M2e-Based Influenza Vaccines with Nucleoprotein: A Review
Abstract
1. Introduction
2. Influenza Pandemics
3. Influenza Vaccination
4. M2e as Immunogen
5. Immunogenicity and Efficacy of Influenza Nucleoprotein (NP) as a Fusion Cargo for M2e
6. Expression of M2e-NP as a Fusion Protein in Heterologous Protein Expression Systems
7. Potentials of M2e-NP Fusion Protein as a Universal Influenza Vaccine
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cardona, C.J.; Xing, Z.; Sandrock, C.E.; Davis, C.E. Avian influenza in birds and mammals. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 255–273. [Google Scholar] [CrossRef] [PubMed]
- Carrat, F.; Flahault, A. Influenza vaccine: The challenge of antigenic drift. Vaccine 2007, 25, 6852–6862. [Google Scholar] [CrossRef] [PubMed]
- Eisfeld, A.J.; Neumann, G.; Kawaoka, Y. Influenza A virus isolation, culture and identification. Nat. Protoc. 2014, 9, 2663. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, E.C. Influenza virus. Trends Microbiol. 2018, 26, 809–810. [Google Scholar] [CrossRef]
- Asha, K.; Kumar, B. Emerging influenza D virus threat: What we know so far! J. Clin. Med. 2019, 8, 192. [Google Scholar] [CrossRef]
- Sautto, G.A.; Kirchenbaum, G.A.; Ross, T.M. Towards a universal influenza vaccine: Different approaches for one goal. Virol. J. 2018, 15, 1–12. [Google Scholar] [CrossRef]
- Vijaykrishna, D.; Holmes, E.C.; Joseph, U.; Fourment, M.; Su, Y.C.; Halpin, R.; Lee, R.T.; Deng, Y.-M.; Gunalan, V.; Lin, X. The contrasting phylodynamics of human influenza B viruses. Elife 2015, 4, e05055. [Google Scholar] [CrossRef]
- Rota, P.A.; Wallis, T.R.; Harmon, M.W.; Rota, J.S.; Kendal, A.P.; Nerome, K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology 1990, 175, 59–68. [Google Scholar] [CrossRef]
- Hause, B.M.; Collin, E.A.; Liu, R.; Huang, B.; Sheng, Z.; Lu, W.; Wang, D.; Nelson, E.A.; Li, F. Characterization of a novel influenza virus in cattle and swine: Proposal for a new genus in the Orthomyxoviridae family. MBio 2014, 5, e00031-14. [Google Scholar] [CrossRef]
- Grohskopf, L.A.; Sokolow, L.Z.; Olsen, S.J.; Bresee, J.S.; Broder, K.R.; Karron, R.A. Prevention and control of influenza with vaccines: Recommendations of the Advisory Committee on Immunization Practices, United States, 2015–2016 influenza season. MMWR. Morb. Mortal. Wkly. Rep. 2015, 64, 818. [Google Scholar] [CrossRef]
- Kim, H.; Webster, R.G.; Webby, R.J. Influenza virus: Dealing with a drifting and shifting pathogen. Viral Immunol. 2018, 31, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Both, G.W.; Sleigh, M.; Cox, N.; Kendal, A. Antigenic drift in influenza virus H3 hemagglutinin from 1968 to 1980: Multiple evolutionary pathways and sequential amino acid changes at key antigenic sites. J. Virol. 1983, 48, 52–60. [Google Scholar] [CrossRef] [PubMed]
- De Jong, J.; Rimmelzwaan, G.; Fouchier, R.; Osterhaus, A. Influenza virus: A master of metamorphosis. J. Infect. 2000, 40, 218–228. [Google Scholar] [CrossRef]
- Webster, R.; Laver, W.; Air, G.; Schild, G. Molecular mechanisms of variation in influenza viruses. Nature 1982, 296, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Treanor, J. Influenza vaccine—outmaneuvering antigenic shift and drift. N. Engl. J. Med. 2004, 350, 218–220. [Google Scholar] [CrossRef]
