An Update on mRNA-Based Viral Vaccines
Abstract
:1. Introduction
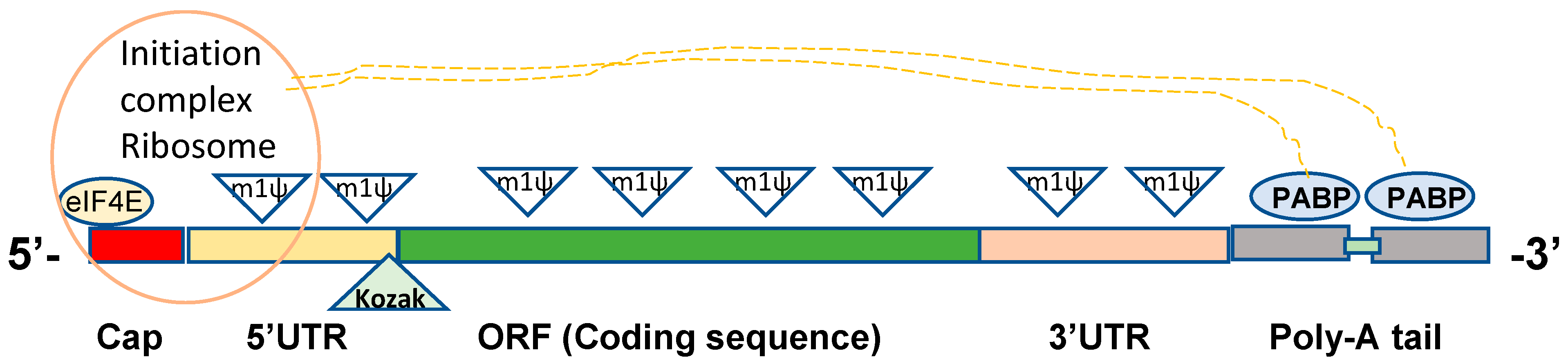
2. Structural Elements in Constructing DNA Template for In Vitro mRNA Synthesis
2.1. 5’ Cap
2.2. 5′ UTRs
2.3. 3′ UTRs
2.4. ORFs
2.5. Poly-A Tail
3. In Vitro Transcription and Modification of Synthetic mRNA
4. Lipid Nanoparticle Delivery of Synthetic mRNAs
5. Immunogenicity of mRNA Vaccines
6. SARS-CoV-2 Synthetic mRNA Vaccines
| Company /Sponsor | mRNA Type | Immunogens | mRNA Dose (μg) | Clinic Data | Preclinical Data |
|---|---|---|---|---|---|
| Moderna | Base-modified mRNA | mRNA-1273: prefusion stabilized Spike-TM | 100 | FDA approved [9] | [30,31] |
| Pfizer/BioNTech | Base-modified mRNA | BNT162b2: prefusion stabilized Spike-TM | 30 | FDA approved [10] | [32] |
| Pfizer/BioNTech | Base-modified mRNA | BNT162b1: receptor binding domain | 30 | phase 2 [114] | |
| CureVac | Unmodified mRNA | CVnCoV: prefusion stabilized Spike-TM | 12 | phase 2b/3 [116] | [105] |
| Arcturus | Self-amplifying mRNA | ARCT-021: full length Spike | 1–10 | phase ½ | [117] |
| Imperial College | Self-amplifying mRNA | full length Spike | 1–10 | [118] | |
| TranslateBio /Sanofi | Unmodified mRNA | prefusion stabilized Spike-TM (MRT55500) | 7.5 | [109] |
7. Influenza mRNA Vaccines
8. Other Viral mRNA Vaccines
9. Safety of COVID-19 Vaccines
10. Conclusions and Perspectives on mRNA-Based Viral Vaccines
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koirala, A.; Joo, Y.J.; Khatami, A.; Chiu, C.; Britton, P.N. Vaccines for COVID-19: The current state of play. Paediatr. Respir. Rev. 2020, 35, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.U.; Kim, Y.; Kumar, S.; Seo, D.; Ashraf, M.; Bae, Y.S. COVID-19 Vaccines (Revisited) and Oral-Mucosal Vector System as a Potential Vaccine Platform. Vaccines 2021, 9, 171. [Google Scholar] [CrossRef]
- Gebre, M.S.; Brito, L.A.; Tostanoski, L.H.; Edwards, D.K.; Carfi, A.; Barouch, D.H. Novel approaches for vaccine development. Cell 2021, 184, 1589–1603. [Google Scholar] [CrossRef] [PubMed]
- NIAID. Available online: https://www.niaid.nih.gov/news-events/nih-clinical-trial-investigational-vaccine-covid-19-begins (accessed on 7 June 2021).
