Oral Immunization of Larvae and Juvenile of Lumpfish (Cyclopterus lumpus) against Vibrio anguillarum Does Not Influence Systemic Immunity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Vibrio anguillarum J360 Culture Conditions
2.2. Bacterin Preparation
2.3. V. anguillarum Bacterin Fluorescent Labeling
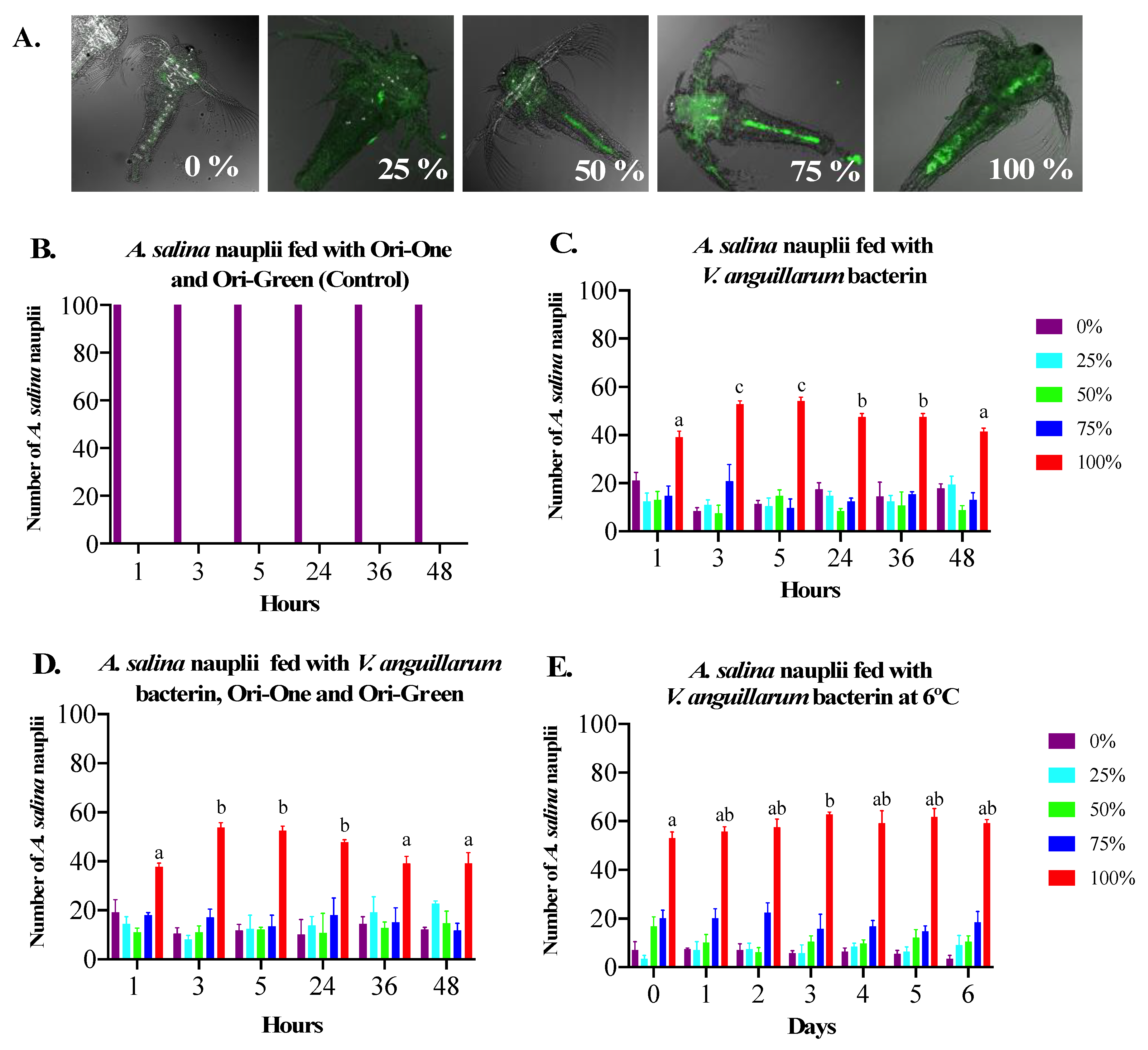
2.4. V. anguillarum Bacterin Bio-Encapsulation in A. salina
2.5. Aquafeed Coating with V. anguillarum Bacterin
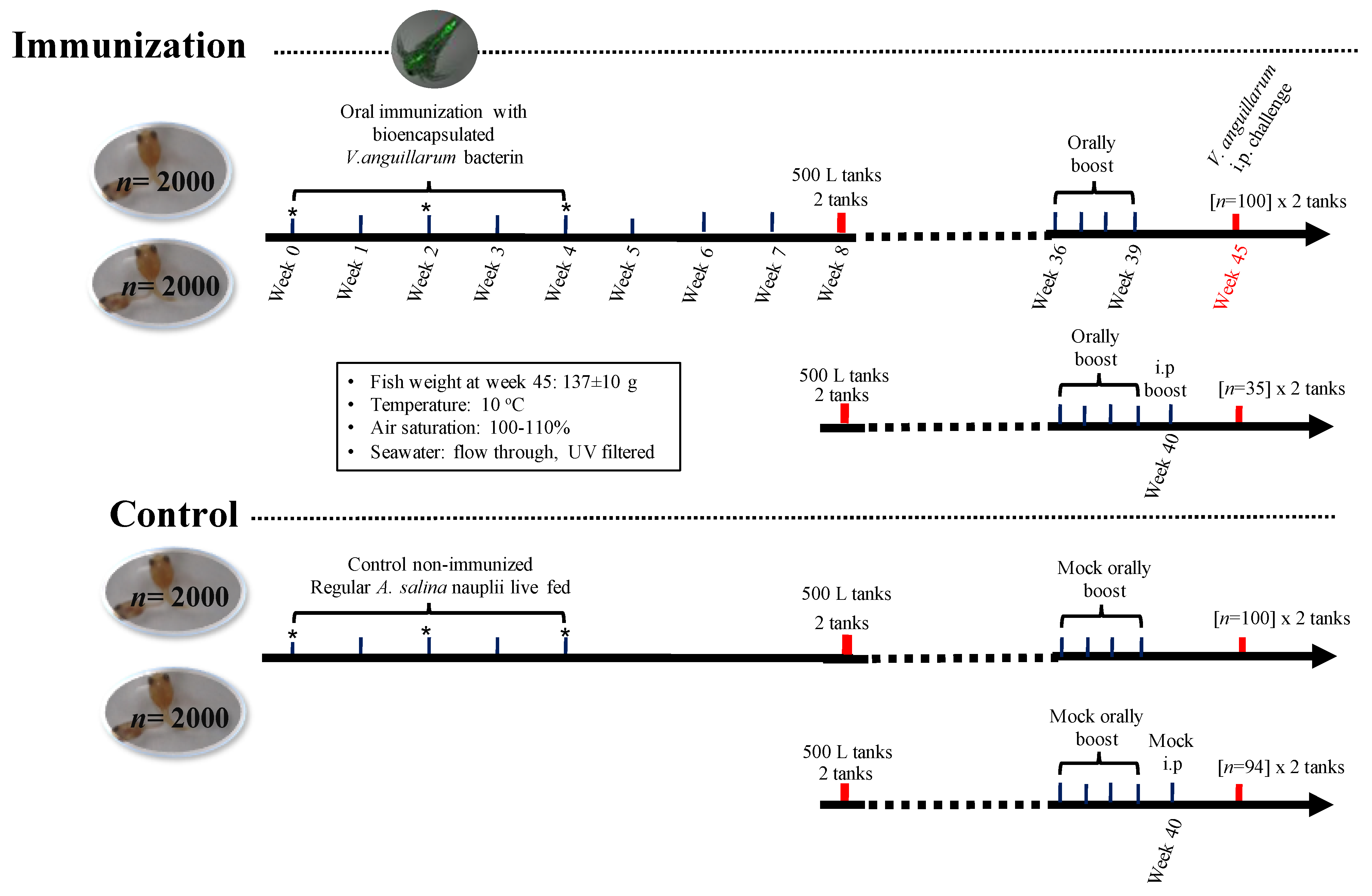
2.6. Fish Culture Conditions
2.7. Lumpfish Immunization Assays
2.8. V. anguillarum J360 Challenge Assays in Lumpfish
2.9. Total RNA Extraction
2.10. cDNA Synthesis and qPCR Parameters
2.11. qPCR Primer Quality Assurance Testing
2.12. Endogenous Control (Normalizer) Selection
2.13. Experimental qPCR Analyses
2.14. Statistical Analysis
3. Results
3.1. Bio-Encapsulation of V. anguillarum Bacterin in A. salina Nauplii
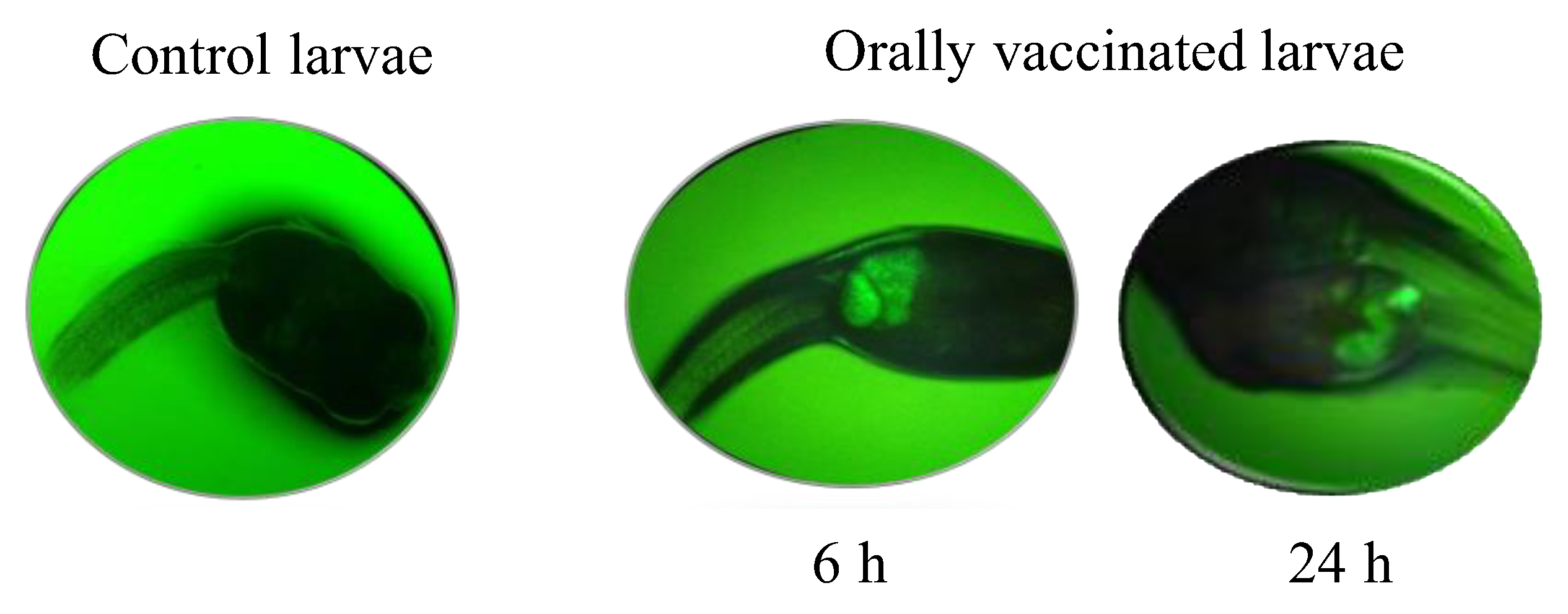
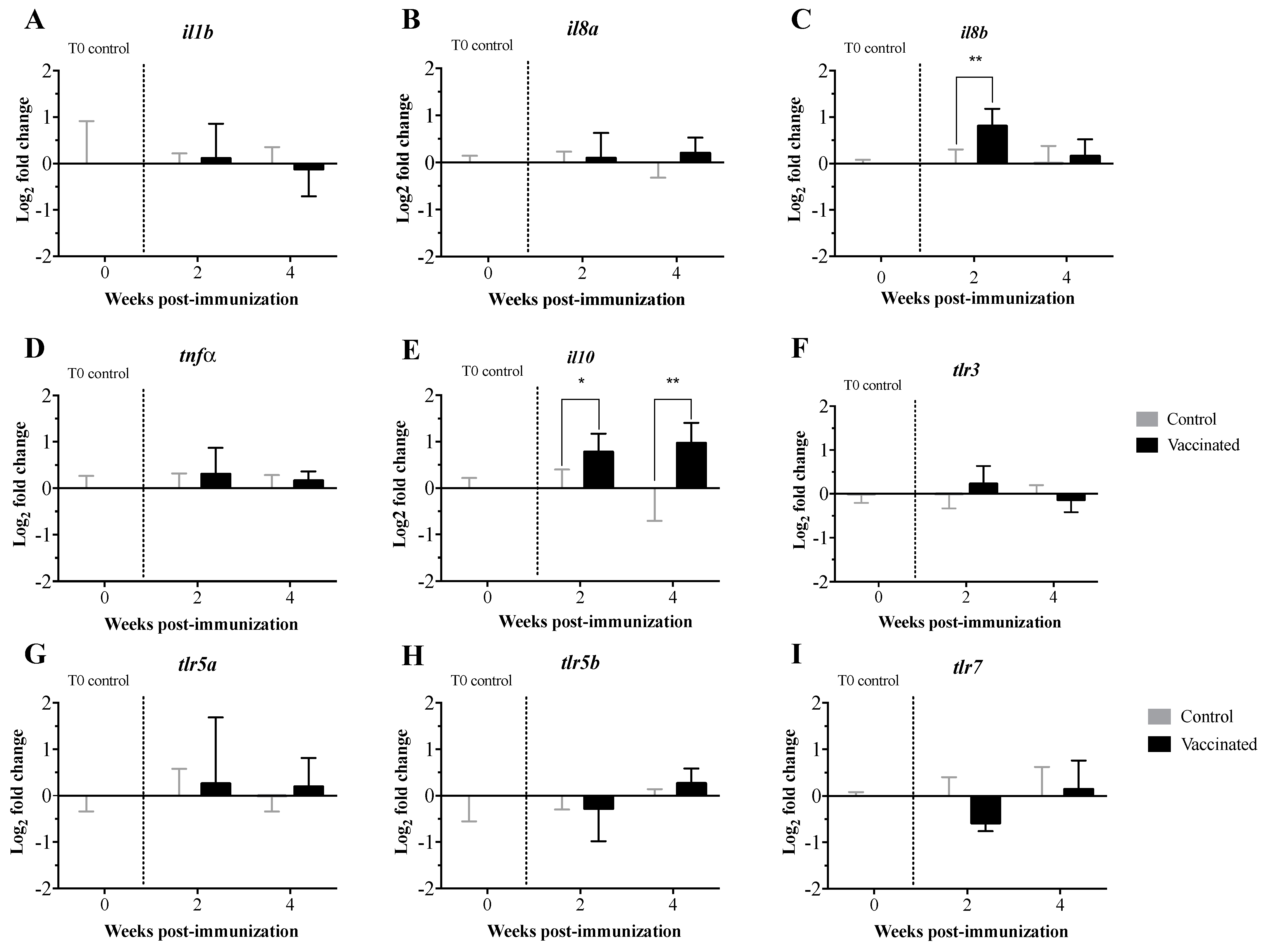
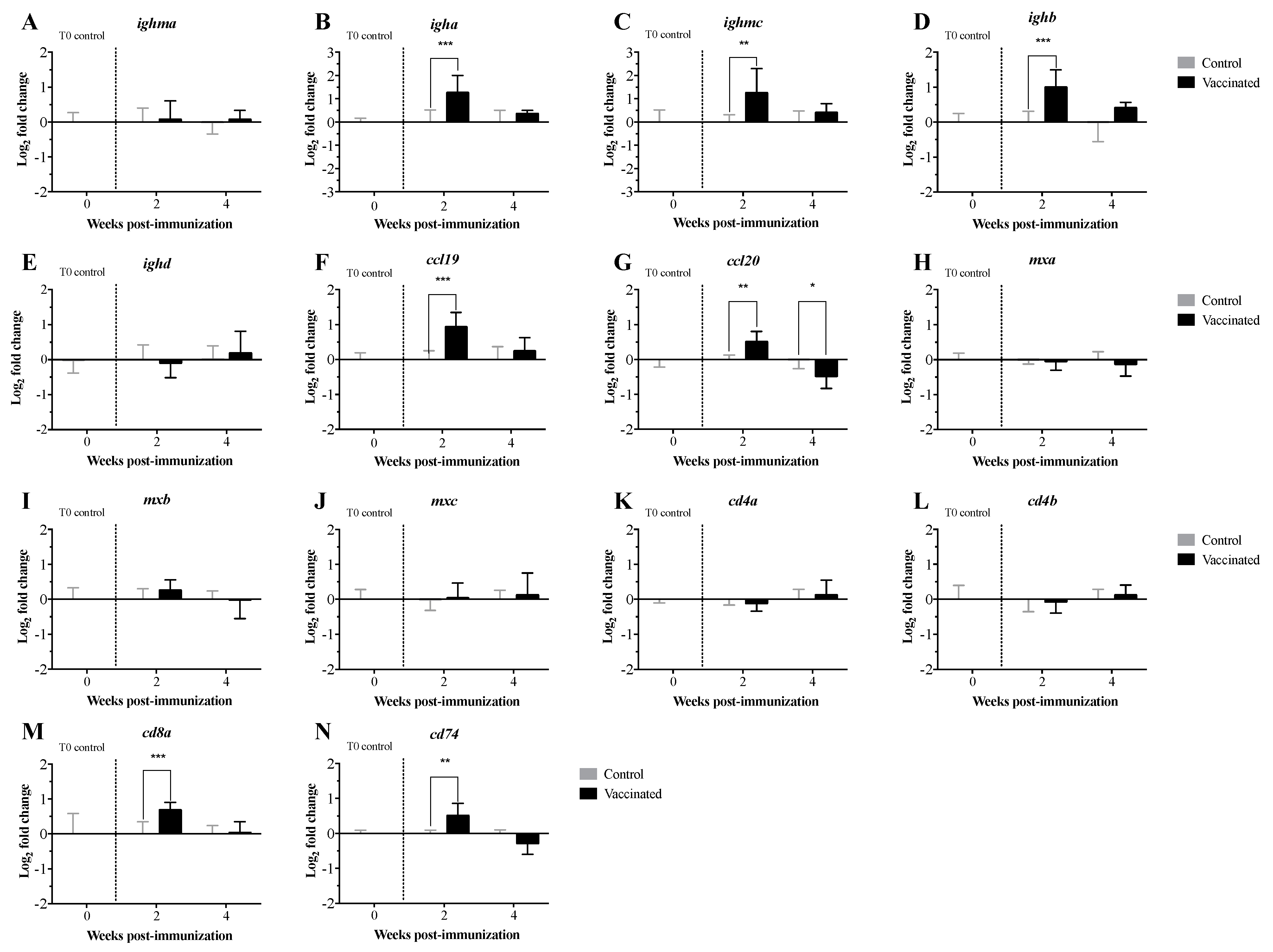
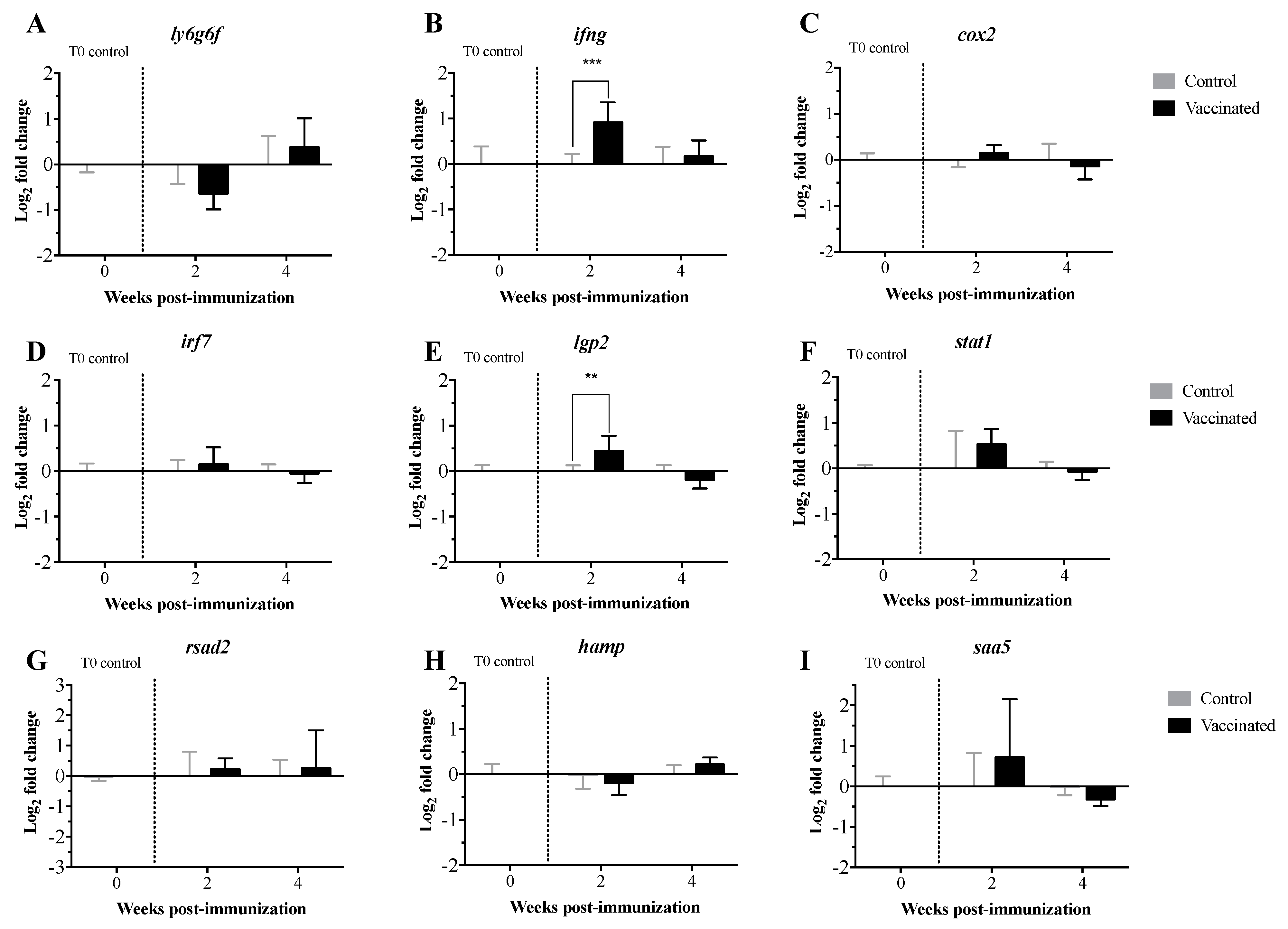
3.2. Transcript Expression Profile of the Immunome of Orally Immunized Lumpfish Larvae