- Garten, R.J.; Davis, C.T.; Russell, C.A.; Shu, B.; Lindstrom, S.; Balish, A.; Sessions, W.M.; Xu, X.; Skepner, E.; Deyde, V. Antigenic and genetic characteristics of swine-origin 2009 A (H1N1) influenza viruses circulating in humans. Science 2009, 325, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Chowell, G.; Bertozzi, S.M.; Colchero, M.A.; Lopez-Gatell, H.; Alpuche-Aranda, C.; Hernandez, M.; Miller, M.A. Severe respiratory disease concurrent with the circulation of H1N1 influenza. N. Engl. J. Med. 2009, 361, 674–679. [Google Scholar] [CrossRef]
- Xu, R.; Ekiert, D.C.; Krause, J.C.; Hai, R.; Crowe, J.E.; Wilson, I.A. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 2010, 328, 357–360. [Google Scholar] [CrossRef]
- Xing, Z.; Cardona, C.J. Preexisting immunity to pandemic (H1N1) 2009. Emerg. Infect. Dis. 2009, 15, 1847. [Google Scholar] [CrossRef]
- Bonmarin, I.; Belchior, E.; Lévy-Bruhl, D. Impact of influenza vaccination on mortality in the French elderly population during the 2000–2009 period. Vaccine 2015, 33, 1099–1101. [Google Scholar] [CrossRef]
- Costantino, C.; Vitale, F. Influenza vaccination in high-risk groups: A revision of existing guidelines and rationale for an evidence-based preventive strategy. J. Prev. Med. Hyg. 2016, 57, E13. [Google Scholar] [PubMed]
- Guan, Y.; Vijaykrishna, D.; Bahl, J.; Zhu, H.; Wang, J.; Smith, G.J. The emergence of pandemic influenza viruses. Prot. Cell 2010, 1, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Martini, M.; Gazzaniga, V.; Bragazzi, N.; Barberis, I. The Spanish Influenza Pandemic: A lesson from history 100 years after 1918. J. Prev. Med. Hyg. 2019, 60, E64–E67. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, S.; Kawaoka, Y. The Pathogenesis of Influenza Virus Infections: The Contributions of Virus and Host Factors. Curr. Opin. Immunol. 2011, 23, 48–486. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Reid, A.H.; Krafft, A.E.; Bijwaard, K.E.; Fanning, T.G. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science 1997, 275, 1793–1796. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, J.K.; Reid, A.H.; Lourens, R.M.; Wang, R.; Jin, G.; Fanning, T.G. Characterization of the 1918 influenza virus polymerase genes. Nature 2005, 437, 889–893. [Google Scholar] [CrossRef]
- Taubenberger, J.K.; Morens, D.M. 1918 Influenza: The mother of all pandemics. Rev. Biomed. 2006, 17, 69–79. [Google Scholar] [CrossRef]
- MacKellar, L. Pandemic influenza: A review. Popul. Dev. Rev. 2007, 33, 429–451. [Google Scholar] [CrossRef]
- Lindstrom, S.E.; Cox, N.J.; Klimov, A. Genetic Analysis of Human H2n2 and Early H3n2 Influenza Viruses, 1957–1972: Evidence for Genetic Divergence and Multiple Reassortment Events. Virology 2004, 328, 101–119. [Google Scholar] [CrossRef]
- Tricco, A.C.; Chit, A.; Soobiah, C.; Hallett, D.; Meier, G.; Chen, M.H.; Tashkandi, M.; Bauch, C.T.; Loeb, M. Comparing influenza vaccine efficacy against mismatched and matched strains: A systematic review and meta-analysis. BMC Med. 2013, 11, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, L.; Spreeuwenberg, P.; Lustig, R.; Taylor, R.J.; Fleming, D.M.; Kroneman, M.; Van Kerkhove, M.D.; Mounts, A.W.; Paget, W.J. Global mortality estimates for the 2009 Influenza Pandemic from the GLaMOR project: A modeling study. PLoS Med 2013, 10, e1001558. [Google Scholar] [CrossRef]