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2-Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Abbasi, J. COVID-19 and mRNA Vaccines-First Large Test for a New Approach. JAMA 2020, 324, 1125–1127. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Haas, E.J.; Angulo, F.J.; McLaughlin, J.M.; Anis, E.; Singer, S.R.; Khan, F.; Brooks, N.; Smaja, M.; Mircus, G.; Pan, K.; et al. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: An observational study using national surveillance data. Lancet 2021, 397, 1819–1829. [Google Scholar] [CrossRef]
- Topol, E.J. Messenger RNA vaccines against SARS-CoV-2. Cell 2021, 184, 1401. [Google Scholar] [CrossRef]
- Wolff, J.A.; Malone, R.W.; Williams, P.; Chong, W.; Acsadi, G.; Jani, A.; Felgner, P.L. Direct gene transfer into mouse muscle in vivo. Science 1990, 247, 1465–1468. [Google Scholar] [CrossRef] [PubMed]
- Martinon, F.; Krishnan, S.; Lenzen, G.; Magne, R.; Gomard, E.; Guillet, J.G.; Levy, J.P.; Meulien, P. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur. J. Immunol. 1993, 23, 1719–1722. [Google Scholar] [CrossRef] [PubMed]
- Kariko, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, B.R.; Muramatsu, H.; Nallagatla, S.R.; Bevilacqua, P.C.; Sansing, L.H.; Weissman, D.; Kariko, K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010, 38, 5884–5892. [Google Scholar] [CrossRef] [Green Version]
- Kariko, K.; Muramatsu, H.; Keller, J.M.; Weissman, D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther. 2012, 20, 948–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kariko, K.; Muramatsu, H.; Ludwig, J.; Weissman, D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011, 39, e142. [Google Scholar] [CrossRef] [Green Version]
- Kariko, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008, 16, 1833–1840. [Google Scholar] [CrossRef] [PubMed]
- Semple, S.C.; Akinc, A.; Chen, J.; Sandhu, A.P.; Mui, B.L.; Cho, C.K.; Sah, D.W.; Stebbing, D.; Crosley, E.J.; Yaworski, E.; et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010, 28, 172–176. [Google Scholar] [CrossRef]
- Hassett, K.J.; Benenato, K.E.; Jacquinet, E.; Lee, A.; Woods, A.; Yuzhakov, O.; Himansu, S.; Deterling, J.; Geilich, B.M.; Ketova, T.; et al. Optimization of Lipid Nanoparticles for Intramuscular Administration of mRNA Vaccines. Mol. Ther. Nucleic Acids 2019, 15, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Bahl, K.; Senn, J.J.; Yuzhakov, O.; Bulychev, A.; Brito, L.A.; Hassett, K.J.; Laska, M.E.; Smith, M.; Almarsson, O.; Thompson, J.; et al. Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses. Mol. Ther. 2017, 25, 1316–1327. [Google Scholar] [CrossRef] [Green Version]
- Feldman, R.A.; Fuhr, R.; Smolenov, I.; Mick Ribeiro, A.; Panther, L.; Watson, M.; Senn, J.J.; Smith, M.; Almarsson, Ö.; Pujar, H.S.; et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine 2019, 37, 3326–3334. [Google Scholar] [CrossRef]
- Nance, K.D.; Meier, J.L. Modifications in an Emergency: The Role of N1-Methylpseudouridine in COVID-19 Vaccines. ACS Cent. Sci. 2021, 7, 748–756. [Google Scholar] [CrossRef]
- Kwon, H.; Kim, M.; Seo, Y.; Moon, Y.S.; Lee, H.J.; Lee, K.; Lee, H. Emergence of synthetic mRNA: In vitro synthesis of mRNA and its applications in regenerative medicine. Biomaterials 2018, 156, 172–193. [Google Scholar] [CrossRef]
- Svitkin, Y.V.; Cheng, Y.M.; Chakraborty, T.; Presnyak, V.; John, M.; Sonenberg, N. N1-methyl-pseudouridine in mRNA enhances translation through eIF2alpha-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 2017, 45, 6023–6036. [Google Scholar] [CrossRef] [Green Version]
- Sonenberg, N.; Gingras, A.C. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 1998, 10, 268–275. [Google Scholar] [CrossRef]
- Asrani, K.H.; Farelli, J.D.; Stahley, M.R.; Miller, R.L.; Cheng, C.J.; Subramanian, R.R.; Brown, J.M. Optimization of mRNA untranslated regions for improved expression of therapeutic mRNA. RNA Biol. 2018, 15, 756–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mignone, F.; Gissi, C.; Liuni, S.; Pesole, G. Untranslated regions of mRNAs. Genome Biol. 2002, 3. [Google Scholar] [CrossRef]
- Roncati, L.; Corsi, L. Nucleoside-modified messenger RNA COVID-19 vaccine platform. J. Med. Virol. 2021, 93, 4054–4057. [Google Scholar] [CrossRef]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schafer, A.; Ziwawo, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 586, 567–571. [Google Scholar] [CrossRef]
- Corbett, K.S.; Flynn, B.; Foulds, K.E.; Francica, J.R.; Boyoglu-Barnum, S.; Werner, A.P.; Flach, B.; O’Connell, S.; Bock, K.W.; Minai, M.; et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020, 383, 1544–1555. [Google Scholar] [CrossRef] [PubMed]