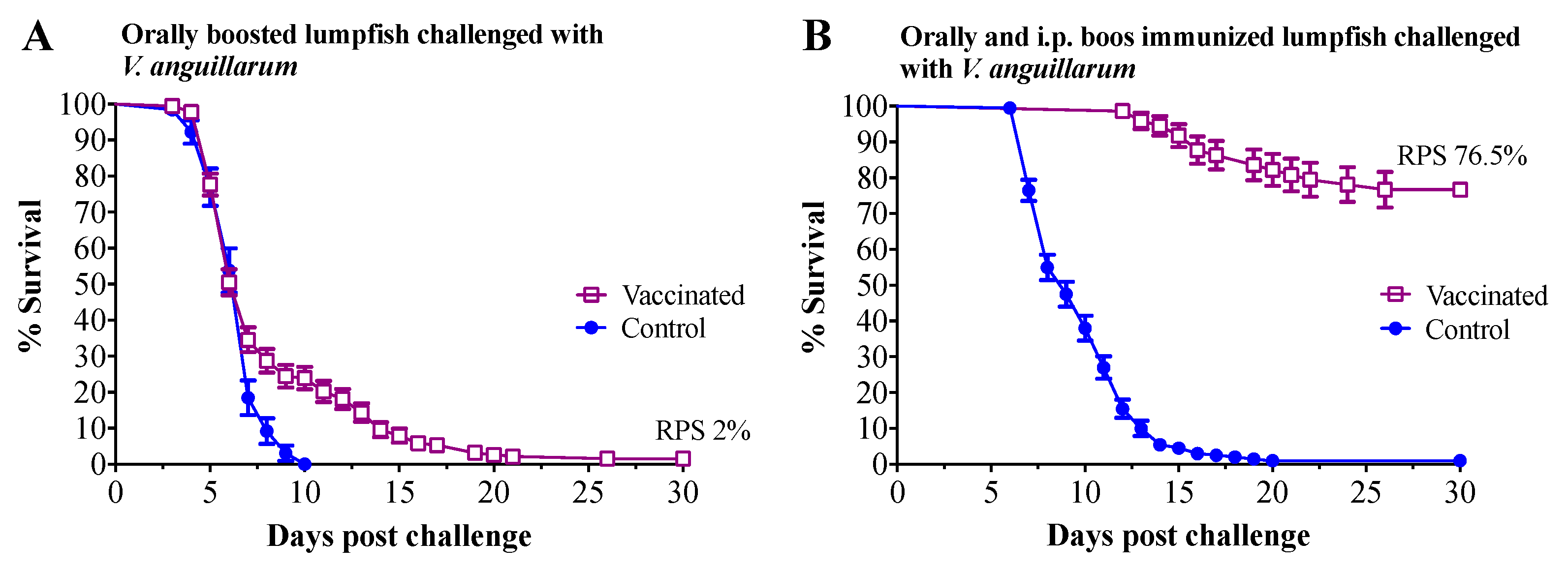
3.3. Vaccine Challenge
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costello, M.J. The global economic cost of sea lice to the salmonid farming industry. J. Fish Dis. 2009, 32, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Fast, M.D. Fish immune responses to parasitic copepod (namely sea lice) infection. Dev. Comp. Immunol. 2014, 43, 300–312. [Google Scholar] [CrossRef]
- Aaen, S.M.; Helgesen, K.O.; Bakke, M.J.; Kaur, K.; Horsberg, T.E. Drug resistance in sea lice: A threat to salmonid aquaculture. Trends Parasitol. 2015, 31, 72–81. [Google Scholar] [CrossRef]
- Overton, K.; Barrett, L.T.; Oppedal, F.; Kristiansen, T.S.; Dempster, T. Sea lice removal by cleaner fish in salmon aquaculture: A review of the evidence base. Aquacult. Environ. Interact. 2020, 12, 31–44. [Google Scholar] [CrossRef] [Green Version]
- Brooks, K.M. Considerations in developing an integrated pest management programme for control of sea lice on farmed salmon in Pacific Canada. J. Fish Dis. 2009, 32, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Brooker, A.J.; Papadopoulou, A.; Gutierrez, C.; Rey, S.; Davie, A.; Migaud, H. Sustainable production and use of cleaner fish for the biological control of sea lice: Recent advances and current challenges. Vet. Rec. 2018, 183, 383. [Google Scholar] [CrossRef] [Green Version]
