Bioinformatics Prediction of SARS-CoV-2 Epitopes as Vaccine Candidates for the Colombian Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
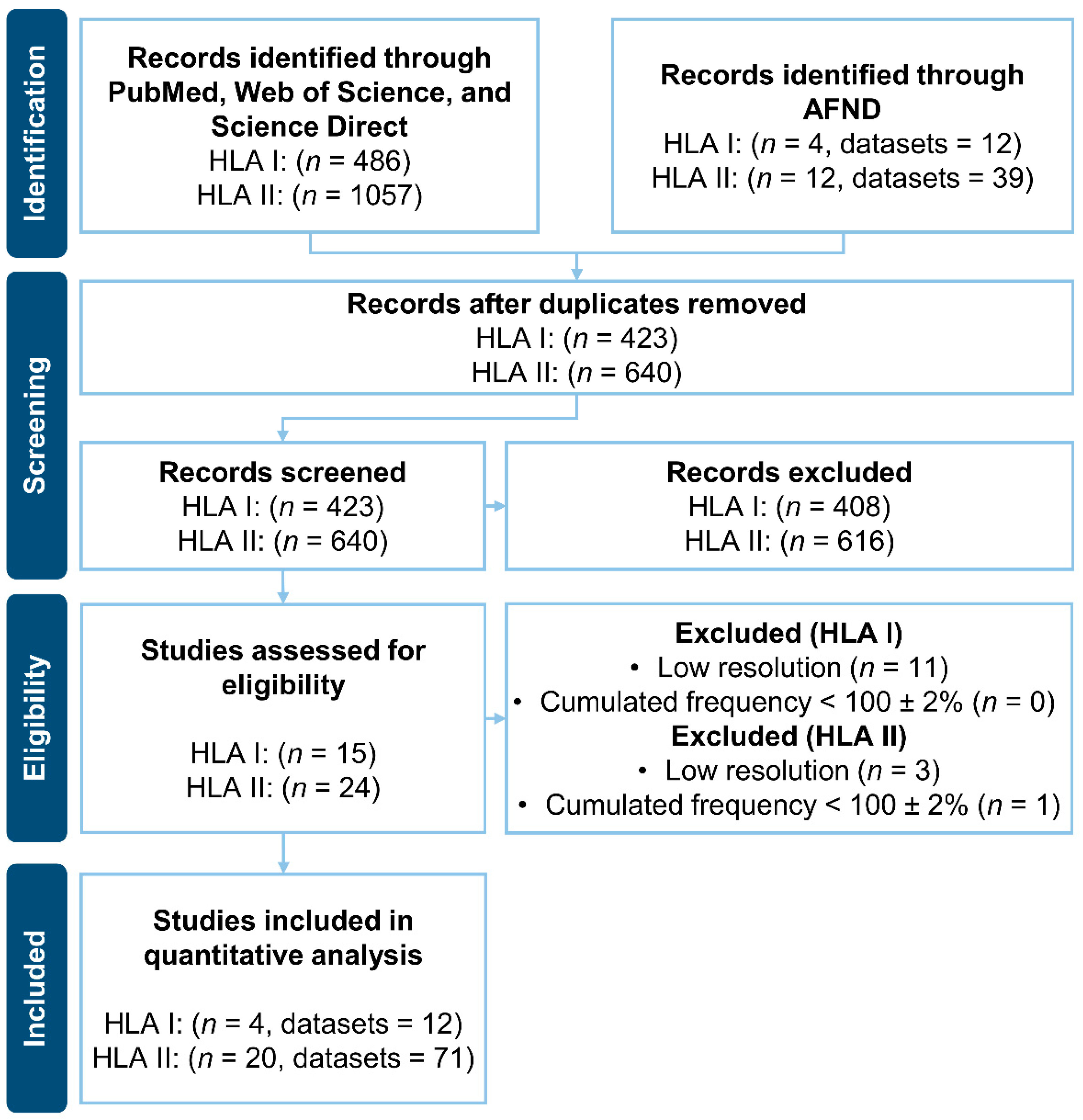
2.2. Literature Search of HLAs Frequencies
2.3. HLAs Selection for Epitope Prediction
2.4. T-Cell Epitope Prediction
2.5. Peptide-Protein Docking Studies
2.6. Interactions Analysis and Molecular Dynamics
3. Results
3.1. Literature Search of HLAs Frequencies
3.2. HLAs Selection for Epitope Prediction
3.2.1. HLA I
3.2.2. HLA II
3.3. T-Cell Epitope Prediction
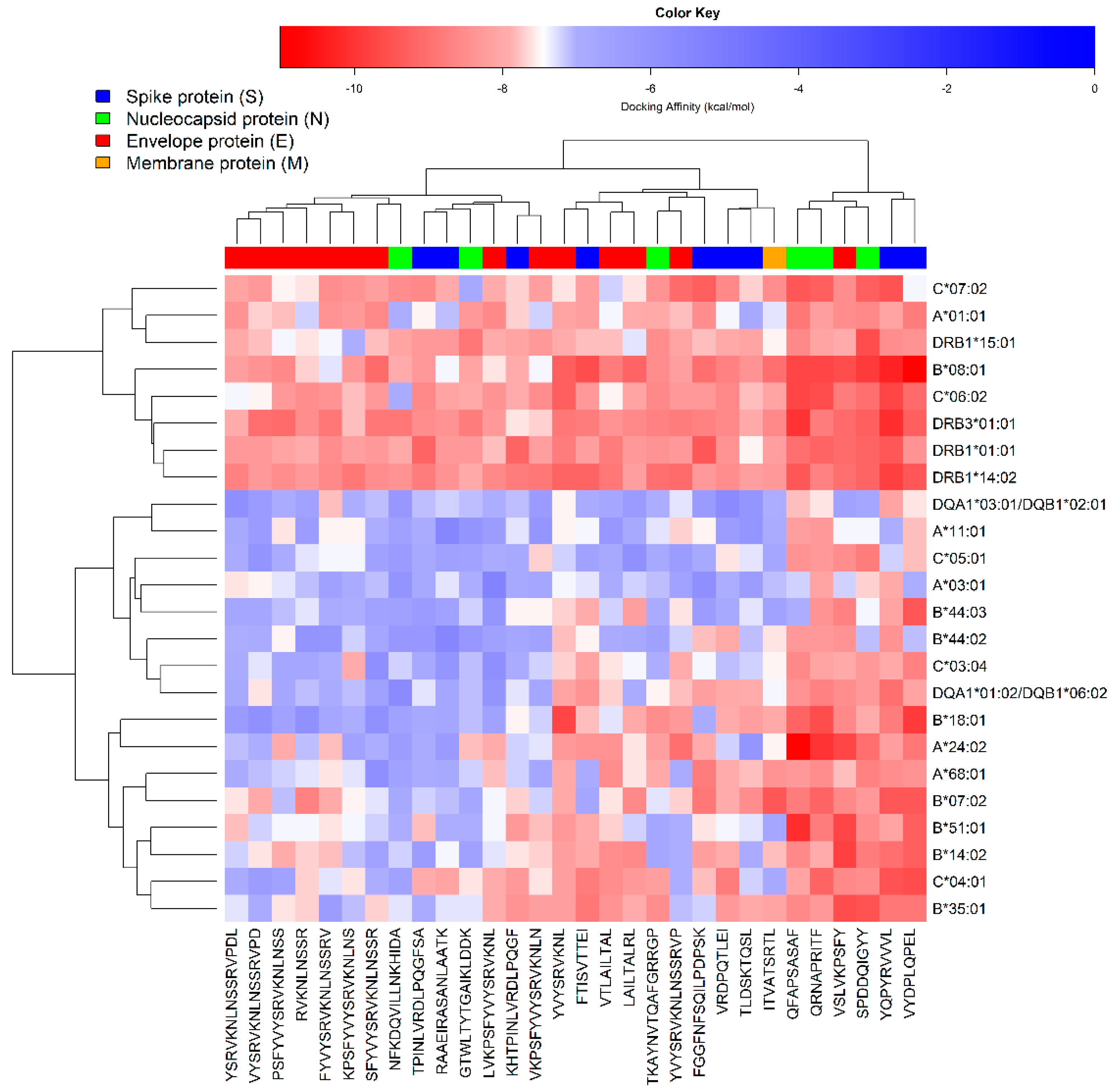
3.4. Peptide-Protein Docking Studies
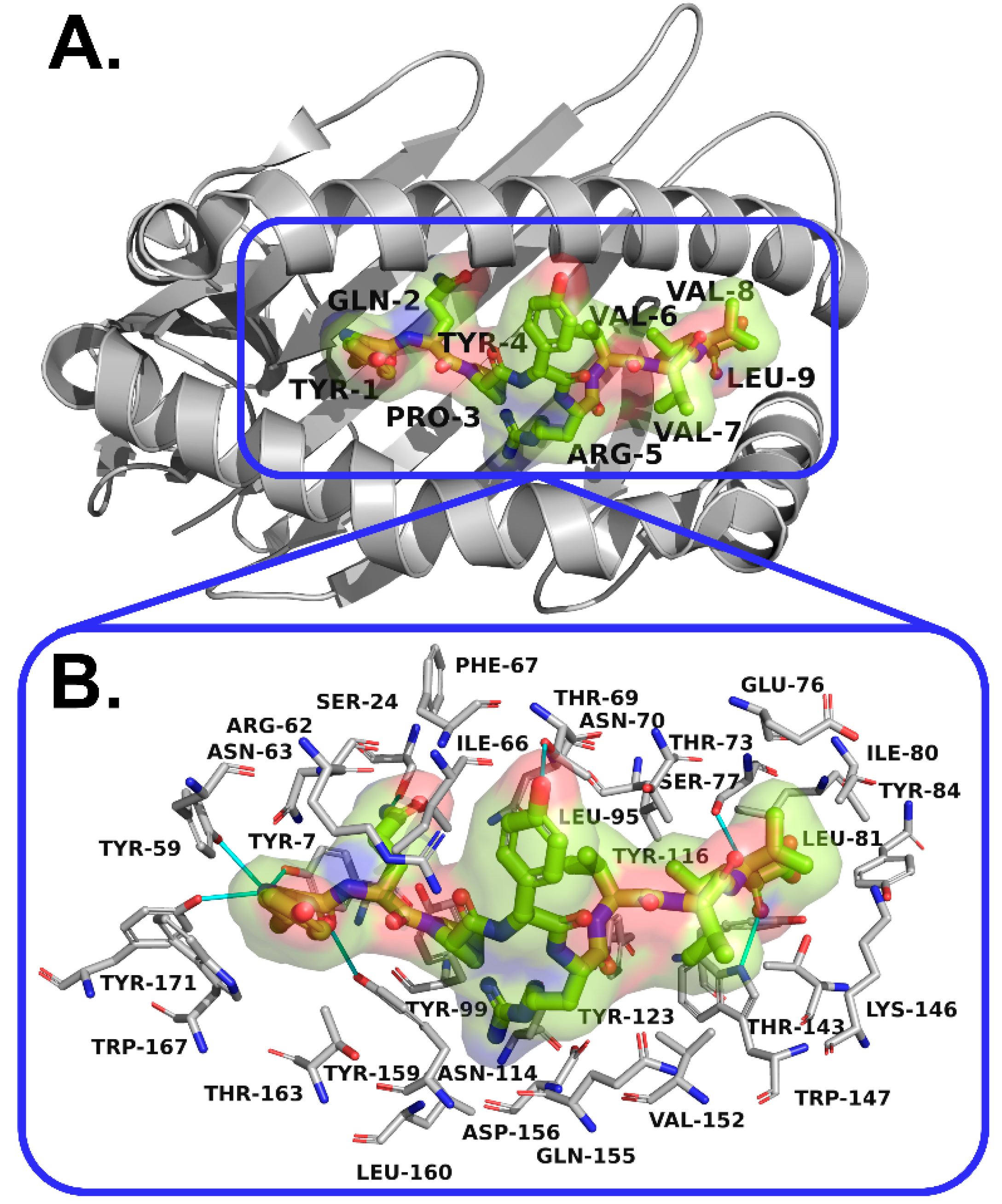
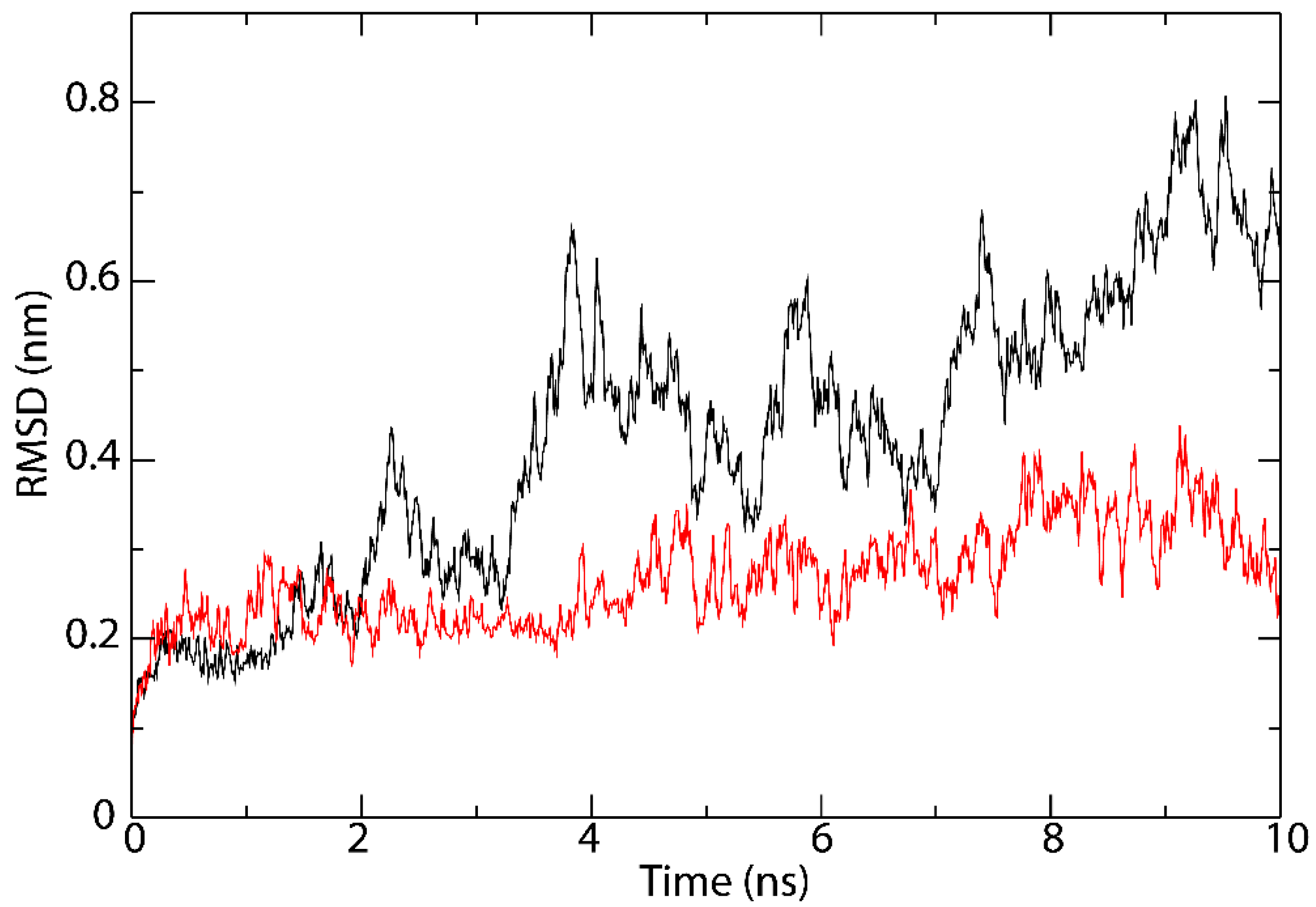
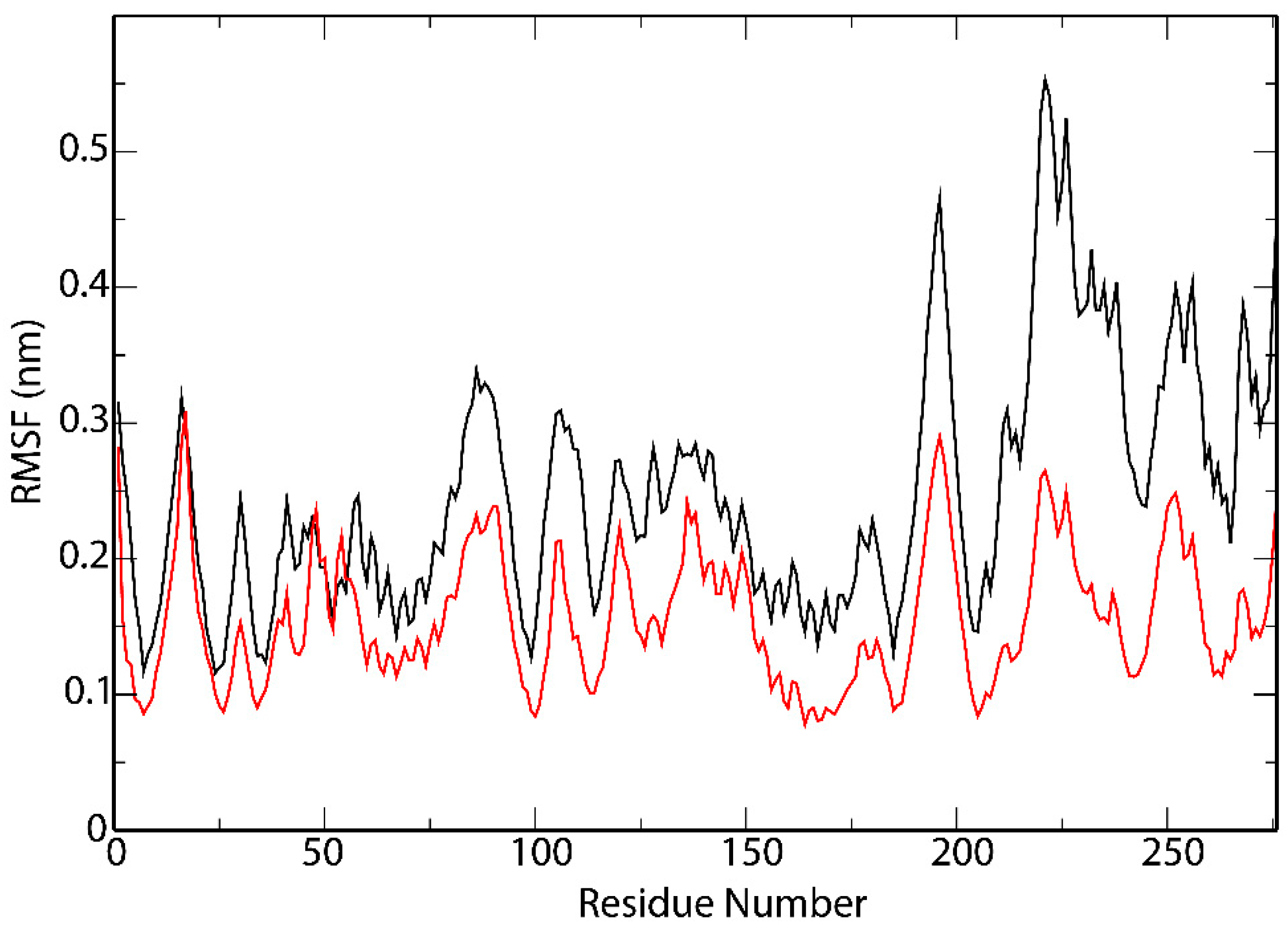
3.5. Interactions Analysis and Molecular Dynamics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galanakis, C.M. The food systems in the era of the Coronavirus (COVID-19) pandemic crisis. Foods 2020, 9, 523. [Google Scholar] [CrossRef]
- World Health Organization. Weekly Epidemiological Update on COVID-19—6 July 2021. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---6-july-2021 (accessed on 6 July 2021).
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard|WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 24 March 2021).
- Acter, T.; Uddin, N.; Das, J.; Akhter, A.; Choudhury, T.R.; Kim, S. Evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as coronavirus disease 2019 (COVID-19) pandemic: A global health emergency. Sci. Total Environ. 2020, 730, 138996. [Google Scholar] [CrossRef]
- Al-Rohaimi, A.H.; Al Otaibi, F. Novel SARS-CoV-2 outbreak and COVID19 disease; a systemic review on the global pandemic. Genes Dis. 2020, 7, 491–501. [Google Scholar] [CrossRef]
