Cellular and Humoral Immunogenicity of a Candidate DNA Vaccine Expressing SARS-CoV-2 Spike Subunit 1
Abstract
:1. Introduction
2. Materials and Methods
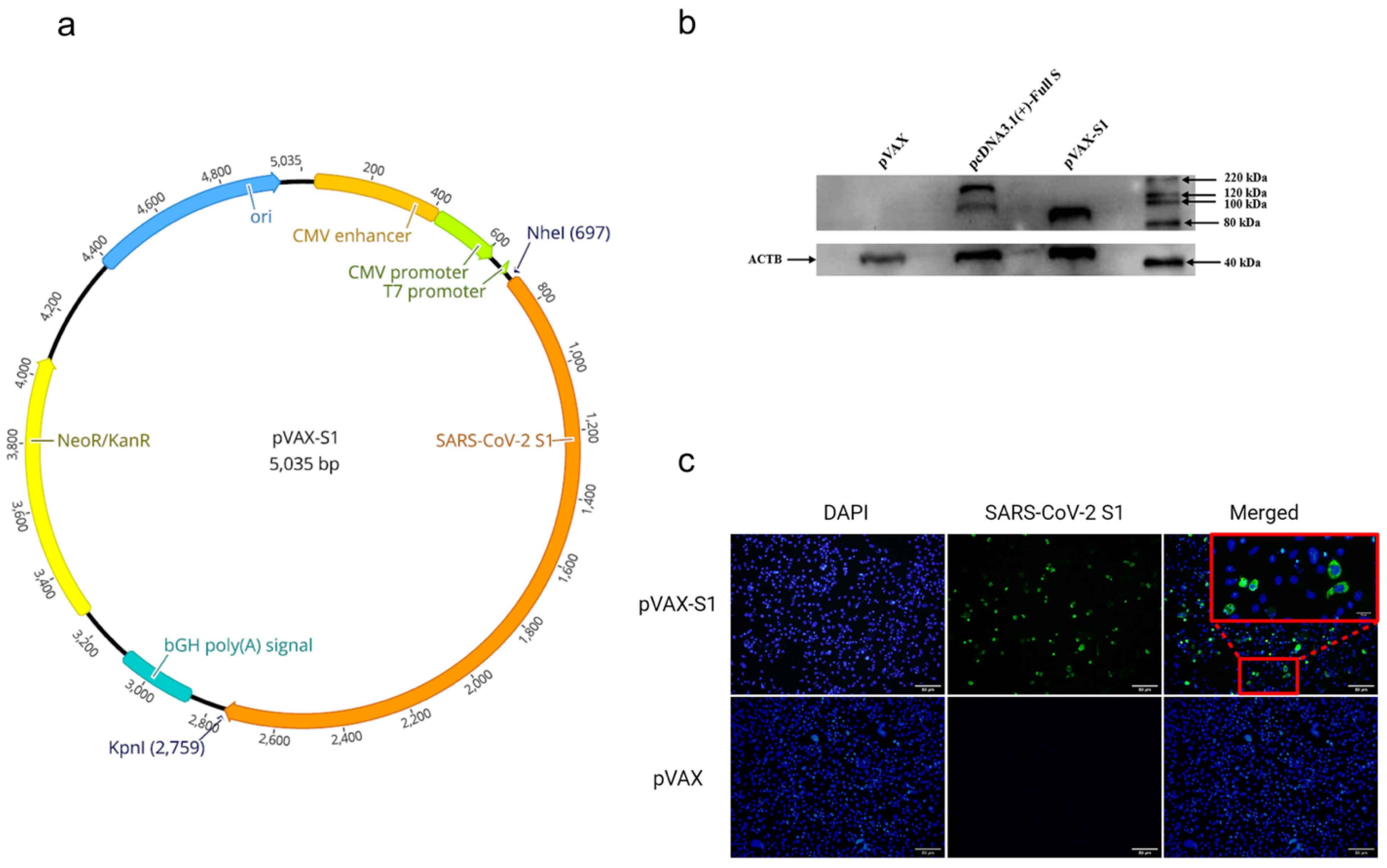
2.1. Generation of DNA Construct
2.2. Detection of SARS-CoV-2 S1 Protein Expression by Western Blot
2.3. Detection of SARS-CoV-2 S1 Protein Expression by Immunofluorescence
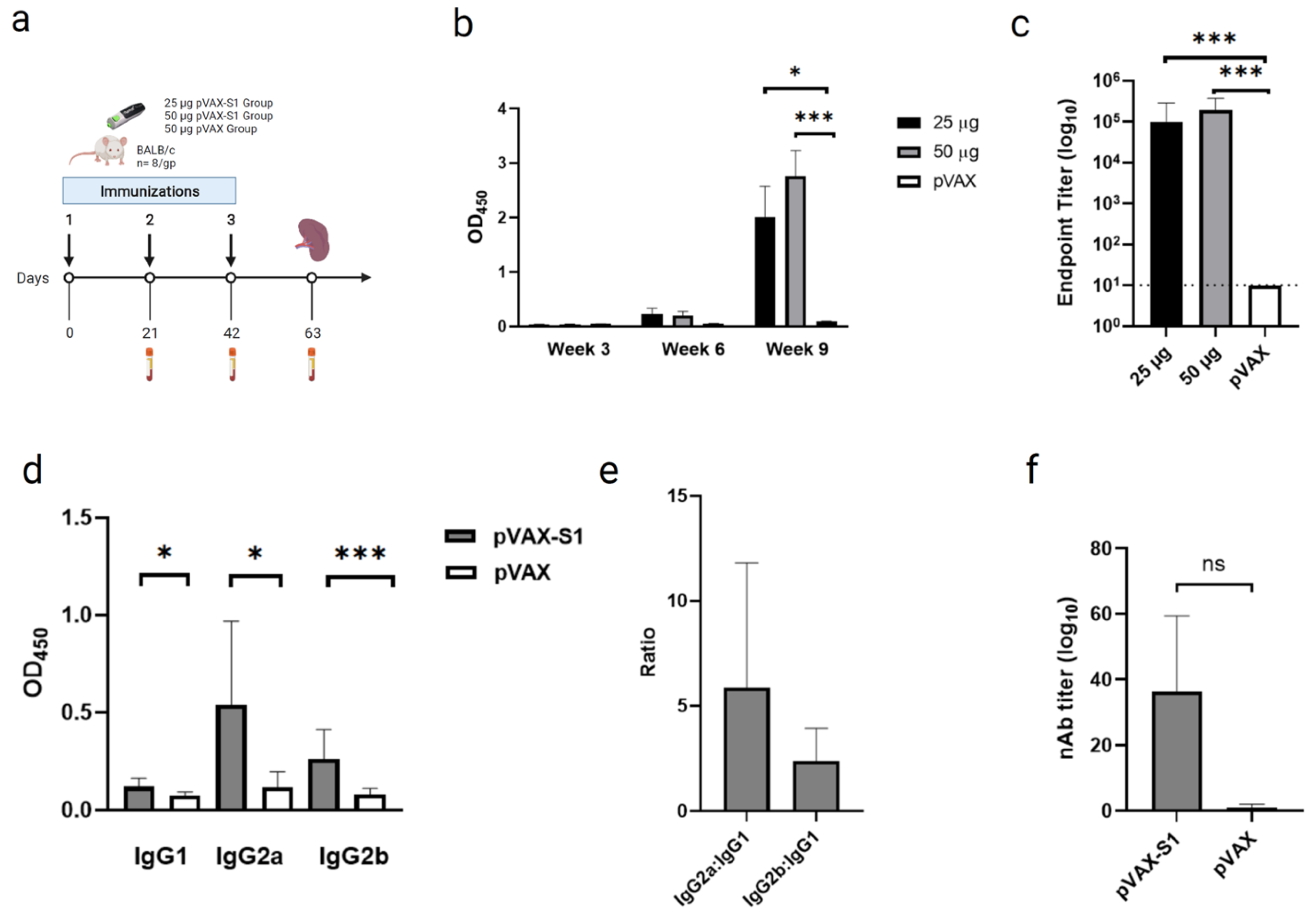
2.4. Immunization and Samples Collection
2.5. Binding Antibodies Measurement by Indirect ELISA
2.6. Neutralizing Antibodies Measurement by Pseudovirus Neutralization Assay
2.7. Determination of T Cell Response by Flow Cytometry
2.8. Statistical Analysis
3. Results
3.1. In Vitro Confirmation of Protein Expression from the Candidate Vaccine
3.2. Evaluation of Binding and Neutralizing Antibodies in Immunized Mice
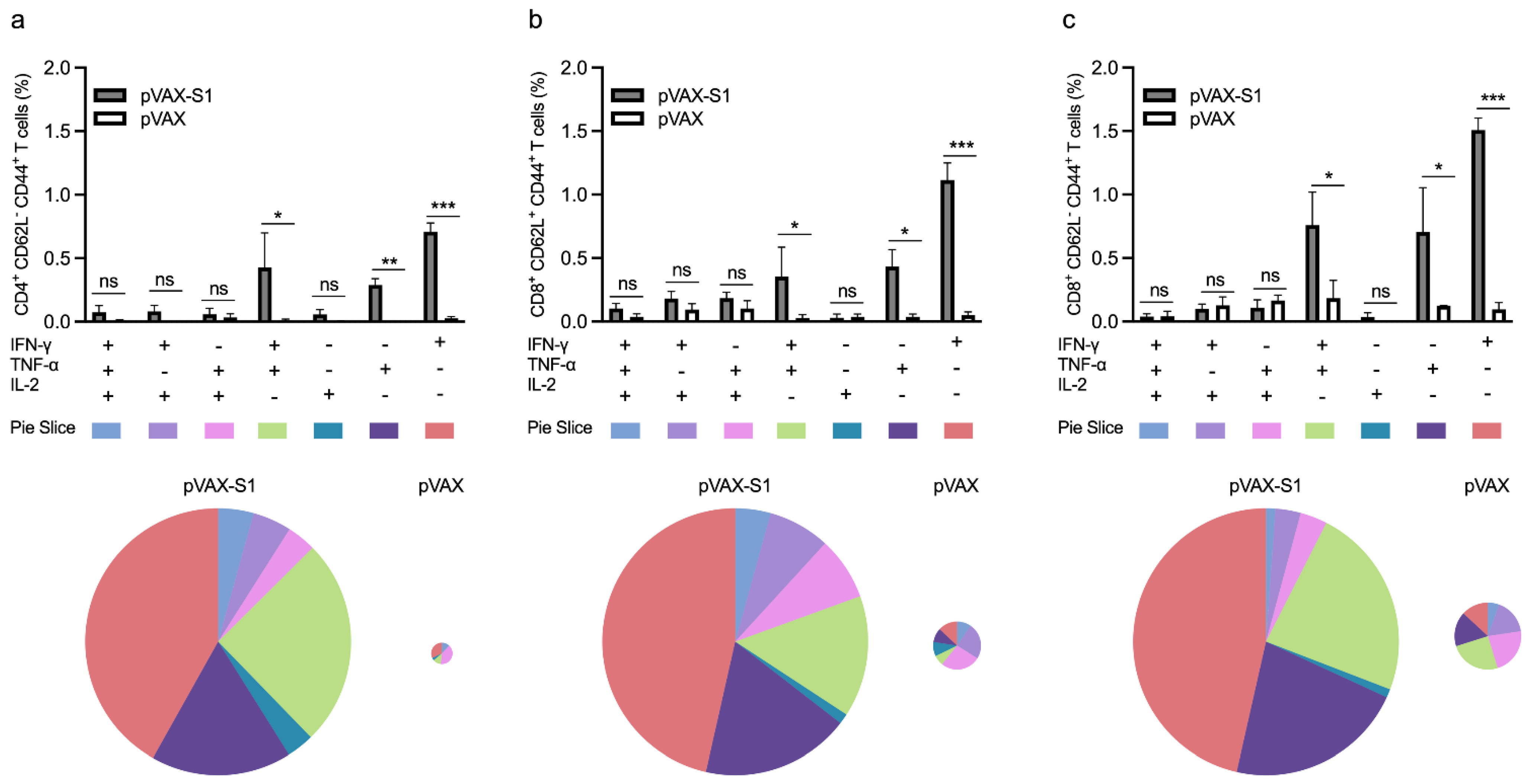
3.3. Evaluation of Cellular Immune Response in Immunized Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hui, D.S.; Azhar, E.I.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; Mchugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, B.J.; Wong, K.H.; Zhou, J.; Wong, K.L.; Young, B.W.; Lu, L.W.; Lee, S.S. SARS-related virus predating SARS outbreak, Hong Kong. Emerg. Infect. Dis. 2004, 10, 176–178. [Google Scholar] [CrossRef]
- Oboho, I.K.; Tomczyk, S.M.; Al-Asmari, A.M.; Banjar, A.A.; Al-Mugti, H.; Aloraini, M.S.; Alkhaldi, K.Z.; Almohammadi, E.L.; Alraddadi, B.M.; Gerber, S.I.; et al. 2014 MERS-CoV outbreak in Jeddah—A link to health care facilities. N. Engl. J. Med. 2015, 372, 846–854. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Coronavirus Disease (COVID-19) Pandemic. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 12 June 2021).