- Treasurer, J.W. A review of potential pathogens of sea lice and the application of cleaner fish in biological control. Pest Manag. Sci. 2002, 58, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Boyce, D.; Ang, K.P.; Prickett, R. Cunner and lumpfish as cleaner fish species in Canada. In Cleaner Fish Biology and Aquaculture Applications; Treasurer, J.W., Ed.; 5M Publishing Ltd.: Sheffield, UK, 2018. [Google Scholar]
- Umasuthan, N.; Valderrama, K.; Vasquez, I.; Segovia, C.; Hossain, A.; Cao, T.; Gnanagobal, H.; Monk, J.; Boyce, D.; Santander, J. A novel marine pathogen isolated from wild cunners (Tautogolabrus adspersus): Comparative genomics and transcriptome profiling of Pseudomonas sp. strain J380. Microorganisms 2021, 9, 812. [Google Scholar] [CrossRef]
- Monk, J.; Boyce, D.L.; Ang, K.P.; George, S.; Tucker, D.; Jeannot, K.; Fry, J.; Gianasi, B.; Hickey, S.; O’Brien, N. Cleaner fish research and production in Newfoundland. Bull. Aqaucult. Assoc. Can. 2016, 2, 39–45. [Google Scholar]
- Charmley, K.J. Habitat selection by two species of cleaner fishes that may be beneficial in removing sea lice from cultured salmon. SURG J. 2019, 11. [Google Scholar] [CrossRef]
- Marcos-Lopez, M.; Donald, K.; Stagg, H.; McCarthy, U. Clinical Vibrio anguillarum infection in lumpsucker Cyclopterus lumpus in Scotland. Vet. Rec. 2013, 173, 319. [Google Scholar] [CrossRef]
- Imsland, A.K.; Reynolds, P.; Eliassen, G.; Hangstad, T.A.; Foss, A.; Vikingstad, E.; Elvegård, T.A. The use of lumpfish (Cyclopterus lumpus L.) to control sea lice (Lepeophtheirus salmonis krøyer) infestations in intensively farmed atlantic salmon (Salmo salar L.). Aquaculture 2014, 424, 18–23. [Google Scholar] [CrossRef]
- Whittaker, B.A.; Consuegra, S.; Garcia de Leaniz, C. Genetic and phenotypic differentiation of lumpfish (Cyclopterus lumpus) across the North Atlantic: Implications for conservation and aquaculture. PeerJ 2018, 6, e5974. [Google Scholar] [CrossRef] [Green Version]
- Toffan, A.; De Salvador, M.; Scholz, F.; Pretto, T.; Buratin, A.; Rodger, H.D.; Toson, M.; Cuenca, A.; Vendramin, N. Lumpfish (Cyclopterus lumpus, Linnaeus) is susceptible to viral nervous necrosis: Result of an experimental infection with different genotypes of Betanodavirus. J. Fish Dis. 2019, 42, 1667–1676. [Google Scholar] [CrossRef] [PubMed]