- Bahrami, M.; Kamalinejad, M.; Latifi, S.A.; Seif, F.; Dadmehr, M. Cytokine storm inCOVID-19 and parthenolide: Preclinical evidence. Phyther. Res. 2020, 34, 2429–2430. [Google Scholar] [CrossRef]
- Karwaciak, I.; Sałkowska, A.; Karaś, K.; Dastych, J.; Ratajewski, M. Nucleocapsid and spike proteins of the Coronavirus SARS-CoV-2 induce IL6 in monocytes and macrophages—Potential implications for cytokine storm syndrome. Vaccines 2021, 9, 54. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Mohsen, M.O.; Zha, L.; Vogel, M.; Speiser, D.E. SARS-CoV-2 structural features may explain limited neutralizing-antibody responses. NPJ Vaccines 2021, 6, 1–5. [Google Scholar] [CrossRef]
- Veldhoen, M.; Simas, J.P. Endemic SARS-CoV-2 will maintain post-pandemic immunity. Nat. Rev. Immunol. 2021, 21, 131–132. [Google Scholar] [CrossRef]
- Rastogi, M.; Pandey, N.; Shukla, A.; Singh, S.K. SARS coronavirus 2: From genome to infectome. Respir. Res. 2020, 21, 1–15. [Google Scholar] [CrossRef]
- Pooladanda, V.; Thatikonda, S.; Godugu, C. The current understanding and potential therapeutic options to combat COVID-19. Life Sci. 2020, 254, 117765. [Google Scholar] [CrossRef]
- Vellingiri, B.; Jayaramayya, K.; Iyer, M.; Narayanasamy, A.; Govindasamy, V.; Giridharan, B.; Ganesan, S.; Venugopal, A.; Venkatesan, D.; Ganesan, H.; et al. COVID-19: A promising cure for the global panic. Sci. Total Environ. 2020, 725, 138277. [Google Scholar] [CrossRef]
- Tahirul Qamar, M.; Alqahtani, S.M.; Alamri, M.A.; Chen, L.L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020, 10, 313–319. [Google Scholar] [CrossRef]
- Satarker, S.; Nampoothiri, M. Structural proteins in severe acute respiratory syndrome Coronavirus-2. Arch. Med. Res. 2020, 51, 482–491. [Google Scholar] [CrossRef]
- Mandala, V.S.; McKay, M.J.; Shcherbakov, A.A.; Dregni, A.J.; Kolocouris, A.; Hong, M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 2020, 27, 1202–1208. [Google Scholar] [CrossRef]