- Adhikari, S.P.; Meng, S.; Wu, Y.J.; Mao, Y.P.; Ye, R.X.; Wang, Q.Z.; Sun, C.; Sylvia, S.; Rozelle, S.; Raat, H.; et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: A scoping review. Infect. Dis. Poverty 2020, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.Y.; Chen, L.; Wang, M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA 2020, 323, 1406–1407. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Zhang, J.; Zeng, H.; Gu, J.; Li, H.; Zheng, L.; Zou, Q. Progress and Prospects on Vaccine Development against SARS-CoV-2. Vaccines 2020, 8, 153. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Dai, T.; Wei, Y.; Zhang, L.; Zheng, M.; Zhou, F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target. Ther. 2020, 5. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Zhu, F.C.; Guan, X.H.; Li, Y.H.; Huang, J.Y.; Jiang, T.; Hou, L.H.; Li, J.X.; Yang, B.F.; Wang, L.; Wang, W.J.; et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 2020, 396, 479–488. [Google Scholar] [CrossRef]
- Sadoff, J.; Le Gars, M.; Shukarev, G.; Heerwegh, D.; Truyers, C.; de Groot, A.M.; Stoop, J.; Tete, S.; Van Damme, W.; Leroux-Roels, I.; et al. Interim Results of a Phase 1-2a Trial of Ad26.COV2.S Covid-19 Vaccine. N. Engl. J. Med. 2021, 384, 1824–1835. [Google Scholar] [CrossRef] [PubMed]
- Logunov, D.Y.; Dolzhikova, I.V.; Shcheblyakov, D.V.; Tukhvatulin, A.I.; Zubkova, O.V.; Dzharullaeva, A.S.; Kovyrshina, A.V.; Lubenets, N.L.; Grousova, D.M.; Erokhova, A.S.; et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: An interim analysis of a randomised controlled phase 3 trial in Russia. Lancet 2021, 397, 671–681. [Google Scholar] [CrossRef]