- Rueness, E.; Berg, P.R.; Gulla, S.; Halvorsen, K.; Järnegren, J.; Malmstrøm, M.; Mo, T.A.; Rimstad, E.; de Boer, H.; Eldegard, K.; et al. Assessment of The Risk to Norwegian Biodiversity from Import of Wrasses and Other Cleaner Fish for Use in Aquaculture. Norwegian Scientific Committee for Food and Environment (VKM). 2019, pp. 2535–4019. Available online: https://vkm.no/download/18.22fe061816d90c026b4a890d/1570782153468/Assessment%20of%20the%20risk%20to%20Norwegian%20biodiversity%20from%20import%20of%20wrasses%20and%20other%20cleaner%20fish%20for%20use%20in%20aquaculture.pdf (accessed on 15 June 2021).
- Powell, A.; Treasurer, J.W.; Pooley, C.L.; Keay, A.J.; Lloyd, R.; Imsland, A.K.; Garcia de Leaniz, C. Use of lumpfish for sea-lice control in salmon farming:challenges and opportunities. Rev. Aquac. 2017, 10, 1–20. [Google Scholar] [CrossRef] [Green Version]
- NDF Fisheries. Norwegian Directorate for Fisheries. 2019. Available online: https://www.fiskeridir.no/English/Aquaculture/Statistics/Cleanerfish-Lumpfish-and-Wrasse (accessed on 15 June 2021).