- Laczko, D.; Hogan, M.J.; Toulmin, S.A.; Hicks, P.; Lederer, K.; Gaudette, B.T.; Castano, D.; Amanat, F.; Muramatsu, H.; Oguin, T.H., III; et al. A Single Immunization with Nucleoside-Modified mRNA Vaccines Elicits Strong Cellular and Humoral Immune Responses against SARS-CoV-2 in Mice. Immunity 2020, 53, 724–732.e727. [Google Scholar] [CrossRef] [PubMed]
- Goldstrohm, A.C.; Wickens, M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008, 9, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Kahvejian, A.; Svitkin, Y.V.; Sukarieh, R.; M’Boutchou, M.N.; Sonenberg, N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005, 19, 104–113. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Day, N.; Trifillis, P.; Kiledjian, M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell Biol. 1999, 19, 4552–4560. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Alvin Chew, B.L.; Lai, Y.; Dong, H.; Xu, L.; Balamkundu, S.; Cai, W.M.; Cui, L.; Liu, C.F.; Fu, X.Y.; et al. Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic Acids Res. 2019, 47, e130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, J.M.; Ujita, A.; Hill, E.; Yousif-Rosales, S.; Smith, C.; Ko, N.; McReynolds, T.; Cabral, C.R.; Escamilla-Powers, J.R.; Houston, M.E. Cap 1 Messenger RNA Synthesis with Co-transcriptional CleanCap((R)) Analog by In Vitro Transcription. Curr. Protoc. 2021, 1, e39. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, A.; Robb, G.B.; Chan, S.H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016, 44, 7511–7526. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Dahlberg, J.E.; Lund, E. Reverse 5′ caps in RNAs made in vitro by phage RNA polymerases. RNA 1995, 1, 957–967. [Google Scholar]
- Jemielity, J.; Fowler, T.; Zuberek, J.; Stepinski, J.; Lewdorowicz, M.; Niedzwiecka, A.; Stolarski, R.; Darzynkiewicz, E.; Rhoads, R.E. Novel “anti-reverse” cap analogs with superior translational properties. RNA 2003, 9, 1108–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardi, N.; Hogan, M.J.; Weissman, D. Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 2020, 65, 14–20. [Google Scholar] [CrossRef]
- Sahin, U.; Muik, A.; Derhovanessian, E.; Vogler, I.; Kranz, L.M.; Vormehr, M.; Baum, A.; Pascal, K.; Quandt, J.; Maurus, D.; et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 2020, 586, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G.; Ivanov, I.P.; Sonenberg, N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 2016, 352, 1413–1416. [Google Scholar] [CrossRef]
- Babendure, J.R.; Babendure, J.L.; Ding, J.H.; Tsien, R.Y. Control of mammalian translation by mRNA structure near caps. RNA 2006, 12, 851–861. [Google Scholar] [CrossRef] [Green Version]
- Kozak, M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 2005, 361, 13–37. [Google Scholar] [CrossRef]
- Komar, A.A.; Hatzoglou, M. Cellular IRES-mediated translation: The war of ITAFs in pathophysiological states. Cell Cycle 2011, 10, 229–240. [Google Scholar] [CrossRef] [Green Version]
- Gallie, D.R. Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J. Virol. 2001, 75, 12141–12152. [Google Scholar] [CrossRef] [Green Version]
- Tan, X.; Wan, Y. Enhanced protein expression by internal ribosomal entry site-driven mRNA translation as a novel approach for in vitro loading of dendritic cells with antigens. Hum. Immunol. 2008, 69, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 1987, 196, 947–950. [Google Scholar] [CrossRef]
- Matoulkova, E.; Michalova, E.; Vojtesek, B.; Hrstka, R. The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012, 9, 563–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orlandini von Niessen, A.G.; Poleganov, M.A.; Rechner, C.; Plaschke, A.; Kranz, L.M.; Fesser, S.; Diken, M.; Lower, M.; Vallazza, B.; Beissert, T.; et al. Improving mRNA-Based Therapeutic Gene Delivery by Expression-Augmenting 3′ UTRs Identified by Cellular Library Screening. Mol. Ther. 2019, 27, 824–836. [Google Scholar] [CrossRef] [Green Version]
- Schwanhausser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Xu, X.S.; Russell, J.E. A nucleolin-binding 3′ untranslated region element stabilizes beta-globin mRNA in vivo. Mol. Cell Biol. 2006, 26, 2419–2429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Zoghaibi, F.; Ashour, T.; Al-Ahmadi, W.; Abulleef, H.; Demirkaya, O.; Khabar, K.S. Bioinformatics and experimental derivation of an efficient hybrid 3′ untranslated region and use in expression active linear DNA with minimum poly(A) region. Gene 2007, 391, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, H.; Najafabadi, H.S.; Oikonomou, P.; Greco, T.M.; Fish, L.; Salavati, R.; Cristea, I.M.; Tavazoie, S. Systematic discovery of structural elements governing stability of mammalian messenger RNAs. Nature 2012, 485, 264–268. [Google Scholar] [CrossRef] [Green Version]
- Mauger, D.M.; Cabral, B.J.; Presnyak, V.; Su, S.V.; Reid, D.W.; Goodman, B.; Link, K.; Khatwani, N.; Reynders, J.; Moore, M.J.; et al. mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. USA 2019, 116, 24075–24083. [Google Scholar] [CrossRef] [Green Version]