- Gatherer, D. The 2009 H1N1 influenza outbreak in its historical context. J. Clin. Virol. 2009, 45, 174–178. [Google Scholar] [CrossRef]
- Baldo, V.; Bertoncello, C.; Cocchio, S.; Fonzo, M.; Pillon, P.; Buja, A.; Baldovin, T. The new pandemic influenza A/(H1N1)pdm09 virus: Is it really “new”? J. Prev. Med. Hyg. 2016, 57, E19–E22. [Google Scholar] [PubMed]
- Christman, M.C.; Kedwaii, A.; Xu, J.; Donis, R.O.; Lu, G. Pandemic (H1N1) 2009 virus revisited: An evolutionary retrospective. Infect. Genet. Evol. 2011, 11, 803–811. [Google Scholar] [CrossRef]
- Mena, I.; Nelson, M.I.; Quezada-Monroy, F.; Dutta, J.; Cortes-Fernández, R.; Lara-Puente, J.H.; Castro-Peralta, F.; Cunha, L.F.; Trovão, N.S.; Lozano-Dubernard, B.; et al. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. eLife 2016, 5, 16777. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.J.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef] [PubMed]
- Smetana, J.; Chlibek, R.; Shaw, J.; Splino, M.; Prymula, R. Influenza vaccination in the elderly. Hum. Vacc. Immunother. 2018, 14, 540–549. [Google Scholar] [CrossRef]
- McLean, H.Q.; Thompson, M.G.; Sundaram, M.E.; Kieke, B.A.; Gaglani, M.; Murthy, K.; Piedra, P.A.; Zimmerman, R.K.; Nowalk, M.P.; Raviotta, J.M. Influenza vaccine effectiveness in the United States during 2012–2013: Variable protection by age and virus type. J. Infect. Dis. 2015, 211, 1529–1540. [Google Scholar] [CrossRef]
- Mohn, K.G.-I.; Smith, I.; Sjursen, H.; Cox, R.J. Immune responses after live attenuated influenza vaccination. Hum. Vacc. Immunother. 2018, 14, 571–578. [Google Scholar] [CrossRef]
- Cox, M.M.J.; Izikson, R.; Post, P.; Dunkle, L. Safety, efficacy, and immunogenicity of Flublok in the prevention of seasonal influenza in adults. Ther. Adv. Vacc. 2015, 3, 97–108. [Google Scholar]
- Sridhar, S.; Brokstad, K.A.; Cox, R.J. Influenza vaccination strategies: Comparing inactivated and live attenuated influenza vaccines. Vaccines 2015, 3, 373–389. [Google Scholar] [CrossRef]
- Wong, T.; Ross, T.M. Steps toward a Universal Influenza Vaccine: Research Models and Comparison of Current Approaches. In Steps Forwards in Diagnosing and Controlling Influenza; InTech: London, UK, 2016; pp. 87–118. [Google Scholar]
- Cox, M.M.J.; Patriarca, P.A.; Treanor, J. FluBlok, a recombinant hemagglutinin influenza vaccine. Influ. Other Respir. Viruses 2008, 2, 211–219. [Google Scholar] [CrossRef]
- Woo, E.J.; Moro, P.L. Postmarketing safety surveillance of quadrivalent recombinant influenza vaccine: Reports to the vaccine adverse event reporting system. Vaccine 2021, 39, 1812–1817. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, A.L.; Kazmin, D.; Napolitani, G.; Clutterbuck, E.A.; Pulendran, B.; Siegrist, C.-A.; Pollard, A.J. AS03-and MF59-adjuvanted influenza vaccines in children. Front. Immunol. 2017, 8, 1760. [Google Scholar] [CrossRef]