- Foss, A.; Imsland, A.K.D.; Roth, B.; Nytrø, A.V. Catch me if you can: How to recapture lumpfish using light as an attractant. Aquac. Eng. 2020, 90, 102074. [Google Scholar] [CrossRef]
- Bolton-Warberg, M. An overview of cleaner fish use in Ireland. J. Fish Dis. 2018, 41, 935–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronneseth, A.; Haugland, G.T.; Colquhoun, D.J.; Brudal, E.; Wergeland, H.I. Protection and antibody reactivity following vaccination of lumpfish (Cyclopterus lumpus L.) against atypical Aeromonas salmonicida. Fish Shellfish Immunol. 2017, 64, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Ellul, R.M.; Bulla, J.; Brudal, E.; Colquhoun, D.; Wergeland, H.; Ronneseth, A. Protection and antibody reactivity in lumpsucker (Cyclopterus lumpus L.) following vaccination against Pasteurella sp. Fish Shellfish Immunol. 2019, 95, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Vasquez, I.; Cao, T.; Chakraborty, S.; Gnanagobal, H.; O’Brien, N.; Monk, J.; Boyce, D.; Westcott, J.D.; Santander, J. Comparative Genomics Analysis of Vibrio anguillarum Isolated from Lumpfish (Cyclopterus lumpus) in Newfoundland Reveal Novel Chromosomal Organizations. Microorganisms 2020, 8, 1666. [Google Scholar] [CrossRef] [PubMed]
- Erkinharju, T.; Dalmo, R.A.; Hansen, M.; Seternes, T. Cleaner fish in aquaculture: Review on diseases and vaccination. Rev. Aquac. 2021, 13, 189–237. [Google Scholar] [CrossRef]
- Frans, I.; Michiels, C.W.; Bossier, P.; Willems, K.A.; Lievens, B.; Rediers, H. Vibrio anguillarum as a fish pathogen: Virulence factors, diagnosis and prevention. J. Fish Dis. 2011, 34, 643–661. [Google Scholar] [CrossRef]
- Holm, K.O.; Nilsson, K.; Hjerde, E.; Willassen, N.P.; Milton, D.L. Complete genome sequence of Vibrio anguillarum strain NB10, a virulent isolate from the Gulf of Bothnia. Stand. Genomic Sci. 2015, 10, 60. Available online: https://www.ncbi.nlm.nih.gov/pubmed/26380645 (accessed on 15 June 2021). [CrossRef] [Green Version]
- Mikkelsen, H.; Lund, V.; Larsen, R.; Seppola, M. Vibriosis vaccines based on various sero-subgroups of Vibrio anguillarum O2 induce specific protection in Atlantic cod (Gadus morhua L.) juveniles. Fish Shellfish Immunol. 2011, 30, 330–339. [Google Scholar] [CrossRef]
- Chakraborty, S.; Cao, T.; Hossain, A.; Gnanagobal, H.; Vasquez, I.; Boyce, D.; Santander, J. Vibrogen-2 vaccine trial in lumpfish (Cyclopterus lumpus) against Vibrio anguillarum. J. Fish Dis. 2019, 42, 1057–1064. [Google Scholar] [CrossRef]
- Dadar, M.; Chakraborty, S.; Dhama, K.; Prasad, M.; Khandia, R.; Hassan, S.; Munjal, A.; Tiwari, R.; Karthik, K.; Kumar, D.; et al. Advances in designing and developing vaccines, drugs and therapeutic Aapproaches to counter human papilloma virus. Front. Immunol. 2018, 9, 2478. [Google Scholar] [CrossRef]
- Plant, K.P.; Lapatra, S.E. Advances in fish vaccine delivery. Dev. Comp. Immunol. 2011, 35, 1256–1262. [Google Scholar] [CrossRef] [PubMed]
- Mutoloki, S.; Munangandu, H.M.; Evensen, O. Oral Vaccination of fish-antigen preparations, uptake, and immune induction. Front. Immunol. 2015, 6, 519. [Google Scholar] [CrossRef] [Green Version]
- Campbell, R.; Adams, A.; Tatner, M.F.; Chair, M.; Sorgeloos, P. Uptake of Vibrio anguillarum vaccine by artemia salina as a potential oral delivery system to fish fry. Fish Shellfish Immunol. 1993, 3, 451–459. [Google Scholar] [CrossRef]
- Lin, C.C.; Lin, J.H.Y.; Chen, M.S.; Yang, H.L. An oral nervous necrosis virus vaccine that induces protective immunity in larvae of grouper (Epinephelus coioides). Aquaculture 2007, 268, 265–273. [Google Scholar] [CrossRef]
- Chen, Y.M.; Shih, C.H.; Liu, H.C.; Wu, C.L.; Lin, C.C.; Wang, H.C.; Chen, T.Y.; Yang, H.L.; Lin, J.H.Y. An oral nervous necrosis virus vaccine using Vibrio anguillarum as an expression host provides early protection. Aquaculture 2011, 321, 26–33. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, W. Molecular Cloning; A Laboratory Manual, 3rd ed.; Cold Spring Harbor Press: New York, NY, USA, 2001. [Google Scholar]
- Leboffe, M.J.; Pierce, B.E. Microbiology: Laboratory Theory & Application; Morton Publishing: Englewood, CO, USA, 2019. [Google Scholar]
- Eslamloo, K.; Kumar, S.; Caballero-Solares, A.; Gnanagobal, H.; Santander, J.; Rise, M.L. Profiling the transcriptome response of Atlantic salmon head kidney to formalin-killed Renibacterium salmoninarum. Fish Shellfish Immunol. 2020, 98, 937–949. [Google Scholar] [CrossRef] [PubMed]
- Santander, J.; Golden, G.; Wanda, S.Y.; Curtiss, R., 3rd. Fur-regulated iron uptake system of Edwardsiella ictaluri and its influence on pathogenesis and immunogenicity in the catfish host. Infect. Immun. 2012, 80, 2689–2703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasquez, I.; Cao, T.; Hossain, A.; Valderrama, K.; Gnanagobal, H.; Dang, M.; Leeuwis, R.H.J.; Ness, M.; Campbell, B.; Gendron, R.; et al. Aeromonas salmonicida infection kinetics and protective immune response to vaccination in sablefish (Anoplopoma fimbria). Fish Shellfish Immunol. 2020, 104, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Valderrama, K.; Soto-Davila, M.; Segovia, C.; Vasquez, I.; Dang, M.; Santander, J. Aeromonas salmonicida infects Atlantic salmon (Salmo salar) erythrocytes. J. Fish Dis. 2019, 42, 1601–1608. [Google Scholar] [CrossRef] [PubMed]
- Wessman, P.; Hakansson, S.; Leifer, K.; Rubino, S. Formulations for freeze-drying of bacteria and their influence on cell survival. J. Vis. Exp. 2013. [Google Scholar] [CrossRef] [Green Version]
- Fissell, W.H.; Manley, S.; Dubnisheva, A.; Glass, J.; Magistrelli, J.E.A.N.; Fleischman, A.J.; Zydney, A.L.; Roy, S. Ficoll is not a rigid sphere. Am. J. Physiol. Renal Physiol. 2007, 293, F1209–F1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inman, J.K. Thymus-independent antigens: The preparation of covalent, hapten-ficoll conjugates. J. Immunol. 1975, 114 Pt 1, 704–709. [Google Scholar]
- Amlot, P.L.; Hayes, A.E.; Gray, D.; Gordon-Smith, E.C.; Humphrey, J.H. Human immune responses in vivo to protein (KLH) and polysaccharide (DNP-Ficoll) neoantigens: Normal subjects compared with bone marrow transplant patients on cyclosporine. Clin. Exp. Immunol. 1986, 64, 125–135. [Google Scholar]
- Anderson, D.P.; Merchant, B.; Dixon, O.W.; Schott, C.F.; Lizzio, E.F. Flush exposure and injection immunization of rainbow trout to selected DNP conjugates. Dev. Comp. Immunol. 1983, 7, 261–268. [Google Scholar] [CrossRef]
- Milley, B.; Kiwan, R.; Ott, G.S.; Calacsan, C.; Kachura, M.; Campbell, J.D.; Kanzler, H.; Coffman, R.L. Optimization, Production, and Characterization of a CpG-Oligonucleotide-Ficoll Conjugate Nanoparticle Adjuvant for Enhanced Immunogenicity of Anthrax Protective Antigen. Bioconjug. Chem. 2016, 27, 1293–1304. [Google Scholar] [CrossRef]
- Amend, D.F. Potency Testing of Fish Vaccines. Fish Biologics: Serodiagnostics and Vaccines. 1981, Volume 49, pp. 447–454. Available online: https://www.scirp.org/(S(vtj3fa45qm1ean45vvffcz55))/reference/ReferencesPapers.aspx?ReferenceID=1919013 (accessed on 15 June 2021).