- Lu, S.; Ye, Q.; Singh, D.; Cao, Y.; Diedrich, J.K.; Yates, J.R.; Villa, E.; Cleveland, D.W.; Corbett, K.D. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat. Commun. 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Thomas, S. The structure of the membrane protein of sars-cov-2 resembles the sugar transporter semisweet. Pathog. Immun. 2020, 5, 342–363. [Google Scholar] [CrossRef]
- Ashik, A.I.; Hasan, M.; Tasnim, A.T.; Chowdhury, M.B.; Hossain, T.; Ahmed, S. An immunoinformatics study on the spike protein of SARS-CoV-2 revealing potential epitopes as vaccine candidates. Heliyon 2020, 6, e04865. [Google Scholar] [CrossRef]
- Sohail, M.S.; Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. In silico T cell epitope identification for SARS-CoV-2: Progress and perspectives. Adv. Drug Deliv. Rev. 2021, 171, 29–47. [Google Scholar] [CrossRef]
- Ita, K. Coronavirus disease (COVID-19): Current status and prospects for drug and vaccine development. Arch. Med. Res. 2021, 52, 15–24. [Google Scholar] [CrossRef]
- Requena, D.; Médico, A.; Chacón, R.D.; Ramírez, M.; Marín-Sánchez, O. Identification of novel candidate epitopes on SARS-CoV-2 proteins for South America: A review of HLA frequencies by country. Front. Immunol. 2020, 11, 2008. [Google Scholar] [CrossRef]
- Tan, H.-X.; Juno, J.A.; Lee, W.S.; Barber-Axthelm, I.; Kelly, H.G.; Wragg, K.M.; Esterbauer, R.; Amarasena, T.; Mordant, F.L.; Subbarao, K.; et al. Immunogenicity of prime-boost protein subunit vaccine strategies against SARS-CoV-2 in mice and macaques. Nat. Commun. 2021, 12, 1403. [Google Scholar] [CrossRef]