- Vasireddy, D.; Atluri, P.; Malayala, S.V.; Vanaparthy, R.; Mohan, G. Review of COVID-19 Vaccines Approved in the United States of America for Emergency Use. J. Clin. Med. Res. 2021, 13, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Keech, C.; Albert, G.; Cho, I.; Robertson, A.; Reed, P.; Neal, S.; Plested, J.S.; Zhu, M.; Cloney-Clark, S.; Zhou, H.; et al. Phase 1–2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020, 383, 2320–2332. [Google Scholar] [CrossRef] [PubMed]
- Poland, G.A.; Ovsyannikova, I.G.; Kennedy, R.B. SARS-CoV-2 immunity: Review and applications to phase 3 vaccine candidates. Lancet 2020, 396, 1595–1606. [Google Scholar] [CrossRef]
- Tebas, P.; Yang, S.; Boyer, J.D.; Reuschel, E.L.; Patel, A.; Christensen-Quick, A.; Andrade, V.M.; Morrow, M.P.; Kraynyak, K.; Agnes, J.; et al. Safety and immunogenicity of INO-4800 DNA vaccine against SARS-CoV-2: A preliminary report of an open-label, Phase 1 clinical trial. EClinicalMedicine 2021, 31. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef]
- McNaughton, A.L.; Paton, R.S.; Edmans, M.; Youngs, J.; Wellens, J.; Phalora, P.; Fyfe, A.; Belij-Rammerstorfer, S.; Bolton, J.S.; Ball, J.; et al. Fatal COVID-19 outcomes are associated with an antibody response targeting epitopes shared with endemic coronaviruses. medRxiv 2021. [Google Scholar] [CrossRef]
- Fang, Y.T.; Lin, C.F.; Liao, P.C.; Kuo, Y.M.; Wang, S.; Yeh, T.M.; Shieh, C.C.; Su, I.J.; Lei, H.Y.; Lin, Y.S. Annexin A2 on lung epithelial cell surface is recognized by severe acute respiratory syndrome-associated coronavirus spike domain 2 antibodies. Mol. Immunol. 2010, 47, 1000–1009. [Google Scholar] [CrossRef]
- Weingartl, H.; Czub, M.; Czub, S.; Neufeld, J.; Marszal, P.; Gren, J.; Smith, G.; Jones, S.; Proulx, R.; Deschambault, Y.; et al. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J. Virol. 2004, 78, 12672–12676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaume, M.; Yip, M.S.; Kam, Y.W.; Cheung, C.Y.; Kien, F.; Roberts, A.; Li, P.H.; Dutry, I.; Escriou, N.; Daeron, M.; et al. SARS CoV subunit vaccine: Antibody-mediated neutralisation and enhancement. Hong Kong Med. J. 2012, 18 (Suppl. 2), 31–36. [Google Scholar]