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Soto-Davila, M.; Hossain, A.; Chakraborty, S.; Rise, M.L.; Santander, J. Aeromonas salmonicida subsp. salmonicida early infection and immune response of Atlantic cod (Gadus morhua L.) primary macrophages. Front. Immunol. 2019, 10, 1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villumsen, K.R.; Neumann, L.; Ohtani, M.; Strom, H.K.; Raida, M.K. Oral and anal vaccination confers full protection against enteric redmouth disease (ERM) in rainbow trout. PLoS ONE 2014, 9, e93845. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.H.; Yu, C.C.; Lin, C.C.; Yang, H.L. An oral delivery system for recombinant subunit vaccine to fish. Dev. Biol 2005, 121, 175–180. [Google Scholar]
- Min, W.; Lillehoj, H.S.; Burnside, J.; Weining, K.C.; Staeheli, P.; Zhu, J.J. Adjuvant effects of IL-1beta, IL-2, IL-8, IL-15, IFN-alpha, IFN-gamma TGF-beta4 and lymphotactin on DNA vaccination against Eimeria acervulina. Vaccine 2001, 20, 267–274. [Google Scholar] [CrossRef]
- Xu, C.; Guo, T.C.; Mutoloki, S.; Haugland, O.; Evensen, O. Gene expression studies of host response to Salmonid alphavirus subtype 3 experimental infections in Atlantic salmon. Vet. Res. 2012, 43, 78. [Google Scholar] [CrossRef] [Green Version]
- Kumari, J.; Bogwald, J.; Dalmo, R.A. Vaccination of Atlantic salmon, Salmo salar L.; with Aeromonas salmonicida and infectious pancreatic necrosis virus (IPNV) showed a mixed Th1/Th2/Treg response. J. Fish Dis. 2013, 36, 881–886. [Google Scholar] [CrossRef]
- Ohtani, M.; Hikima, J.; Kondo, H.; Hirono, I.; Jung, T.S.; Aoki, T. Evolutional conservation of molecular structure and antiviral function of a viral RNA receptor, LGP2, in Japanese flounder, Paralichthys olivaceus. J. Immunol. 2010, 185, 7507–7517. [Google Scholar] [CrossRef] [Green Version]
- Chang, M.; Collet, B.; Nie, P.; Lester, K.; Campbell, S.; Secombes, C.J.; Zou, J. Expression and functional characterization of the RIG-I-like receptors MDA5 and LGP2 in Rainbow trout (Oncorhynchus mykiss). J. Virol. 2011, 85, 8403–8412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Veen, A.G.; Maillard, P.V.; Schmidt, J.M.; Lee, S.A.; Deddouche-Grass, S.; Borg, A.; Kjaer, S.; Snijders, A.P.; Reis e Sousa, C. The RIG-I-like receptor LGP2 inhibits Dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. EMBO J. 2018, 37. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.M.; Zhao, X.; Li, Z.; Wu, M.; Gui, J.F.; Zhang, Y.B. Alternative splicing transcripts of Zebrafish LGP2 Ggene Ddifferentially contribute to IFN antiviral response. J. Immunol. 2018, 200, 688–703. [Google Scholar] [CrossRef]
- Arancibia, S.A.; Beltran, C.J.; Aguirre, I.M.; Silva, P.; Peralta, A.L.; Malinarich, F.; Hermoso, M.A. Toll-like receptors are key participants in innate immune responses. Biol. Res. 2007, 40, 97–112. [Google Scholar] [CrossRef] [Green Version]
- Jayaramu, P.K.; Tripathi, G.; Pavan Kumar, A.; Keezhedath, J.; Pathan, M.K.; Kurcheti, P.P. Studies on expression pattern of toll-like receptor 5 (TLR5) in Edwardsiella tarda infected Pangasianodon hypophthalmus. Fish Shellfish Immunol. 2017, 63, 68–73. [Google Scholar] [CrossRef]
- Ji, J.; Rao, Y.; Wan, Q.; Liao, Z.; Su, J. Teleost-Specific TLR19 localizes to endosome, recognizes dsRNA, recruits TRIF, triggers both IFN and NF-kappaB pathways, and protects cells from grass carp reovirus infection. J. Immunol. 2018, 200, 573–585. [Google Scholar] [CrossRef]
- Barton, G.M.; Medzhitov, R. Control of adaptive immune responses by Toll-like receptors. Curr. Opin. Immunol. 2002, 14, 380–383. [Google Scholar] [CrossRef]
- Eggestol, H.O.; Lunde, H.S.; Ronneseth, A.; Fredman, D.; Petersen, K.; Mishra, C.K.; Furmanek, T.; Colquhoun, D.J.; Wergeland, H.I.; Haugland, G.T. Transcriptome-wide mapping of signaling pathways and early immune responses in lumpfish leukocytes upon in vitro bacterial exposure. Sci. Rep. 2018, 8, 5261. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, G. Emerging role of lymphocyte Aantigen-6 family of genes in cancer and immune cells. Front. Immunol. 2019, 10, 819. [Google Scholar] [CrossRef]
- MacNeil, I.; Kennedy, J.; Godfrey, D.I.; Jenkins, N.A.; Masciantonio, M.; Mineo, C.; Gilbert, D.J.; Copeland, N.G.; Boyd, R.L.; Zlotnik, A. Isolation of a cDNA encoding thymic shared antigen-1. A new member of the Ly6 family with a possible role in T cell development. J. Immunol. 1993, 151, 6913–6923. [Google Scholar] [PubMed]
- Mallya, M.; Campbell, R.D.; Aguado, B. Characterization of the five novel Ly-6 superfamily members encoded in the MHC, and detection of cells expressing their potential ligands. Protein Sci. 2006, 15, 2244–2256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loughner, C.L.; Bruford, E.A.; McAndrews, M.S.; Delp, E.E.; Swamynathan, S.; Swamynathan, S.K. Organization, evolution and functions of the human and mouse Ly6/uPAR family genes. Hum. Genom. 2016, 10, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danilova, N.; Bussmann, J.; Jekosch, K.; Steiner, L.A. The immunoglobulin heavy-chain locus in zebrafish: Identification and expression of a previously unknown isotype, immunoglobulin Z. Nat. Immunol. 2005, 6, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Savan, R.; Aman, A.; Sato, K.; Yamaguchi, R.; Sakai, M. Discovery of a new class of immunoglobulin heavy chain from fugu. Eur. J. Immunol. 2005, 35, 3320–3331. [Google Scholar] [CrossRef]
- Yasuike, M.; de Boer, J.; von Schalburg, K.R.; Cooper, G.A.; McKinnel, L.; Messmer, A.; So, S.; Davidson, W.S.; Koop, B.F. Evolution of duplicated IgH loci in Atlantic salmon, Salmo salar. BMC Genom. 2010, 11, 486. [Google Scholar] [CrossRef] [Green Version]
