Tamburini, S.; Bentley, W.E.; Jousson, O.; et al. Two-Way Chemical Communication between Artificial and Natural Cells. ACS Cent. Sci. 2017, 3, 117–123. [Google Scholar] [CrossRef]
- Tang, T.-Y.D.; Cecchi, D.; Fracasso, G.; Accardi, D.; Coutable-Pennarun, A.; Mansy, S.S.; Perriman, A.W.; Anderson, J.L.R.; Mann, S. Gene-Mediated Chemical Communication in Synthetic Protocell Communities. ACS Synth. Biol. 2017, 7, 339–346. [Google Scholar] [CrossRef]
- Yang, Q.; Guo, Z.; Liu, H.; Peng, R.; Xu, L.; Bi, C.; He, Y.; Liu, Q.; Tan, W. A Cascade Signaling Network between Artificial Cells Switching Activity of Synthetic Transmembrane Channels. J. Am. Chem. Soc. 2020, 143, 232–240. [Google Scholar] [CrossRef]
- Chakraborty, T.; Wegner, S.V. Cell to Cell Signaling through Light in Artificial Cell Communities: Glowing Predator Lures Prey. ACS Nano 2021, 15, 9434–9444. [Google Scholar] [CrossRef]
- Chakraborty, T.; Bartelt, S.M.; Steinkühler, J.; Dimova, R.; Wegner, S.V. Light controlled cell-to-cell adhesion and chemical communication in minimal synthetic cells. Chem. Commun. 2019, 55, 9448–9451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Booth, M.J.; Schild, V.R.; Graham, A.D.; Olof, S.N.; Bayley, H. Light-activated communication in synthetic tissues. Sci. Adv. 2016, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, Y.; Li, M.; Booth, R.; Mann, S. Predatory behaviour in synthetic protocell communities. Nat. Chem. 2016 92 2016, 9, 110–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, B.; Ma, Y.; Ferguson, A.L.; Liu, A.P. In search of a novel chassis material for synthetic cells: Emergence of synthetic peptide compartment. Soft Matter 2020, 16, 10769–10780. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Ma, Y.; Hiraki, H.L.; Baker, B.; Ferguson, A.L.; Liu, A. Facile Formation of Giant Elastin-like Polypeptide Vesicles as Synthetic Cells. Chem. Commun. 2021. [Google Scholar] [CrossRef]
- Lentini, R.; Yeh Martín, N.; Mansy, S.S. Communicating artificial cells. Curr. Opin. Chem. Biol. 2016, 34, 53–61. [Google Scholar] [CrossRef]
- Jahnke, K.; Ritzmann, N.; Fichtler, J.; Nitschke, A.; Dreher, Y.; Abele, T.; Hofhaus, G.; Platzman, I.; Schröder, R.R.; Müller, D.J.; et al. Proton gradients from light-harvesting E. coli control DNA assemblies for synthetic cells. Nat. Commun. 2021, 12, 3967. [Google Scholar] [CrossRef]
- Schwarz-Schilling, M.; Aufinger, L.; Mückl, A.; Simmel, F.C. Chemical communication between bacteria and cell-free gene expression systems within linear chains of emulsion droplets. Integr. Biol. 2016, 8, 564–570. [Google Scholar] [CrossRef] [Green Version]
- Qian, X.; Westensee, I.N.; Fernandes, C.C.; Städler, B. Enzyme Mimic Facilitated Artificial Cell to Mammalian Cell Signal Transfer. Angew. Chemie Int. Ed. 2021, 60, 18704–18711. [Google Scholar] [CrossRef]
- Toparlak, Ö.D.; Zasso, J.; Bridi, S.; Serra, M.D.; Macchi, P.; Conti, L.; Baudet, M.-L.; Mansy, S.S. Artificial cells drive neural differentiation. Sci. Adv. 2020, 6, eabb4920. [Google Scholar] [CrossRef]
- Yandrapalli, N.; Petit, J.; Bäumchen, O.; Robinson, T. Surfactant-free production of biomimetic giant unilamellar vesicles using PDMS-based microfluidics. Commun. Chem. 2021, 4, 100. [Google Scholar] [CrossRef]
- Elani, Y. Interfacing Living and Synthetic Cells as an Emerging Frontier in Synthetic Biology. Angew. Chemie Int. Ed. 2021, 60, 5602–5611. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, B.; Moghimianavval, H.; Hwang, S.-W.; Liu, A.P. Synthetic Cell as a Platform for Understanding Membrane-Membrane Interactions. Membranes 2021, 11, 912. https://doi.org/10.3390/membranes11120912
Sharma B, Moghimianavval H, Hwang S-W, Liu AP. Synthetic Cell as a Platform for Understanding Membrane-Membrane Interactions. Membranes. 2021; 11(12):912. https://doi.org/10.3390/membranes11120912
Chicago/Turabian StyleSharma, Bineet, Hossein Moghimianavval, Sung-Won Hwang, and Allen P. Liu. 2021. "Synthetic Cell as a Platform for Understanding Membrane-Membrane Interactions" Membranes 11, no. 12: 912. https://doi.org/10.3390/membranes11120912
APA StyleSharma, B., Moghimianavval, H., Hwang, S.-W., & Liu, A. P. (2021). Synthetic Cell as a Platform for Understanding Membrane-Membrane Interactions. Membranes, 11(12), 912. https://doi.org/10.3390/membranes11120912