- Mustoe, A.M.; Corley, M.; Laederach, A.; Weeks, K.M. Messenger RNA Structure Regulates Translation Initiation: A Mechanism Exploited from Bacteria to Humans. Biochemistry 2018, 57, 3537–3539. [Google Scholar] [CrossRef] [Green Version]
- Sample, P.J.; Wang, B.; Reid, D.W.; Presnyak, V.; McFadyen, I.J.; Morris, D.R.; Seelig, G. Human 5′ UTR design and variant effect prediction from a massively parallel translation assay. Nat. Biotechnol. 2019, 37, 803–809. [Google Scholar] [CrossRef]
- Al-Saif, M.; Khabar, K.S. UU/UA dinucleotide frequency reduction in coding regions results in increased mRNA stability and protein expression. Mol. Ther. 2012, 20, 954–959. [Google Scholar] [CrossRef] [Green Version]
- Vaidyanathan, S.; Azizian, K.T.; Haque, A.; Henderson, J.M.; Hendel, A.; Shore, S.; Antony, J.S.; Hogrefe, R.I.; Kormann, M.S.D.; Porteus, M.H.; et al. Uridine Depletion and Chemical Modification Increase Cas9 mRNA Activity and Reduce Immunogenicity without HPLC Purification. Mol. Ther. Nucleic Acids 2018, 12, 530–542. [Google Scholar] [CrossRef]
- Thess, A.; Grund, S.; Mui, B.L.; Hope, M.J.; Baumhof, P.; Fotin-Mleczek, M.; Schlake, T. Sequence-engineered mRNA Without Chemical Nucleoside Modifications Enables an Effective Protein Therapy in Large Animals. Mol. Ther. 2015, 23, 1456–1464. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Yu, C.H.; Zhao, F.; Dang, Y.; Wu, C.; Xie, P.; Sachs, M.S.; Liu, Y. eRF1 mediates codon usage effects on mRNA translation efficiency through premature termination at rare codons. Nucleic Acids Res. 2019, 47, 9243–9258. [Google Scholar] [CrossRef] [Green Version]
- Benteyn, D.; Anguille, S.; Van Lint, S.; Heirman, C.; Van Nuffel, A.M.; Corthals, J.; Ochsenreither, S.; Waelput, W.; Van Beneden, K.; Breckpot, K.; et al. Design of an Optimized Wilms’ Tumor 1 (WT1) mRNA Construct for Enhanced WT1 Expression and Improved Immunogenicity In Vitro and In Vivo. Mol. Ther. Nucleic Acids 2013, 2, e134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Victor, M.P.; Acharya, D.; Begum, T.; Ghosh, T.C. The optimization of mRNA expression level by its intrinsic properties-Insights from codon usage pattern and structural stability of mRNA. Genomics 2019, 111, 1292–1297. [Google Scholar] [CrossRef] [PubMed]
- Griswold, K.E.; Mahmood, N.A.; Iverson, B.L.; Georgiou, G. Effects of codon usage versus putative 5’-mRNA structure on the expression of Fusarium solani cutinase in the Escherichia coli cytoplasm. Protein Expr. Purif. 2003, 27, 134–142. [Google Scholar] [CrossRef]
- Mitarai, N.; Sneppen, K.; Pedersen, S. Ribosome collisions and translation efficiency: Optimization by codon usage and mRNA destabilization. J. Mol. Biol. 2008, 382, 236–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weissman, D. mRNA transcript therapy. Expert Rev. Vaccin. 2015, 14, 265–281. [Google Scholar] [CrossRef] [PubMed]
- Holtkamp, S.; Kreiter, S.; Selmi, A.; Simon, P.; Koslowski, M.; Huber, C.; Tureci, O.; Sahin, U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 2006, 108, 4009–4017. [Google Scholar] [CrossRef]
- Trepotec, Z.; Geiger, J.; Plank, C.; Aneja, M.K.; Rudolph, C. Segmented poly(A) tails significantly reduce recombination of plasmid DNA without affecting mRNA translation efficiency or half-life. RNA 2019, 25, 507–518. [Google Scholar] [CrossRef]
- Chamberlin, M.; McGrath, J.; Waskell, L. New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature 1970, 228, 227–231. [Google Scholar] [CrossRef]
- Chamberlin, M.; Kingston, R.; Gilman, M.; Wiggs, J.; de Vera, A. Isolation of bacterial and bacteriophage RNA polymerases and their use in synthesis of RNA in vitro. Methods Enzymol. 1983, 101, 540–568. [Google Scholar]
- Li, B.; Luo, X.; Dong, Y. Effects of Chemically Modified Messenger RNA on Protein Expression. Bioconjug. Chem. 2016, 27, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Meyer, A.J.; Hirao, I.; Ellington, A.D. Genetic alphabet expansion transcription generating functional RNA molecules containing a five-letter alphabet including modified unnatural and natural base nucleotides by thermostable T7 RNA polymerase variants. Chem. Commun. 2017, 53, 12309–12312. [Google Scholar] [CrossRef]
- Milisavljevic, N.; Perlikova, P.; Pohl, R.; Hocek, M. Enzymatic synthesis of base-modified RNA by T7 RNA polymerase. A systematic study and comparison of 5-substituted pyrimidine and 7-substituted 7-deazapurine nucleoside triphosphates as substrates. Org. Biomol. Chem. 2018, 16, 5800–5807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akira, S. TLR signaling. Curr Top. Microbiol. Immunol. 2006, 311, 1–16. [Google Scholar]