- Schroeder, H.W.; Cavacini, L., Jr. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 2010, 125 (Suppl. 2), S41–S52. [Google Scholar] [CrossRef] [Green Version]
- Bromley, S.K.; Mempel, T.R.; Luster, A.D. Orchestrating the orchestrators: Chemokines in control of T cell traffic. Nat. Immunol. 2008, 9, 970–980. [Google Scholar] [CrossRef]
- Yan, Y.; Chen, R.; Wang, X.; Hu, K.; Huang, L.; Lu, M.; Hu, Q. CCL19 and CCR7 expression, signaling pathways, and adjuvant functions in viral Iinfection and prevention. Front. Cell Dev. Biol. 2019, 7, 212. [Google Scholar] [CrossRef]
- Liu, F.; Wang, T.; Hu, Y.; Tian, G.; Secombes, C.J.; Wang, T. Expansion of fish CCL20_like chemokines by genome and local gene duplication: Characterisation and expression analysis of 10 CCL20_like chemokines in rainbow trout (Oncorhynchus mykiss). Dev. Comp. Immunol. 2020, 103, 103502. [Google Scholar] [CrossRef]
- Buonocore, F.; Randelli, E.; Bird, S.; Secombes, C.J.; Costantini, S.; Facchiano, A.; Mazzini, M.; Scapigliati, G. The CD8alpha from sea bass (Dicentrarchus labrax L.): Cloning, expression and 3D modelling. Fish Shellfish Immunol. 2006, 20, 637–646. [Google Scholar] [CrossRef]
- Moldenhauer, G.; Henne, C.; Karhausen, J.; Moller, P. Surface-expressed invariant chain (CD74) is required for internalization of human leucocyte antigen-DR molecules to early endosomal compartments. Immunology 1999, 96, 473–484. [Google Scholar] [CrossRef]
- Beswick, E.J.; Reyes, V.E. CD74 in antigen presentation, inflammation, and cancers of the gastrointestinal tract. World, J. Gastroenterol. 2009, 15, 2855–2861. [Google Scholar] [CrossRef]
- Star, B.; Nederbragt, A.J.; Jentoft, S.; Grimholt, U.; Malmstrom, M.; Gregers, T.F.; Rounge, T.B.; Paulsen, J.; Solbakken, M.H.; Sharma, A.; et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature 2011, 477, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Trowsdale, J.; Knight, J.C. Major histocompatibility complex genomics and human disease. Annu. Rev. Genom. Hum. Genet. 2013, 14, 301–323. [Google Scholar] [CrossRef] [Green Version]
- Dijkstra, J.M.; Grimholt, U. Major histocompatibility complex (MHC) fragment numbers alone-in Atlantic cod and in general-do not represent functional variability. F1000Research 2018, 7, 963. [Google Scholar] [CrossRef] [PubMed]
- Dubin, A.; Jorgensen, T.E.; Moum, T.; Johansen, S.D.; Jakt, L.M. Complete loss of the MHC II pathway in an anglerfish, Lophius piscatorius. Biol. Lett. 2019, 15, 20190594. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.A. The development of feeding behaviour in the lumpfish, Cyclopterus lumpus. J. Fish Biol. 1986, 29, 171–178. [Google Scholar] [CrossRef]
- Imsland, A.K.D.; Danielsen, M.; Jonassend, T.M.; Hangstadd, T.A.; Falk-Petersenc, I.B. Effect of incubation temperature on eggs and larvae of lumpfish (Cyclopterus lumpus). Aquaculture 2019, 498, 217–222. [Google Scholar] [CrossRef]
- Galeotti, M.; Romano, N.; Volpatti, D.; Bulfon, C.; Brunetti, A.; Tiscar, P.G.; Mosca, F.; Bertoni, F.; Marchetti, M.G.; Abelli, L. Innovative vaccination protocol against vibriosis in Dicentrarchus labrax (L.) juveniles: Improvement of immune parameters and protection to challenge. Vaccine 2013, 31, 1224–1230. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.A.; Flynn, J.K.; Amend, D.F. Duration of immunity in salmonids vaccinated by direct immersion with Yersinia ruckeri and Vibrio anguillarum bacterins. J. Fish Dis. 1983, 5, 207–213. [Google Scholar] [CrossRef]
- Johnson, K.A.; Amend, D.F. Comparison of efficacy of several delivery methods using Yersinia ruckeri bacterin on rainbow trout, Salmo gairdneri Richardson. J. Fish Dis. 1983, 6, 331–336. [Google Scholar] [CrossRef]
- Chettri, J.K.; Jaafar, R.M.; Skov, J.; Kania, P.W.; Dalsgaard, I.; Buchmann, K. Booster immersion vaccination using diluted Yersinia ruckeri bacterin confers protection against ERM in rainbow trout. Aquaculture 2015, 440, 1–5. [Google Scholar] [CrossRef]
- Johnson, K.A.; Amend, D.F. Efficacy of Vibrio anguillarum and Yersinia ruckeri bacterins applied by oral and anal intubation of salmonids. J. Fish Dis. 1983, 6, 473–476. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dang, M.; Cao, T.; Vasquez, I.; Hossain, A.; Gnanagobal, H.; Kumar, S.; Hall, J.R.; Monk, J.; Boyce, D.; Westcott, J.; et al. Oral Immunization of Larvae and Juvenile of Lumpfish (Cyclopterus lumpus) against Vibrio anguillarum Does Not Influence Systemic Immunity. Vaccines 2021, 9, 819. https://doi.org/10.3390/vaccines9080819
Dang M, Cao T, Vasquez I, Hossain A, Gnanagobal H, Kumar S, Hall JR, Monk J, Boyce D, Westcott J, et al. Oral Immunization of Larvae and Juvenile of Lumpfish (Cyclopterus lumpus) against Vibrio anguillarum Does Not Influence Systemic Immunity. Vaccines. 2021; 9(8):819. https://doi.org/10.3390/vaccines9080819
Chicago/Turabian StyleDang, My, Trung Cao, Ignacio Vasquez, Ahmed Hossain, Hajarooba Gnanagobal, Surendra Kumar, Jennifer R. Hall, Jennifer Monk, Danny Boyce, Jillian Westcott, and et al. 2021. "Oral Immunization of Larvae and Juvenile of Lumpfish (Cyclopterus lumpus) against Vibrio anguillarum Does Not Influence Systemic Immunity" Vaccines 9, no. 8: 819. https://doi.org/10.3390/vaccines9080819
APA StyleDang, M., Cao, T., Vasquez, I., Hossain, A., Gnanagobal, H., Kumar, S., Hall, J. R., Monk, J., Boyce, D., Westcott, J., & Santander, J. (2021). Oral Immunization of Larvae and Juvenile of Lumpfish (Cyclopterus lumpus) against Vibrio anguillarum Does Not Influence Systemic Immunity. Vaccines, 9(8), 819. https://doi.org/10.3390/vaccines9080819