- Dobaño, C.; Santano, R.; Jiménez, A.; Vidal, M.; Chi, J.; Melero, N.R.; Popovic, M.; López-Aladid, R.; Fernández-Barat, L.; Tortajada, M.; et al. Immunogenicity and crossreactivity of antibodies to the nucleocapsid protein of SARS-CoV-2: Utility and limitations in seroprevalence and immunity studies. Transl. Res. 2021, 232, 60–74. [Google Scholar] [CrossRef]
- Diao, B.; Wen, K.; Zhang, J.; Chen, J.; Han, C.; Chen, Y.; Wang, S.; Deng, G.; Zhou, H.; Wu, Y. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin. Microbiol. Infect. 2021, 27, 289.e1–289.e4. [Google Scholar] [CrossRef]
- Peng, Y.; Mentzer, A.J.; Liu, G.; Yao, X.; Yin, Z.; Dong, D.; Dejnirattisai, W.; Rostron, T.; Supasa, P.; Liu, C.; et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020, 21, 1336–1345. [Google Scholar] [CrossRef]
- Nelde, A.; Bilich, T.; Heitmann, J.S.; Maringer, Y.; Salih, H.R.; Roerden, M.; Lübke, M.; Bauer, J.; Rieth, J.; Wacker, M.; et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021, 22, 74–85. [Google Scholar] [CrossRef]
- Kiyotani, K.; Toyoshima, Y.; Nemoto, K.; Nakamura, Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J. Hum. Genet. 2020, 65, 569–575. [Google Scholar] [CrossRef]
- Gonzalez-Galarza, F.F.; McCabe, A.; dos Santos, E.J.M.; Jones, J.; Takeshita, L.; Ortega-Rivera, N.D.; Cid-Pavon, G.M.D.; Ramsbottom, K.; Ghattaoraya, G.; Alfirevic, A.; et al. Allele frequency net database (AFND) 2020 update: Gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020, 48, D783–D788. [Google Scholar] [CrossRef]
- Arnaiz-Villena, A.; Palacio-Gruber, J.; Juarez, I.; Hernandez, E.; Muniz, E.; Bayona, B.; Campos, C.; Nieto, J.; Martin-Villa, M.; Silvera, C. HLA in North Colombia Chimila Amerindians. Hum. Immunol. 2018, 79, 189–190. [Google Scholar] [CrossRef]