- Jaume, M.; Yip, M.S.; Cheung, C.Y.; Leung, H.L.; Li, P.H.; Kien, F.; Dutry, I.; Callendret, B.; Escriou, N.; Altmeyer, R.; et al. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent FcγR pathway. J. Virol. 2011, 85, 10582–10597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, R.C.; Scott, F.W. Antibody-mediated enhancement of disease in feline infectious peritonitis: Comparisons with dengue hemorrhagic fever. Comp. Immunol. Microbiol. Infect. Dis. 1981, 4, 175–189. [Google Scholar] [CrossRef]
- Olsen, C.W. A review of feline infectious peritonitis virus: Molecular biology, immunopathogenesis, clinical aspects, and vaccination. Vet. Microbiol. 1993, 36, 1–37. [Google Scholar] [CrossRef]
- Al-Amri, S.S.; Abbas, A.T.; Siddiq, L.A.; Alghamdi, A.; Sanki, M.A.; Al-Muhanna, M.K.; Alhabbab, R.Y.; Azhar, E.I.; Li, X.; Hashem, A.M. Immunogenicity of Candidate MERS-CoV DNA Vaccines Based on the Spike Protein. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Almahboub, S.A.; Algaissi, A.; Alfaleh, M.A.; ElAssouli, M.Z.; Hashem, A.M. Evaluation of Neutralizing Antibodies against Highly Pathogenic Coronaviruses: A Detailed Protocol for a Rapid Evaluation of Neutralizing Antibodies Using Vesicular Stomatitis Virus (Vsv) Pseudovirus-Based Assay. Front. Microbiol. 2020, 4. [Google Scholar] [CrossRef]