- Kariko, K.; Ni, H.; Capodici, J.; Lamphier, M.; Weissman, D. mRNA is an endogenous ligand for Toll-like receptor 3. J. Biol. Chem. 2004, 279, 12542–12550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhang, Z.; Luo, J.; Han, X.; Wei, Y.; Wei, X. mRNA vaccine: A potential therapeutic strategy. Mol. Cancer 2021, 20, 33. [Google Scholar] [CrossRef]
- Barral, P.M.; Sarkar, D.; Su, Z.Z.; Barber, G.N.; DeSalle, R.; Racaniello, V.R.; Fisher, P.B. Functions of the cytoplasmic RNA sensors RIG-I and MDA-5: Key regulators of innate immunity. Pharmacol. Ther. 2009, 124, 219–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryter, J.M.; Schultz, S.C. Molecular basis of double-stranded RNA-protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998, 17, 7505–7513. [Google Scholar] [CrossRef] [Green Version]
- Balachandran, S.; Roberts, P.C.; Brown, L.E.; Truong, H.; Pattnaik, A.K.; Archer, D.R.; Barber, G.N. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 2000, 13, 129–141. [Google Scholar] [CrossRef] [Green Version]
- Garcia, M.A.; Meurs, E.F.; Esteban, M. The dsRNA protein kinase PKR: Virus and cell control. Biochimie 2007, 89, 799–811. [Google Scholar] [CrossRef]
- Weissman, D.; Kariko, K. mRNA: Fulfilling the Promise of Gene Therapy. Mol. Ther. 2015, 23, 1416–1417. [Google Scholar] [CrossRef] [Green Version]
- Raper, S.E.; Chirmule, N.; Lee, F.S.; Wivel, N.A.; Bagg, A.; Gao, G.P.; Wilson, J.M.; Batshaw, M.L. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol. Genet. Metab. 2003, 80, 148–158. [Google Scholar] [CrossRef]
- Boros, G.; Miko, E.; Muramatsu, H.; Weissman, D.; Emri, E.; Rozsa, D.; Nagy, G.; Juhasz, A.; Juhasz, I.; van der Horst, G.; et al. Transfection of pseudouridine-modified mRNA encoding CPD-photolyase leads to repair of DNA damage in human keratinocytes: A new approach with future therapeutic potential. J. Photochem. Photobiol. B 2013, 129, 93–99. [Google Scholar] [CrossRef] [Green Version]
- Andries, O.; Mc Cafferty, S.; De Smedt, S.C.; Weiss, R.; Sanders, N.N.; Kitada, T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 2015, 217, 337–344. [Google Scholar] [CrossRef]
- Nelson, J.; Sorensen, E.W.; Mintri, S.; Rabideau, A.E.; Zheng, W.; Besin, G.; Khatwani, N.; Su, S.V.; Miracco, E.J.; Issa, W.J.; et al. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 2020, 6, eaaz6893. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Greenwald, E.; Ahmad, S.; Hur, S. An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res. 2018, 46, 5239–5249. [Google Scholar] [CrossRef]
- Durbin, A.F.; Wang, C.; Marcotrigiano, J.; Gehrke, L. RNAs Containing Modified Nucleotides Fail To Trigger RIG-I Conformational Changes for Innate Immune Signaling. mBio 2016, 7, e00833-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freund, I.; Eigenbrod, T.; Helm, M.; Dalpke, A.H. RNA Modifications Modulate Activation of Innate Toll-Like Receptors. Genes 2019, 10, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, M.A.; Xu, Y.; Tao, W.; Ubellacker, J.M.; Lim, M.; Aum, D.; Lee, G.Y.; Zhou, K.; Zope, H.; Yu, M.; et al. Restoration of tumour-growth suppression in vivo via systemic nanoparticle-mediated delivery of PTEN mRNA. Nat. Biomed. Eng. 2018, 2, 850–864. [Google Scholar] [CrossRef]
- Lorenz, C.; Fotin-Mleczek, M.; Roth, G.; Becker, C.; Dam, T.C.; Verdurmen, W.P.; Brock, R.; Probst, J.; Schlake, T. Protein expression from exogenous mRNA: Uptake by receptor-mediated endocytosis and trafficking via the lysosomal pathway. RNA Biol. 2011, 8, 627–636. [Google Scholar] [CrossRef]
- Buschmann, M.D.; Carrasco, M.J.; Alishetty, S.; Paige, M.; Alameh, M.G.; Weissman, D. Nanomaterial Delivery Systems for mRNA Vaccines. Vaccines 2021, 9, 65. [Google Scholar] [CrossRef]
- Geall, A.J.; Verma, A.; Otten, G.R.; Shaw, C.A.; Hekele, A.; Banerjee, K.; Cu, Y.; Beard, C.W.; Brito, L.A.; Krucker, T.; et al. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA 2012, 109, 14604–14609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. [Google Scholar] [CrossRef]
- Liang, F.; Lindgren, G.; Lin, A.; Thompson, E.A.; Ols, S.; Rohss, J.; John, S.; Hassett, K.; Yuzhakov, O.; Bahl, K.; et al. Efficient Targeting and Activation of Antigen-Presenting Cells In Vivo after Modified mRNA Vaccine Administration in Rhesus Macaques. Mol. Ther. 2017, 25, 2635–2647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Perez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.J.; Naradikian, M.S.; Parkhouse, K.; Cain, D.W.; Jones, L.; Moody, M.A.; Verkerke, H.P.; Myles, A.; Willis, E.; et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018, 215, 1571–1588. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, G.; Ols, S.; Liang, F.; Thompson, E.A.; Lin, A.; Hellgren, F.; Bahl, K.; John, S.; Yuzhakov, O.; Hassett, K.J.; et al. Induction of Robust B Cell Responses after Influenza mRNA Vaccination Is Accompanied by Circulating Hemagglutinin-Specific ICOS+ PD-1+ CXCR3+ T Follicular Helper Cells. Front. Immunol. 2017, 8, 1539. [Google Scholar] [CrossRef] [Green Version]