- Arnaiz-Villena, A.; Palacio-Gruber, J.; Juarez, I.; Muniz, E.; Hernandez, E.; Bayona, B.; Campos, C.; Nunez, J.; Lopez-Nares, A.; Martin-Villa, M.; et al. Study of Colombia North Wiwa el encanto Amerindians HLA- genes: Pacific Islanders relatedness. Hum. Immunol. 2018, 79, 530–531. [Google Scholar] [CrossRef]
- Single, R.M.; Meyer, D.; Nunes, K.; Francisco, R.S.; Hunemeier, T.; Maiers, M.; Hurley, C.K.; Bedoya, G.; Gallo, C.; Hurtado, A.M.; et al. Demographic history and selection at HLA loci in Native Americans. PLoS ONE 2020, 15, e0241282. [Google Scholar] [CrossRef]
- Páez-Gutiérrez, I.A.; Hernández-Mejía, D.G.; Vanegas, D.; Camacho-Rodríguez, B.; Perdomo-Arciniegas, A.M. HLA-A, -B, -C, -DRB1 and -DQB1 allele and haplotype frequencies of 1463 umbilical cord blood units typed in high resolution from Bogotá, Colombia. Hum. Immunol. 2019, 80, 425–426. [Google Scholar] [CrossRef]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2021, 48, W449–W454. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Dimitrov, I.; Flower, D.R.; Doytchinova, I. AllerTOP—A server for in silico prediction of allergens. BMC Bioinform. 2013, 14, S4. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef] [Green Version]
- Thévenet, P.; Shen, Y.; Maupetit, J.; Guyon, F.; Derreumaux, P.; Tufféry, P. PEP-FOLD: An updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012, 40, W288–W293. [Google Scholar] [CrossRef] [Green Version]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; di Costanzo, L.; Christie, C.; Dalenberg, K.; Duarte, J.M.; Dutta, S.; et al. RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019, 47, D464–D474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and AutoDockTools4: Automated docking with selective receptor flexiblity. J. Comput. Chem. 2009, 16, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Development Team R, version 3.6.3; R Foundation for Statistical Computing: Vienna, Austria, 2019.