- Goodman, A.G.; Heinen, P.P.; Guerra, S.; Vijayan, A.; Sorzano, C.O.; Gomez, C.E.; Esteban, M. A human multi-epitope recombinant vaccinia virus as a universal T cell vaccine candidate against influenza virus. PLoS ONE 2011, 6, e25938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez, P.; Martín-Acebes, M.A.; Poderoso, T.; Lázaro-Frías, A.; Saiz, J.C.; Sorzano, C.; Esteban, M.; García-Arriaza, J. The combined vaccination protocol of DNA/MVA expressing Zika virus structural proteins as efficient inducer of T and B cell immune responses. Emerg. Microbes Infect. 2021, 10, 1441–1456. [Google Scholar] [CrossRef] [PubMed]
- Muthumani, K.; Falzarano, D.; Reuschel, E.L.; Tingey, C.; Flingai, S.; Villarreal, D.O.; Wise, M.; Patel, A.; Izmirly, A.; Aljuaid, A.; et al. A synthetic consensus anti-spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci. Transl. Med. 2015, 7, 301ra132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, J.A. Vector Design for Improved DNA Vaccine Efficacy, Safety and Production. Vaccines 2013, 1, 225–249. [Google Scholar] [CrossRef]
- Fernández, A. SARS-CoV-2 Glycosylation Suggests That Vaccines Should Have Adopted the S1 Subunit as Antigen. ACS Pharmacol. Transl. Sci. 2021, 4, 1016–1017. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Cao, H.; Liu, C. SARS-CoV-2 S1 is superior to the RBD as a COVID-19 subunit vaccine antigen. J. Med. Virol. 2021, 93, 892–898. [Google Scholar] [CrossRef] [PubMed]
- Ababneh, M.; Alrwashdeh, M.; Khalifeh, M. Recombinant adenoviral vaccine encoding the spike 1 subunit of the Middle East Respiratory Syndrome Coronavirus elicits strong humoral and cellular immune responses in mice. Vet. World 2019, 12, 1554–1562. [Google Scholar] [CrossRef] [Green Version]
- Hashem, A.M.; Algaissi, A.; Agrawal, A.S.; Al-Amri, S.S.; Alhabbab, R.Y.; Sohrab, S.S.; Almasoud, A.S.; Alharbi, N.K.; Peng, B.H.; Russell, M.; et al. A Highly Immunogenic, Protective, and Safe Adenovirus-Based Vaccine Expressing Middle East Respiratory Syndrome Coronavirus S1-CD40L Fusion Protein in a Transgenic Human Dipeptidyl Peptidase 4 Mouse Model. J. Infect. Dis. 2019, 8, 1558–1567. [Google Scholar] [CrossRef]
