Genetic Variability of PRRSV Vaccine Strains Used in the National Eradication Programme, Hungary
Abstract
1. Introduction
2. Materials and Methods
2.1. Swine Herds
2.2. Vaccines
2.3. Sample Collection
2.4. Diagnostic Examinations
2.5. Sequence Analysis
3. Results
3.1. Porcilis PRRS Vaccine
3.1.1. Sequence Analysis of ORF5
3.1.2. Sequence Analysis of ORF7
3.2. Unistrain PRRS Vaccine (Former Amervac)
3.2.1. Sequence Analysis of ORF5
3.2.2. Sequence Analysis of ORF7
3.3. ReproCyc PRRS EU Vaccine (and Its Non Adjuvanted Variant Ingelvac PRRSFLEX EU Vaccine)
3.3.1. Sequence Analysis of ORF5
3.3.2. Sequence Analysis of ORF7
4. Discussion
- (1)
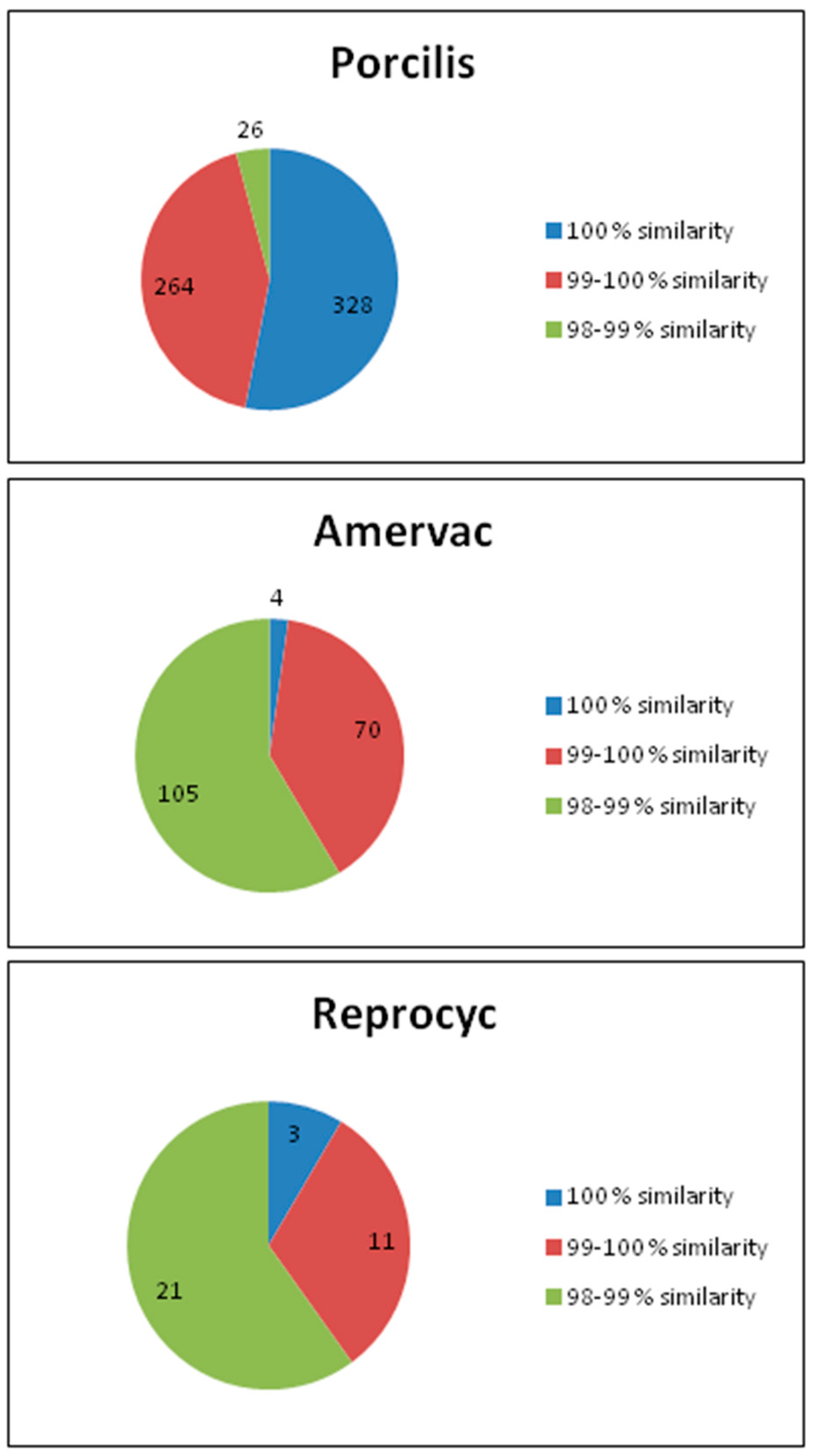
- The ORF5 region of the strain of the Porcilis PRRS vaccine showed that irrespective of the way the vaccine virus entering the body of the pig (vaccination or “infection” with after vaccination shed virus), it is consistently stable and such genetic change does not occur during the period of entry that would interfere with virus vaccine sequencing. The PRRSV strain found in the Porcilis PRRS vaccine does not seem to be prone to significant genetic changes. If the PRRSV ORF5 or ORF7 sequence is at least 98% similar to the Porcilis PRRSV strain, it is safe to say that Porcilis PRRSV strain was detected (Figure 1 and Figure 3).