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S. Gplots: Various R programming tools for plotting data. R Packag. 2009, 2, 1. [Google Scholar]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- MacKerell, A.D.; Bashford, D.; Bellott, M.; Dunbrack, R.L.; Evanseck, J.D.; Field, M.J.; Fischer, S.; Gao, J.; Guo, H.; Ha, S.; et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 1998, 102, 3586–3616. [Google Scholar] [CrossRef] [PubMed]
- Painter, C.A.; Cruz, A.; López, G.E.; Stern, L.J.; Zavala-Ruiz, Z. Model for the peptide-free conformation of class II MHC proteins. PLoS ONE 2008, 3, e2403. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.; Tworowski, D.; Detroja, R.; Mukherjee, S.B.; Frenkel-Morgenstern, M. Immunoinformatics and structural analysis for identification of immunodominant epitopes in SARS-CoV-2 as potential vaccine targets. Vaccines 2020, 8, 290. [Google Scholar] [CrossRef]
- Smith, K.D.; Kurago, Z.B.; Lutz, C.T. Conformational changes in MHC class I molecules. Antibody, T-cell receptor, and NK cell recognition in an HLA-B7 model system. Immunol. Res. 1997, 16, 243–259. [Google Scholar] [CrossRef]
- Chen, Z.; Ruan, P.; Wang, L.; Nie, X.; Ma, X.; Tan, Y. T and B cell Epitope analysis of SARS-CoV-2 S protein based on immunoinformatics and experimental research. J. Cell. Mol. Med. 2021, 25, 1274–1289. [Google Scholar] [CrossRef]
- Chen, H.-Z.; Tang, L.-L.; Yu, X.-L.; Zhou, J.; Chang, Y.-F.; Wu, X. Bioinformatics analysis of epitope-based vaccine design against the novel SARS-CoV-2. Infect. Dis. Poverty 2020, 9, 1–10. [Google Scholar] [CrossRef]
- World Health Organization. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 7 July 2021).
- Poran, A.; Harjanto, D.; Malloy, M.; Arieta, C.M.; Rothenberg, D.A.; Lenkala, D.; van Buuren, M.M.; Addona, T.A.; Rooney, M.S.; Srinivasan, L.; et al. Sequence-based prediction of SARS-CoV-2 vaccine targets using a mass spectrometry-based bioinformatics predictor identifies immunogenic T cell epitopes. Genome Med. 2020, 12, 1–15. [Google Scholar] [CrossRef]
- Ambrose, J.M.; Veeraraghavan, V.P.; Kullappan, M.; Chellapandiyan, P.; Mohan, S.K.; Manivel, V.A. Comparison of immunological profiles of SARS-CoV-2 variants in the COVID-19 pandemic trends: An immunoinformatics approach. Antibiotics 2021, 10, 535. [Google Scholar] [CrossRef]
- Anand, R.; Biswal, S.; Bhatt, R.; Tiwary, B.N. Computational perspectives revealed prospective vaccine candidates from five structural proteins of novel SARS corona virus 2019 (SARS-CoV-2). PeerJ 2020, 8, e9855. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.X.; Lim, J.; Jazayeri, S.D.; Poppema, S.; Poh, C.L. Development of multi-epitope peptide-based vaccines against SARS-CoV-2. Biomed. J. 2021, 44, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Van Deutekom, H.W.M.; Keşmir, C. Zooming into the binding groove of HLA molecules: Which positions and which substitutions change peptide binding most? Immunogenetics 2015, 67, 425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knapp, B.; Deane, C.M. T-cell receptor binding affects the dynamics of the peptide/MHC-I complex. J. Chem. Inf. Model. 2015, 56, 46–53. [Google Scholar] [CrossRef]

| Search | Query |
|---|---|
| HLA Class I | (“MHC Class I” OR “MHC I” OR “HLA Class I” OR “HLA I” OR “HLA-A” OR “HLA-B” OR “HLA-C”) AND “Colombia” |
| HLA Class II | Query #1: (“MHC Class II” OR “MHC II” OR “HLA Class II” OR “HLA II”) AND “Colombia” Query #2: (“DRB1” OR “DRB3” OR “DRB4” OR “DRB5” OR “DQB1” OR “DQA1”) AND “HLA” AND “Colombia” Query #3: (“DPA1” OR “DPB1”) AND “HLA” AND “Colombia” |
| Protein | Length | NCBI Reference Sequence |
|---|---|---|
| Spike protein (S) | 1273 aa | YP_009724390.1 |
| Envelope protein (E) | 75 aa | YP_009724392.1 |
| Membrane glycoprotein (M) | 222 aa | YP_009724393.1 |
| Nucleocapsid phosphoprotein (N) | 419 aa | YP_009724397.2 |
| HLA I | Results |
|---|---|
| PubMed | 78 |
| Web of Science | 116 |
| Science Direct | 292 |