- Vogel, A.B.; Kanevsky, I.; Che, Y.; Swanson, K.A.; Muik, A.; Vormehr, M.; Kranz, L.M.; Walzer, K.C.; Hein, S.; Guler, A.; et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 2021, 592, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Lederer, K.; Castano, D.; Gomez Atria, D.; Oguin, T.H., III; Wang, S.; Manzoni, T.B.; Muramatsu, H.; Hogan, M.J.; Amanat, F.; Cherubin, P.; et al. SARS-CoV-2 mRNA Vaccines Foster Potent Antigen-Specific Germinal Center Responses Associated with Neutralizing Antibody Generation. Immunity 2020, 53, 1281–1295-e1285. [Google Scholar] [CrossRef]
- Tai, W.; Zhang, X.; Drelich, A.; Shi, J.; Hsu, J.C.; Luchsinger, L.; Hillyer, C.D.; Tseng, C.K.; Jiang, S.; Du, L. A novel receptor-binding domain (RBD)-based mRNA vaccine against SARS-CoV-2. Cell Res. 2020, 30, 932–935. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.R.; Qu, Y.; Zhu, H.; Yang, Y.; Vogel, A.B.; Sahin, U.; Qin, C.; Hui, A. BNT162b2 Vaccine Encoding the SARS-CoV-2 P2 S Protects Transgenic hACE2 Mice against COVID-19. Vaccines 2021, 9, 324. [Google Scholar] [CrossRef]
- Bettini, E.; Locci, M. SARS-CoV-2 mRNA Vaccines: Immunological Mechanism and Beyond. Vaccines 2021, 9, 147. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Ji, K.; Tian, S.; Wang, F.; Huang, B.; Tong, Z.; Tan, S.; Hao, J.; Wang, Q.; Tan, W.; et al. A single-dose mRNA vaccine provides a long-term protection for hACE2 transgenic mice from SARS-CoV-2. Nat. Commun. 2021, 12, 776. [Google Scholar] [CrossRef] [PubMed]
- Rauch, S.; Roth, N.; Schwendt, K.; Fotin-Mleczek, M.; Mueller, S.O.; Petsch, B. mRNA-based SARS-CoV-2 vaccine candidate CVnCoV induces high levels of virus-neutralising antibodies and mediates protection in rodents. NPJ Vaccines 2021, 6, 57. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.N.; Li, X.F.; Deng, Y.Q.; Zhao, H.; Huang, Y.J.; Yang, G.; Huang, W.J.; Gao, P.; Zhou, C.; Zhang, R.R.; et al. A Thermostable mRNA Vaccine against COVID-19. Cell 2020, 182, 1271–1283-e1216. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Cottrell, C.A.; Wang, N.; Pallesen, J.; Yassine, H.M.; Turner, H.L.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Pre-fusion structure of a human coronavirus spike protein. Nature 2016, 531, 118–121. [Google Scholar] [CrossRef] [Green Version]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef] [Green Version]
- Kalnin, K.V.; Plitnik, T.; Kishko, M.; Zhang, J.; Zhang, D.; Beauvais, A.; Anosova, N.G.; Tibbitts, T.; Di Napoli, J.; Ulinski, G.; et al. Immunogenicity and efficacy of mRNA COVID-19 vaccine MRT5500 in preclinical animal models. NPJ Vaccines 2021, 6, 61. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.H.; Patel, N.; Haupt, R.; Zhou, H.; Weston, S.; Hammond, H.; Logue, J.; Portnoff, A.D.; Norton, J.; Guebre-Xabier, M.; et al. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat. Commun. 2021, 12, 372. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020, 383, 2427–2438. [Google Scholar] [CrossRef]
- Blakney, A.K.; Ip, S.; Geall, A.J. An Update on Self-Amplifying mRNA Vaccine Development. Vaccines 2021, 9, 97. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 2020, 586, 589–593. [Google Scholar] [CrossRef]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef] [PubMed]
- CureVac. 2021, Disappointing Late-Stage Results for CureVac’s COVID-19 mRNA Vaccine Trial. Available online: clinicalomics.com (accessed on 23 June 2021).
- De Alwis, R.; Gan, E.S.; Chen, S.; Leong, Y.S.; Tan, H.C.; Zhang, S.L.; Yau, C.; Low, J.G.H.; Kalimuddin, S.; Matsuda, D.; et al. A single dose of self-transcribing and replicating RNA-based SARS-CoV-2 vaccine produces protective adaptive immunity in mice. Mol. Ther. 2021, 29, 1970–1983. [Google Scholar] [CrossRef]
- McKay, P.F.; Hu, K.; Blakney, A.K.; Samnuan, K.; Brown, J.C.; Penn, R.; Zhou, J.; Bouton, C.R.; Rogers, P.; Polra, K.; et al. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020, 11, 3523. [Google Scholar] [CrossRef]
- Petsch, B.; Schnee, M.; Vogel, A.B.; Lange, E.; Hoffmann, B.; Voss, D.; Schlake, T.; Thess, A.; Kallen, K.J.; Stitz, L.; et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012, 30, 1210–1216. [Google Scholar] [CrossRef] [PubMed]
- Brazzoli, M.; Magini, D.; Bonci, A.; Buccato, S.; Giovani, C.; Kratzer, R.; Zurli, V.; Mangiavacchi, S.; Casini, D.; Brito, L.M.; et al. Induction of Broad-Based Immunity and Protective Efficacy by Self-amplifying mRNA Vaccines Encoding Influenza Virus Hemagglutinin. J. Virol. 2016, 90, 332–344. [Google Scholar] [CrossRef] [Green Version]