- (2)
- Comparison of the ORF5 and ORF7 regions of PRRSV with the ORF5 and ORF7 regions of the three live vaccines can be estimated that if they are ≥99% similar, it is likely that the sequence detected in the sample is a consequence of a direct application or indirect spread. However, care should be taken with regard to the ORF7 section of the Porcilis PRRS vaccine strain, which is 100% identical to the ORF7 section of the Lelystad PRRS strain.
- (3)
- Based on sequence analysis, the sequencing of the PRRSV ORF5 section alone is sufficient in most cases to determine the PRRSV sequence identity. With care, useful information is also obtained by analyzing the PRRSV ORF7 sequence.
- (4)
- For spreading the infections among herds, epidemiological investigation is greatly facilitated by the sequencing of PRRSV ORF5 and ORF7. During the epidemiological investigation it is important to know the origin of the herd tested and the knowledge of the immunisation applied in the surrounding farms. Knowledge of vaccine(s) used for immunisation against PRRS over several years in the investigated population is also essential. It is also indispensable to know the age of the sampled animals, the type of production and the time of arrival at the farm. In the eradicating process, when using live attenuated virus vaccines, the PCR positivity is very important to determine whether it is caused by the wild type virus or the vaccine virus.
- (5)
- In relation to sequence distribution along the stages of the eradication programme, as the eradication process approached the final stage, more and more pig farms became free from the disease, and stopped vaccination. Therefore, the number of sequences of vaccine origin decreased. In contrast to the wild-type viruses where viral evolution, and emergence of new variants occurred along the long-term eradication process, only limited farm-specific vaccine sequence evolution was observed.
- (6)
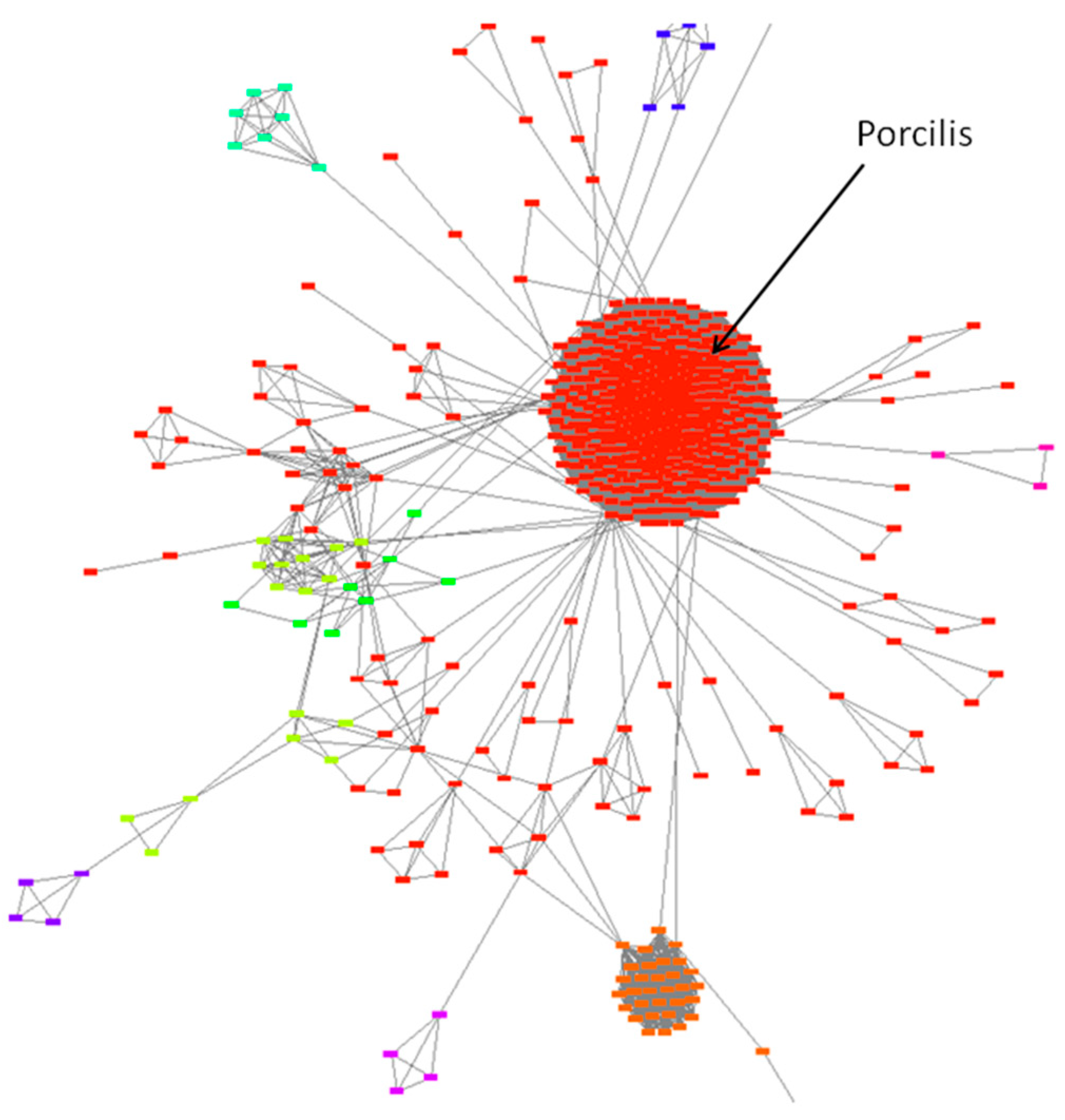