- Yang, Y.; Du, L. SARS-CoV-2 spike protein: A key target for eliciting persistent neutralizing antibodies. Signal Transduct. Target. Ther. 2021, 6. [Google Scholar] [CrossRef]
- Yu, J.; Tostanoski, L.H.; Peter, L.; Mercado, N.B.; McMahan, K.; Mahrokhian, S.H.; Nkolola, J.P.; Liu, J.; Li, Z.; Chandrashekar, A.; et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020, 369, 806–811. [Google Scholar] [CrossRef]
- Sanchez-Felipe, L.; Vercruysse, T.; Sharma, S.; Ma, J.; Lemmens, V.; Van Looveren, D.; Arkalagud Javarappa, M.P.; Boudewijns, R.; Malengier-Devlies, B.; Liesenborghs, L.; et al. A single-dose live-attenuated YF17D vectored SARS-CoV-2 vaccine candidate. Nature 2021, 590, 320–325. [Google Scholar] [CrossRef]
- Ren, W.; Sun, H.; Gao, G.F.; Chen, J.; Sun, S.; Zhao, R.; Gao, G.; Hu, Y.; Zhao, G.; Chen, Y.; et al. Recombinant SARS-CoV-2 spike S1-Fc fusion protein induced high levels of neutralizing responses in nonhuman primates. Vaccine 2020, 38, 5653–5658. [Google Scholar] [CrossRef]
- He, Y.; Zhou, Y.; Wu, H.; Luo, B.; Chen, J.; Li, W.; Jiang, S. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: Implication for developing SARS diagnostics and vaccines. J. Immunol. 2004, 173, 4050–4057. [Google Scholar] [CrossRef] [PubMed]
- Hotez, P.J.; Bottazzi, M.E.; Tseng, C.T.; Zhan, B.; Lustigman, S.; Du, L.; Jiang, S. Calling for rapid development of a safe and effective MERS vaccine. Microbes Infect. 2014, 16, 529–531. [Google Scholar] [CrossRef] [PubMed]
- Pushparajah, D.; Jimenez, S.; Wong, S.; Alattas, H.; Nafissi, N.; Slavcev, R.A. Advances in gene-based vaccine platforms to address the COVID-19 pandemic. Adv. Drug Deliv. Rev. 2021, 170, 113–141. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Vaccine Tracker and Landscape. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 24 June 2021).