- Lutz, J.; Lazzaro, S.; Habbeddine, M.; Schmidt, K.E.; Baumhof, P.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; Hope, M.J.; Heidenreich, R.; et al. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. NPJ Vaccines 2017, 2, 29. [Google Scholar] [CrossRef] [Green Version]
- Pardi, N.; Parkhouse, K.; Kirkpatrick, E.; McMahon, M.; Zost, S.J.; Mui, B.L.; Tam, Y.K.; Kariko, K.; Barbosa, C.J.; Madden, T.D.; et al. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018, 9, 3361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freyn, A.W.; Ramos da Silva, J.; Rosado, V.C.; Bliss, C.M.; Pine, M.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; de Souza Ferreira, L.C.; Weissman, D.; et al. A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol. Ther. 2020, 28, 1569–1584. [Google Scholar] [CrossRef] [PubMed]
- Maruggi, G.; Ulmer, J.B.; Rappuoli, R.; Yu, D. Self-amplifying mRNA-Based Vaccine Technology and Its Mode of Action. Curr. Top. Microbiol. Immunol. 2021. [Google Scholar] [CrossRef]
- Maruggi, G.; Zhang, C.; Li, J.; Ulmer, J.B.; Yu, D. mRNA as a Transformative Technology for Vaccine Development to Control Infectious Diseases. Mol. Ther. 2019, 27, 757–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espeseth, A.S.; Cejas, P.J.; Citron, M.P.; Wang, D.; DiStefano, D.J.; Callahan, C.; Donnell, G.O.; Galli, J.D.; Swoyer, R.; Touch, S.; et al. Modified mRNA/lipid nanoparticle-based vaccines expressing respiratory syncytial virus F protein variants are immunogenic and protective in rodent models of RSV infection. NPJ Vaccines 2020, 5, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aliprantis, A.O.; Shaw, C.A.; Griffin, P.; Farinola, N.; Railkar, R.A.; Cao, X.; Liu, W.; Sachs, J.R.; Swenson, C.J.; Lee, H.; et al. A phase 1, randomized, placebo-controlled study to evaluate the safety and immunogenicity of an mRNA-based RSV prefusion F protein vaccine in healthy younger and older adults. Hum. Vaccines Immunother. 2021, 17, 1248–1261. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Pelc, R.S.; Muramatsu, H.; Andersen, H.; DeMaso, C.R.; Dowd, K.A.; Sutherland, L.L.; Scearce, R.M.; Parks, R.; et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 2017, 543, 248–251. [Google Scholar] [CrossRef]
- Richner, J.M.; Himansu, S.; Dowd, K.A.; Butler, S.L.; Salazar, V.; Fox, J.M.; Julander, J.G.; Tang, W.W.; Shresta, S.; Pierson, T.C.; et al. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell 2017, 168, 1114–1125-e1110. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Z.; Portela Catani, J.P.; Mc Cafferty, S.; Couck, L.; Van Den Broeck, W.; Gorle, N.; Vandenbroucke, R.E.; Devriendt, B.; Ulbert, S.; Cnops, L.; et al. Immunogenicity and Protection Efficacy of a Naked Self-Replicating mRNA-Based Zika Virus Vaccine. Vaccines 2019, 7, 96. [Google Scholar] [CrossRef] [Green Version]
- Schnee, M.; Vogel, A.B.; Voss, D.; Petsch, B.; Baumhof, P.; Kramps, T.; Stitz, L. An mRNA Vaccine Encoding Rabies Virus Glycoprotein Induces Protection against Lethal Infection in Mice and Correlates of Protection in Adult and Newborn Pigs. PLoS Negl. Trop. Dis. 2016, 10, e0004746. [Google Scholar] [CrossRef]
- Stitz, L.; Vogel, A.; Schnee, M.; Voss, D.; Rauch, S.; Mutzke, T.; Ketterer, T.; Kramps, T.; Petsch, B. A thermostable messenger RNA based vaccine against rabies. PLoS Negl. Trop. Dis. 2017, 11, e0006108. [Google Scholar] [CrossRef] [Green Version]
- Aldrich, C.; Leroux-Roels, I.; Huang, K.B.; Bica, M.A.; Loeliger, E.; Schoenborn-Kellenberger, O.; Walz, L.; Leroux-Roels, G.; von Sonnenburg, F.; Oostvogels, L. Proof-of-concept of a low-dose unmodified mRNA-based rabies vaccine formulated with lipid nanoparticles in human volunteers: A phase 1 trial. Vaccine 2021, 39, 1310–1318. [Google Scholar] [CrossRef]
- Saunders, K.O.; Pardi, N.; Parks, R.; Santra, S.; Mu, Z.; Sutherland, L.; Scearce, R.; Barr, M.; Eaton, A.; Hernandez, G.; et al. Lipid nanoparticle encapsulated nucleoside-modified mRNA vaccines elicit polyfunctional HIV-1 antibodies comparable to proteins in nonhuman primates. NPJ Vaccines 2021, 6, 1–4. [Google Scholar] [CrossRef]
- Nelson, C.S.; Jenks, J.A.; Pardi, N.; Goodwin, M.; Roark, H.; Edwards, W.; McLellan, J.S.; Pollara, J.; Weissman, D.; Permar, S.R. Human Cytomegalovirus Glycoprotein B Nucleoside-Modified mRNA Vaccine Elicits Antibody Responses with Greater Durability and Breadth than MF59-Adjuvanted gB Protein Immunization. J. Virol. 2020, 94, e00186-20. [Google Scholar] [CrossRef]
- John, S.; Yuzhakov, O.; Woods, A.; Deterling, J.; Hassett, K.; Shaw, C.A.; Ciaramella, G. Multi-antigenic human cytomegalovirus mRNA vaccines that elicit potent humoral and cell-mediated immunity. Vaccine 2018, 36, 1689–1699. [Google Scholar] [CrossRef]
- Cabanillas, B.; Akdis, C.A.; Novak, N. Allergic reactions to the first COVID-19 vaccine: A potential role of polyethylene glycol? Allergy 2021, 76, 1617–1618. [Google Scholar] [CrossRef] [PubMed]