- Several PRRSV ORF5 and ORF7 sequences may be present at the same time in a single herd. Knowledge of a sufficient number of well-characterised PRRSV ORF5 and ORF7 se-quences available over the years is a pre-requisite for an adequately performed epide-miological investigation. It is necessary to evaluate as many PRRSV ORF5 and ORF7 sequences as possible in an infected herd from several animal groups and from differ-ent age groups. The use of a similarity network method can provide reliable and easy-to-understand information for sequencing. In this way, it is possible to determine with sufficient certainty the origin of the infection and to detect any further infections as soon as possible. In similarity network representation, the Porcilis strains are or-ganised into a single node confirming the genetic stability of this vaccine strain. In contrast, the Amervac and Reprocyc strains do not form a node but a dense network, and develop farm-specific minor lineages. These data can serve as useful information that may contribute to further PRRSV vaccine development.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holtkamp, D.; Kliebenstein, J.B.; Zimmerman, J.; Neumann, E.; Rotto, H.; Yoder, T.; Wang, C.; Yeske, P.; Mowrer, C.L.; Haley, C. Economic impact of porcine reproductive and respiratory syndrome virus on U.S. pork producers. Anim. Ind. Rep. 2012, 658, 3–8. [Google Scholar] [CrossRef]
- Nathues, H.; Alarcon, P.; Rushton, J.; Jolie, R.; Fiebig, K.; Jimenez, M.; Geurts, V.; Nathues, C. Cost of porcine reproductive and respiratory syndrome virus at individual farm level—An economic disease model. Prev. Vet. Med. 2017, 142, 16–29. [Google Scholar] [CrossRef]
- Valdes-Donoso, P.; Alvarez, J.; Jarvis, L.S.; Morrison, R.B.; Perez, A.M. Production losses from an endemic animal disease: Porcine reproductive and respiratory syndrome (PRRS) in selected Midwest US sow farms. Front. Vet. Sci. 2018, 5, 102. [Google Scholar] [CrossRef]
- Brinton, M.A.; Gulyaeva, A.; Balasuriya, U.B.R.; Dunowska, M.; Faaberg, K.S.; Goldberg, T.; Leung, F.C.; Nauwynck, H.J.; Snijder, E.J.; Stadejek, T.; et al. Proposal 2017.012S.A.v1. Expansion of the Rank Structure of the Family Arteriviridae and Renaming Its Taxa. 2018. Available online: https://talk.ictvonline.org/ICTV/proposals/20 17.001S.012-017S.R.Nidovirales.zip (accessed on 20 January 2020).