| HLA II | Results of Query #1 | Results of Query #2 | Results of Query #3 |
|---|---|---|---|
| PubMed | 77 | 105 | 6 |
| Web of Science | 106 | 106 | 6 |
| Science Direct | 345 | 266 | 40 |
| Ethnic Groups | HLA-A | HLA-B | HLA-C | |||
|---|---|---|---|---|---|---|
| Allele | Frequency (%) | Allele | Frequency (%) | Allele | Frequency (%) | |
| Colombia-Bogotá (n = 1463) | A*24:02 | 20.8 | B*35:43 | 8.6 | C*04:01 | 14.9 |
| A*02:01 | 16.1 | B*40:02 | 8.4 | C*01:02 | 11.4 | |
| A*01:01 | 6.1 | B*44:03 | 5.6 | C*07:02 | 9.7 | |
| A*03:01 | 6.1 | B*51:01 | 5.6 | C*07:01 | 8.9 | |
| A*68:01 | 5.2 | B*07:02 | 5.0 | C*03:04 | 8.2 | |
| A*29:02 | 4.5 | B*35:01 | 4.5 | C*05:01 | 5.1 | |
| A*11:01 | 4.3 | B*14:02 | 4.0 | C*06:02 | 5.1 | |
| A*31:01 | 4.0 | B*44:02 | 3.9 | C*16:01 | 5.1 | |
| A*23:01 | 3.3 | B*18:01 | 3.5 | C*08:02 | 4.9 | |
| A*02:22 | 3.2 | B*08:01 | 3.2 | C*12:03 | 4.5 | |
| Ethnic Groups | HLA-A | HLA-B | HLA-C | |||
|---|---|---|---|---|---|---|
| Allele | Frequency | Allele | Frequency | Allele | Frequency | |
| Arhuaco (n = 17) | A*24:02 | 0.441 | B*35:43 | 0.441 | C*01:02 | 0.382 |
| Embera (n = 14) | A*24:02 | 0.536 | B*39:05 | 0.429 | C*07:02 | 0.464 |
| Inga (n = 16) | A*24:02 | 0.367 | B*40:02 | 0.286 | C*01:02 | 0.367 |
| Kogi (n = 15) | A*24:02 | 0.433 | B*35:43 | 0.429 | C*01:02 | 0.571 |
| North Chimila (n = 47) | A*24:02 | 0.457 | B*51:10 | 0.404 | C*15:02 | 0.468 |
| North Wiwa El Encanto (n = 52) | A*24:02 | 0.433 | B*35:43 | 0.385 | C*01:02 | 0.51 |
| Waunana (n = 20) | A*24:02 | 0.6 | B*40:02 | 0.25 | C*03:04 | 0.35 |
| Wayuu (n = 15) | A*24:02 | 0.2 | B*40:02 | 0.2 | C*04:01 | 0.25 |
| Zenu (n = 16) | A*24:02 | 0.417 | B*40:02 | 0.25 | C*15:02 | 0.25 |
| Ticuna Arara (n = 17) | A*24:02 | 0.5 | B*39:03 | 0.286 | C*07:02 | 0.429 |
| Ticuna Tarapaca (n = 19) | A*24:02 | 0.526 | B*40:02 | 0.447 | C*03:04 | 0.529 |
| HLA II Alleles | WAFs (%) | n |
|---|---|---|
| Mestizo | ||
| DQB1*03:02 | 22.47 | 1737 |
| DQB1*03:01 | 19.23 | 1737 |
| DQB1*02:01 | 14.45 | 1737 |
| DRB1*04:07 | 12.39 | 1737 |
| DQB1*05:01 | 11.63 | 1737 |
| DQB1*04:02 | 10.76 | 1737 |
| DRB1*07:01 | 9.29 | 1925 |
| DQB1*06:02 | 7.03 | 1737 |
| DRB1*15:01 | 6.28 | 1925 |
| DRB1*08:02 | 6.25 | 1737 |
| DRB1*03:01 | 5.74 | 1925 |
| DRB1*13:01 | 5.09 | 1925 |
| African Colombian | ||
| DQA1*01:02 | 23.42 | 234 |
| DQA1*05:01 | 19.86 | 140 |
| DQB1*02:01 | 19.79 | 182 |
| DQA1*01:01 | 18.04 | 234 |
| DQB1*05:01 | 17.69 | 182 |
| DQB1*06:02 | 16.21 | 182 |
| DQB1*03:01 | 15.88 | 182 |
| DRB1*15:03 | 13.47 | 182 |
| DQA1*03:01 | 12.77 | 94 |
| DQB1*04:02 | 12.37 | 182 |
| DQA1*04:01 | 11.29 | 140 |
| DRB1*03:01 | 10.98 | 182 |
| DRB1*03:02 | 9.17 | 182 |
| DRB1*07:01 | 8.56 | 182 |
| DRB1*08:01 | 8.30 | 42 |
| DQA1*02:01 | 8.04 | 234 |
| DRB1*08:04 | 6.00 | 42 |
| DRB1*13:04 | 6.00 | 42 |
| DQB1*03:02 | 5.94 | 182 |
| DRB1*13:02 | 5.65 | 182 |
| DRB1*01:01 | 5.14 | 140 |
| Colombian Amerindians | ||
| DPB1*04:02 | 49.99 | 668 |
| DQA1*03:01 | 46.27 | 1573 |
| DPB1*14:01 | 45.67 | 668 |
| DRB4*01:00 | 44.23 | 1300 |
| DQB1*03:02 | 43.49 | 2633 |
| DRB4*01:01 | 38.10 | 34 |
| DQA1*05:01 | 35.05 | 2084 |
| DRB1*04.03 | 32.30 | 48 |
| DQB1*03:01 | 32.06 | 2633 |
| DQA1*05:00 | 19.62 | 321 |
| DQB1*04:02 | 18.53 | 2537 |
| DRB5*01:00 | 18.20 | 77 |
| DRB3*01:01 | 18.11 | 1573 |
| DRB1*14:02 | 18.01 | 2173 |
| DQA1*04:01 | 17.59 | 2348 |
| DRB1*04:07 | 17.32 | 2829 |
| DRB5*02:00 | 17.11 | 1257 |
| DRB1*16:02 | 14.48 | 2777 |
| DRB1*08:022 | 14.29 | 42 |
| DRB1*04:11 | 14.19 | 2691 |
| DRB1*08:02 | 10.47 | 2701 |
| DRB1*08:04 | 7.32 | 2091 |
| DRB1*04:04 | 7.23 | 2722 |
| Type | HLA I and HLA II Alleles |
|---|---|
| HLA-A | HLA-A*24:02, HLA-A*02:01, HLA-A*01:01, HLA-A*03:01, HLA-A*68:01, HLA-A*29:02, HLA-A*11:01, HLA-A*31:01, HLA-A*23:01, and HLA-A*02:22. |
| HLA-B | HLA-B*35:43, HLA-B*40:02, HLA-B*44:03, HLA-B*51:01, HLA-B*07:02, HLA-B*35:01, HLA-B*14:02, HLA-B*44:02, HLA-B*18:01, HLA-B*08:01, HLA-B*39:05, HLA-B*51:10, and HLA-B*39:03. |