- Wylon, K.; Dolle, S.; Worm, M. Polyethylene glycol as a cause of anaphylaxis. Allergy Asthma Clin. Immunol. 2016, 12, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwertz, H.; Rowley, J.W.; Schumann, G.G.; Thorack, U.; Campbell, R.A.; Manne, B.K.; Zimmerman, G.A.; Weyrich, A.S.; Rondina, M.T. Endogenous LINE-1 (Long Interspersed Nuclear Element-1) Reverse Transcriptase Activity in Platelets Controls Translational Events Through RNA-DNA Hybrids. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 801–815. [Google Scholar] [CrossRef]
- Klug, M.E.; Lazareva, O.; Kirmes, K.; Rosenbaum, M.; Lukas, M.; Weidlich, S.; Spinner, C.D.; von Scheidt, M.; Gosetti, R.; Baumbach, J. Platelet expression and reactivity after BNT162b2 vaccine administration. medRxiv 2021. [Google Scholar] [CrossRef]
- Dotan, A.; Shoenfeld, Y. Perspectives on vaccine induced thrombotic thrombocytopenia. J. Autoimmun. 2021, 121, 102663. [Google Scholar] [CrossRef]
- Scully, M.; Singh, D.; Lown, R.; Poles, A.; Solomon, T.; Levi, M.; Goldblatt, D.; Kotoucek, P.; Thomas, W.; Lester, W. Pathologic Antibodies to Platelet Factor 4 after ChAdOx1 nCoV-19 Vaccination. N. Engl. J. Med. 2021, 384, 2202–2211. [Google Scholar] [CrossRef]
- Muir, K.L.; Kallam, A.; Koepsell, S.A.; Gundabolu, K. Thrombotic Thrombocytopenia after Ad26.COV2.S Vaccination. N. Engl. J. Med. 2021, 384, 1964–1965. [Google Scholar] [CrossRef]
- Bongiovanni, D.; Klug, M.; Lazareva, O.; Weidlich, S.; Biasi, M.; Ursu, S.; Warth, S.; Buske, C.; Lukas, M.; Spinner, C.D.; et al. SARS-CoV-2 infection is associated with a pro-thrombotic platelet phenotype. Cell Death Dis. 2021, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.O.; Kafai, N.M.; Dmitriev, I.P.; Fox, J.M.; Smith, B.K.; Harvey, I.B.; Chen, R.E.; Winkler, E.S.; Wessel, A.W.; Case, J.B.; et al. A Single-Dose Intranasal ChAd Vaccine Protects Upper and Lower Respiratory Tracts against SARS-CoV-2. Cell 2020, 183, 169–184-e113. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Purushotham, J.; Schulz, J.; Holbrook, M.; Bushmaker, T.; Carmody, A.; Port, J.; Yinda, K.C.; Okumura, A.; Saturday, G.; et al. Intranasal ChAdOx1 nCoV-19/AZD1222 vaccination reduces shedding of SARS-CoV-2 D614G in rhesus macaques. bioRxiv 2021. [Google Scholar] [CrossRef]
- Phua, K.K.; Staats, H.F.; Leong, K.W.; Nair, S.K. Intranasal mRNA nanoparticle vaccination induces prophylactic and therapeutic anti-tumor immunity. Sci. Rep. 2014, 4, 5128. [Google Scholar] [CrossRef] [PubMed]
- Migliore, A.; Gigliucci, G.; Di Marzo, R.; Russo, D.; Mammucari, M. Intradermal Vaccination: A Potential Tool in the Battle Against the COVID-19 Pandemic? Risk Manag. Healthc. Policy 2021, 14, 2079–2087. [Google Scholar] [CrossRef] [PubMed]
| Immunogens | mRNA Type | Model/Route/mRNA Dose (μg) | References |
|---|---|---|---|
| Hemagglutinin (HA) (H1N1, H3N2, H5N1 virus) | Codon optimized/protamine RNA | Mice: i.d./20 μg, 80 μg. Ferrets: i.m./20, 80, 250 ug Pigs: i.d./250 μg | [119] |
| HA (H1N1, A/Cal09) | Self-amplifying mRNA/Cationic nanoemulsion | Mice: i.m./0.1, 1,10 μg Ferrets: i.m./15, 45 μg | [120] |
| HA (H1N1pdm09) | Codon optimized/unmodified/LNPs | Mice: i.m./0.5, 5 μg NHPs: i.m./10 μg | [121] |
| HA (H10N8) | Nucleoside modified/LNPs | NHPs: i.m./50 μg mRNA +/− GLA (5 μg) adjuvant | [98] |
| HA (H1N1, A/Cal09) | Nucleoside modified (m1ψ-mRNA/LNPs) | Mice: i.d. (3, 10, 30 μg) Mice: i.m. (10, 30, 90 μg) Ferrets: i.m./30 μg | [122] |
| HA (H1, A/PR8) | Nucleoside modified (m1ψ-mRNA/LNPs) | Mice: i.d. (30 μg, single dose) | [97] |
| Mini-HA stalk, NA, M2, NP | Nucleoside modified (m1ψ-mRNA/LNPs) | Mice: i.d. (0.05, 0.5, 5, 10, 20 μg, single dose) | [123] |
| HA (H10N8, H7N9; H10 mRNA in phase 1) | Nucleoside modified/LNPs | Mice: i.d., i.m./10 μg, single dose; 0.4, 2 μg boost dose) Ferrets: i.d./10, 50, 200 μg NHPs: i.d., i.m./200, 400 μg Humans (phase 1): i.m./100 μg | [21] |
| HA (H10N8, H7N9; H10 mRNA, phase 1) | Nucleoside modified/LNPs | Adults: i.m./H7N9 mRNA 10, 25, 50 μg; i.m./H10N8 mRNA 25, 50, 75, 100, 400 μg; i.d./25, 50 μg | [22] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeeva, S.; Kim, K.-H.; Shin, C.H.; Wang, B.-Z.; Kang, S.-M. An Update on mRNA-Based Viral Vaccines. Vaccines 2021, 9, 965. https://doi.org/10.3390/vaccines9090965
Jeeva S, Kim K-H, Shin CH, Wang B-Z, Kang S-M. An Update on mRNA-Based Viral Vaccines. Vaccines. 2021; 9(9):965. https://doi.org/10.3390/vaccines9090965
Chicago/Turabian StyleJeeva, Subbiah, Ki-Hye Kim, Chong Hyun Shin, Bao-Zhong Wang, and Sang-Moo Kang. 2021. "An Update on mRNA-Based Viral Vaccines" Vaccines 9, no. 9: 965. https://doi.org/10.3390/vaccines9090965
APA StyleJeeva, S., Kim, K.-H., Shin, C. H., Wang, B.-Z., & Kang, S.-M. (2021). An Update on mRNA-Based Viral Vaccines. Vaccines, 9(9), 965. https://doi.org/10.3390/vaccines9090965