- Keffaber, K.K. Reproductive failure of unknown etiology. Am. Assoc. Swine Pr. Newsl. 1989, 1, 1–9. [Google Scholar]
- Ahl, R.; Pensaert, M.; Robertson, I.B.; Terpstra, C.; Van der Sande, W.; Walker, K.J.; White, M.E.; Meredith, M. Porcine reproductive and respiratory syndrome (PRRS or blue-eared pig disease). Vet. Rec. 1992, 130, 87–89. [Google Scholar] [CrossRef]
- Hornyák, Á.; Pálfi, V.; Karakas, M. Porcine reproductive and respiratory syndrome szerológiai felmérése Magyarországon [Serological investigation of porcine reproductive and respiratory syndrome in Hungary. In Abstract Book of the Academic Reports of the Veterinary Committee of the Hungarian Academy of Sciences; Hungarian Academy of Sciences: Budapest, Hungary, 1997. [Google Scholar]
- Medveczky, I. Áttekintés a sertés reprodukciós zavarokkal és légzőszervi tünetekkel járó szindrómájának (PRRS) újabb ismereteiről [An overview of recent knowledge on porcine reproductive and respiratory syndrome (PRRS)]. Magy. Állatorv. Lapja 1996, 57, 367–371. [Google Scholar]
- Charerntantanakul, W. Porcine reproductive and respiratory syndrome virus vaccines: Immunogenicity, efficacy and safety aspects. World J. Virol. 2012, 1, 23–30. [Google Scholar] [CrossRef]
- Hanada, K.; Suzuki, Y.; Nakane, T.; Hirode, O.; Gojobori, T. The origin and evolution of porcine reproductive and respiratory viruses. Mol. Biol. Evol. 2005, 22, 1024–1031. [Google Scholar] [CrossRef] [PubMed]
- Diaz, I.; Darwich, L.; Pappaterra, G.; Pujols, J.; Mateu, E. Different European-type vaccines against porcine reproductive and respiratory syndrome virus have different immunological properties and confer different protection to pigs. Virology 2006, 351, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Zuckermann, F.A.; Garcia, E.A.; Luque, I.D.; Christopher-Hennings, J.; Doster, A.; Brito, M.; Osorio, F. Assessment of the efficacy of commercial porcine reproductive and respiratory syndrome virus (PRRSV) vaccines based on measurement of serologic response, frequency of gamma-IFN-producing cells and virological parameters of protection upon challenge. Vet. Microbiol. 2007, 123, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Roca, M.; Gimeno, M.; Bruguera, S.; Segales, J.; Diaz, I.; Galindo-Cardiel, I.J.; Martinez, E.; Darwich, L.; Fang, Y.; Maldonado, R.; et al. Effects of challenge with a virulent genotype II strain of porcine reproductive and respiratory syndrome virus on piglets vaccinated with an attenuated genotype I strain vaccine. Vet. J. 2012, 193, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wu, B.; Amer, S.; Luo, J.; Zhang, H.; Guo, Y.; Dong, G.; Zhao, B.; He, H. Phylogenetic analysis and molecular characteristics of seven variant Chinese field isolates of PRRSV. BMC Microbiol. 2010, 10, 146. [Google Scholar] [CrossRef]
- Murtaugh, M.P.; Genzow, M. Immunological solutions for treatment and prevention of porcine reproductive and respiratory syndrome (PRRS). Vaccine 2011, 29, 8192–8204. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Lobo, F.J.; Carrascosa de Lome, L.; Díez-Fuertes, F.; Segalés, J.; García-Artiga, C.; Simarro, I.; Castro, J.M.; Prieto, C. Safety of porcine reproductive and respiratory syndrome modified live virus (MLV) vaccine strains in a young pig infection model. Vet. Res. 2013, 44, 115. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Xiao, Y.; Opriessnig, T.; Ding, Y.; Yu, Y.; Nan, Y.; Ma, Z.; Halbur, P.G.; Zhang, Y.-J. Enhancing neutralizing antibody production by an interferon-inducing porcine reproductive and respiratory syndrome virus strain. Vaccine 2013, 31, 5537–5543. [Google Scholar] [CrossRef] [PubMed]