- Mor, G.; Singla, M.; Steinberg, A.D.; Hoffman, S.L.; Okuda, K.; Klinman, D.M. Do DNA vaccines induce autoimmune disease? Hum. Gene Ther. 1997, 8, 293–300. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alluhaybi, K.A.; Alharbi, R.H.; Alhabbab, R.Y.; Aljehani, N.D.; Alamri, S.S.; Basabrain, M.; Alharbi, R.; Abdulaal, W.H.; Alfaleh, M.A.; Tamming, L.; et al. Cellular and Humoral Immunogenicity of a Candidate DNA Vaccine Expressing SARS-CoV-2 Spike Subunit 1. Vaccines 2021, 9, 852. https://doi.org/10.3390/vaccines9080852
Alluhaybi KA, Alharbi RH, Alhabbab RY, Aljehani ND, Alamri SS, Basabrain M, Alharbi R, Abdulaal WH, Alfaleh MA, Tamming L, et al. Cellular and Humoral Immunogenicity of a Candidate DNA Vaccine Expressing SARS-CoV-2 Spike Subunit 1. Vaccines. 2021; 9(8):852. https://doi.org/10.3390/vaccines9080852
Chicago/Turabian StyleAlluhaybi, Khalid A., Rahaf H. Alharbi, Rowa Y. Alhabbab, Najwa D. Aljehani, Sawsan S. Alamri, Mohammad Basabrain, Rehaf Alharbi, Wesam H. Abdulaal, Mohamed A. Alfaleh, Levi Tamming, and et al. 2021. "Cellular and Humoral Immunogenicity of a Candidate DNA Vaccine Expressing SARS-CoV-2 Spike Subunit 1" Vaccines 9, no. 8: 852. https://doi.org/10.3390/vaccines9080852
APA StyleAlluhaybi, K. A., Alharbi, R. H., Alhabbab, R. Y., Aljehani, N. D., Alamri, S. S., Basabrain, M., Alharbi, R., Abdulaal, W. H., Alfaleh, M. A., Tamming, L., Zhang, W., Hassanain, M., Algaissi, A., Abuzenadah, A. M., Li, X., & Hashem, A. M. (2021). Cellular and Humoral Immunogenicity of a Candidate DNA Vaccine Expressing SARS-CoV-2 Spike Subunit 1. Vaccines, 9(8), 852. https://doi.org/10.3390/vaccines9080852