| HLA-C | HLA-C*04:01, HLA-C*01:02, HLA-C*07:02, HLA-C*07:01, HLA-C*03:04, HLA-C*05:01, HLA-C*06:02, HLA-C*16:01, HLA-C*08:02, HLA-C*12:03, and HLA-C*15:02. |
| HLA-DRB1 | DRB1*01:01, DRB1*03:01, DRB1*03:02, DRB1*04:03, DRB1*04:04, DRB1*04:07, DRB1*04:11, DRB1*07:01, DRB1*08:01, DRB1*08:02, DRB1*08:04, DRB1*08:22, DRB1*13:01, DRB1*13:02, DRB1*13:04, DRB1*14:02, DRB1*15:01, DRB1*15:03, and DRB1*16:02. |
| Epitopes | Estimated Coverage for Colombian Population (WAF) | Experiments (IEDB) | IEDB ID |
|---|---|---|---|
| Spike Protein (S) | |||
| HLA 1 | |||
| VYDPLQPEL | HLA-A = 50.85%, HLA-B = 34.39%, HLA-C = 82.07%. | ML: (+). TC-IFNg: (−). | 71996 |
| YQPYRVVVL | HLA-A = 43.43%, HLA-B = 18.49%, HLA-C = 82.07%. | TC-QB: (+). | 1334394 |
| TLDSKTQSL | HLA-A = 25.42%, HLA-B = 15.04%, HLA-C = 82.07%. | TC-A: (+). TC-QB: (+). TC-IFNg: (−). | 1075075 |
| VRDPQTLEI | HLA-A = 24.09%, HLA-B = 7.42%, HLA-C = 57.39%. | - | |
| FTISVTTEI | HLA-A = 19.34%, HLA-B = 14.84%, HLA-C = 57.5%. | TC-A: (+). | 1317060 |
| HLA 2 | |||
| RAAEIRASANLAATK | HLA-DRB1 = 48.88%. | ML: (+). TC-IFNg: (−). | 533447 |
| TPINLVRDLPQGFSA | HLA-DRB1 = 33.7%. | ML: (+). | 1330624 |
| FGGFNFSQILPDPSK | HLA-DRB1 = 33.47%. | - | |
| KHTPINLVRDLPQGF | HLA-DRB1 = 31.59%. | TC-A: (+). TC-IFNg: (+). TC-IL5: (−). | 1309123 |
| Envelope Protein (E) | |||
| HLA 1 | |||
| YVYSRVKNL | HLA-A = 19.34%, HLA-B = 27.01%, HLA-C = 82.07%. | TC-A: (+). TC-QB: (+). | 1075128 |
| LAILTALRL | HLA-B = 6.19%, HLA-C = 22.09%. | - | |
| VSLVKPSFY | HLA-A = 14.9%, HLA-B = 8.65%, HLA-C = 27.97%. | - | |
| VTLAILTAL | HLA-A = 16.13%, HLA-C = 33.47%. | - | |
| RVKNLNSSR | HLA-A = 19.54%. | - | |
| HLA 2 | |||
| KPSFYVYSRVKNLNS | HLA-DRB1 = 43.18%. | - | |
| VYSRVKNLNSSRVPD | HLA-DRB1 = 55.99%. | - | |
| VKPSFYVYSRVKNLN | HLA-DRB1 = 43.18%. | - | |
| YSRVKNLNSSRVPDL | HLA-DRB1 = 37.1%. | - | |
| SFYVYSRVKNLNSSR | HLA-DRB1 = 61.92%. | - | |
| PSFYVYSRVKNLNSS | HLA-DRB1 = 55.74%. | - | |
| FYVYSRVKNLNSSRV | HLA-DRB1 = 42.58%. | TC-IFNg: (+). TC-TNF: (+). TC-IL5: (−). TC-A: (−). | 1310430 |
| LVKPSFYVYSRVKNL | HLA-DRB1 = 26.5%. | - | |
| YVYSRVKNLNSSRVP | HLA-DRB1 = 51.81%. | - | |
| Membrane Protein (M) | |||
| HLA 1 | |||
| ITVATSRTL | HLA-C = 47.52%. | - | |
| Nucleocapsid Protein (N) | |||
| HLA 1 | |||
| QFAPSASAF | HLA-A = 49.12%, HLA-B = 25.14%, HLA-C = 47.85%. | - | |
| QRNAPRITF | HLA-A = 47.09%, HLA-B = 19.77%, HLA-C = 24.33%. | TC-IFNg: (+). TC-QB: (+). TC-TNF: (−). TC-A: (−). | 1309136 |
| SPDDQIGYY | HLA-A = 3.24%, HLA-B = 25.38%, HLA-C = 26%. | TC-A: (+). TC-IFNg: (−). | 1310816 |
| HLA 2 | |||
| GTWLTYTGAIKLDDK | HLA-DRB1 = 40.37%. | TC-IFNg: (+). TC-A: (+). TC-TNF: (+). | 1310464 |
| NFKDQVILLNKHIDA | HLA-DRB1 = 24.69%. | - | |
| TKAYNVTQAFGRRGP | HLA-DRB1 = 52.85%. | - | |
| Epitopes | SARS-CoV-2 Protein | IFN Epitope Server | Worldwide Coverage (%) [IEDB] |
|---|---|---|---|
| YQPYRVVVL | S | 0.29285355 | 96.62 |
| RAAEIRASANLAATK | S | 0.29346657 | ND |
| QFAPSASAF | N | 0.82212984 | 77.60 |
| SPDDQIGYY | N | 0.24883001 | 80.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montes-Grajales, D.; Olivero-Verbel, J. Bioinformatics Prediction of SARS-CoV-2 Epitopes as Vaccine Candidates for the Colombian Population. Vaccines 2021, 9, 797. https://doi.org/10.3390/vaccines9070797
Montes-Grajales D, Olivero-Verbel J. Bioinformatics Prediction of SARS-CoV-2 Epitopes as Vaccine Candidates for the Colombian Population. Vaccines. 2021; 9(7):797. https://doi.org/10.3390/vaccines9070797
Chicago/Turabian StyleMontes-Grajales, Diana, and Jesus Olivero-Verbel. 2021. "Bioinformatics Prediction of SARS-CoV-2 Epitopes as Vaccine Candidates for the Colombian Population" Vaccines 9, no. 7: 797. https://doi.org/10.3390/vaccines9070797
APA StyleMontes-Grajales, D., & Olivero-Verbel, J. (2021). Bioinformatics Prediction of SARS-CoV-2 Epitopes as Vaccine Candidates for the Colombian Population. Vaccines, 9(7), 797. https://doi.org/10.3390/vaccines9070797