- Botner, A.; Strandbygaard, B.; Sorensen, K.J.; Have, P.; Madsen, K.G.; Madsen, E.S.; Alexandersen, S. Appearance of acute PRRS-like symptoms in sow herds after vaccination with a modified live PRRS vaccine. Vet. Rec. 1997, 141, 497–499. [Google Scholar] [CrossRef]
- Madsen, K.G.; Hansen, C.M.; Madsen, E.S.; Strandbygaard, B.; Botner, A.; Sorensen, K.J. Sequence analysis of porcine reproductive and respiratory syndrome virus of the American type collected from Danish swine herds. Arch. Virol. 1998, 143, 1683–1700. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Wei, Z.; Zhang, G.; Li, Z.; Ma, J.; Xie, Q.; Sun, B.; Bi, Y. Complete genome sequence of a novel variant porcine reproductive and respiratory syndrome virus (PRRSV) strain: Evidence for recombination between vaccine and wild-type PRRSV strains. J. Virol. 2012, 86, 9543. [Google Scholar] [CrossRef]
- Nielsen, H.S.; Oleksievicz, M.B.; Forsberg, R.; Stadejek, T.; Botner, A.; Storgard, T. Reversion of a live porcine reproductive and respiratory syndrome virus vaccine investigated by parallel mutations. J. Gen. Virol. 2001, 82, 1263–1272. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Szabó, P.M.; Szalay, D.; Kecskeméti, S.; Molnár, T.; Szabó, I.; Bálint, Á. Investigations on spreading of PRRSV among swine herds by improved minimum spanning network analysis. Sci. Rep. 2020, 10, 19217. [Google Scholar] [CrossRef] [PubMed]
- Wensvoort, G. Lelystad virus and the porcine epidemic abortion and respiratory syndrome. Vet. Res. 1993, 24, 117–124. [Google Scholar] [PubMed]
- Ministry of Agriculture. 41/1997. (V. 28.) (Decree of the Minister of Agriculture on issuing the Animal Health Regulation). Available online: https://njt.hu/jogszabaly/1997-41-20-11.49 (accessed on 28 May 1997).
- Szabó, I.; Bognár, L.; Molnár, T.; Nemes, I.; Bálint, Á. PRRS eradication from swine farms in five regions of Hungary. Acta Vet. Hung. 2020, 68, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Bøtner, A.; Strandbygaard, B.; Sørensen, K.J.; Oleksiewicz, M.B.; Storgaard, T. Distinction between infections with European and American/vaccine type PRRS virus after vaccination with a modified-live PRRS virus vaccine. Vet. Res. 2000, 31, 72. [Google Scholar] [CrossRef]


| Vaccine | Marketing Authorisation Holder | % of Applying Farms |
|---|---|---|
| Porcilis | MSD | 29% |
| Progressis * | Ceva-Phylaxia (formerly Merial) | 37% |
| Reprocyc | Boehringer Ingelheim | 9% |
| Amervac, Unistrain | Hipra | 25% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bálint, Á.; Molnár, T.; Kecskeméti, S.; Kulcsár, G.; Soós, T.; Szabó, P.M.; Kaszab, E.; Fornyos, K.; Zádori, Z.; Bányai, K.; et al. Genetic Variability of PRRSV Vaccine Strains Used in the National Eradication Programme, Hungary. Vaccines 2021, 9, 849. https://doi.org/10.3390/vaccines9080849
Bálint Á, Molnár T, Kecskeméti S, Kulcsár G, Soós T, Szabó PM, Kaszab E, Fornyos K, Zádori Z, Bányai K, et al. Genetic Variability of PRRSV Vaccine Strains Used in the National Eradication Programme, Hungary. Vaccines. 2021; 9(8):849. https://doi.org/10.3390/vaccines9080849
Chicago/Turabian StyleBálint, Ádám, Tamás Molnár, Sándor Kecskeméti, Gábor Kulcsár, Tibor Soós, Péter M. Szabó, Eszter Kaszab, Kinga Fornyos, Zoltán Zádori, Krisztián Bányai, and et al. 2021. "Genetic Variability of PRRSV Vaccine Strains Used in the National Eradication Programme, Hungary" Vaccines 9, no. 8: 849. https://doi.org/10.3390/vaccines9080849
APA StyleBálint, Á., Molnár, T., Kecskeméti, S., Kulcsár, G., Soós, T., Szabó, P. M., Kaszab, E., Fornyos, K., Zádori, Z., Bányai, K., & Szabó, I. (2021). Genetic Variability of PRRSV Vaccine Strains Used in the National Eradication Programme, Hungary. Vaccines, 9(8), 849. https://doi.org/10.3390/vaccines9080849