- Tam, J.S.; Capeding, M.R.Z.; Lum, L.C.S.; Chotpitayasunondh, T.; Jiang, Z.; Huang, L.-M.; Lee, B.W.; Qian, Y.; Samakoses, R.; Lolekha, S. Efficacy and safety of a live attenuated, cold-adapted influenza vaccine, trivalent against culture-confirmed influenza in young children in Asia. Ped. Inf. Dis. J. 2007, 26, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Neto, H.B.; Farhat, C.K.; Tregnaghi, M.W.; Madhi, S.A.; Razmpour, A.; Palladino, G.; Small, M.G.; Gruber, W.C.; Forrest, B.D.; Group, D.-P.L.S. Efficacy and safety of 1 and 2 doses of live attenuated influenza vaccine in vaccine-naive children. Ped. Inf. Dis. J. 2009, 28, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.G.; Pierse, N.; Huang, Q.S.; Prasad, N.; Duque, J.; Newbern, E.C.; Baker, M.G.; Turner, N.; McArthur, C. Influenza vaccine effectiveness in preventing influenza-associated intensive care admissions and attenuating severe disease among adults in New Zealand 2012–2015. Vaccine 2018, 36, 5916–5925. [Google Scholar] [CrossRef]
- Villiers, P.J.T.D.; Steele, A.D.; Hiemstra, L.A.; Rappaport, R.; Dunning, A.J.; Gruber, W.C.; Forrest, B.D. Efficacy and safety of a live attenuated influenza vaccine in adults 60 years of age and older. Vaccine 2009, 28, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Praditsuwan, R.; Assantachai, P.; Wasi, C.; Puthavatana, P.; Kositanont, U. The efficacy and effectiveness of influenza vaccination among Thai elderly persons living in the community. J. Med. Assoc. Thail. 2005, 88, 256–264. [Google Scholar]
- Pera, A.; Campos, C.; López, N.; Hassouneh, F.; Alonso, C.; Tarazona, R.; Solana, R. Immunosenescence: Implications for response to infection and vaccination in older people. Maturitas 2015, 82, 50–55. [Google Scholar] [CrossRef]
- Grohskopf, L.A.; Alyanak, E.; Broder, K.R.; Walter, E.B.; Fry, A.M.; Jernigan, D.B. Prevention and Control of Seasonal Influenza with Vaccines: Recommendations of the Advisory Committee on Immunization Practices—United States, 2019–2020 Influenza Season. MMWR Recomm. Rep. 2019, 68, 1. [Google Scholar] [CrossRef]
- Stamboulian, D.; Bonvehi, P.; Nacinovich, F.; Cox, N. Influenza infection. Dis. Clin. N. Am. 2000, 14, 141–166. [Google Scholar] [CrossRef]
- Rondy, M.; El Omeiri, N.; Thompson, M.G.; Levêque, A.; Moren, A.; Sullivan, S.G. Effectiveness of influenza vaccines in preventing severe influenza illness among adults: A systematic review and meta-analysis of test-negative design case-control studies. J. Infect. 2017, 75, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.W.; Cowling, B.J.; Gao, H.Z.; Thompson, M.G. Comparative immunogenicity of enhanced seasonal influenza vaccines in older adults: A systematic review and meta-analysis. J. Infect. Dis. 2019, 219, 1525–1535. [Google Scholar] [CrossRef]
- Jianping, H.; Xin, F.; Changshun, L.; Bo, Z.; Linxiu, G.; Wei, X.; Jiande, S. Assessment of effectiveness of Vaxigrip. Vaccine 1999, 17, S57–S58. [Google Scholar] [CrossRef]
- Lang, P.-O.; Mendes, A.; Socquet, J.; Assir, N.; Govind, S.; Aspinall, R. Effectiveness of influenza vaccine in aging and older adults: Comprehensive analysis of the evidence. Clin. Interv. Aging 2012, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Kilbourne, E.D. Influenza pandemics: Can we prepare for the unpredictable? Viral Immunol. 2004, 17, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Gerdil, C. The annual production cycle for influenza vaccine. Vaccine 2003, 21, 1776–1779. [Google Scholar] [CrossRef]
- Fiers, W.; De Filette, M.; El Bakkouri, K.; Schepens, B.; Roose, K.; Schotsaert, M.; Birkett, A.; Saelens, X. M2e-based universal influenza A vaccine. Vaccine 2009, 27, 6280–6283. [Google Scholar] [CrossRef]
- Yap, W.B.; Toong, S.T.; Hassan, S.S.; Lafi, A.S.A.; Santhanam, J.; Khaithir, T.M.N.; Musa, N.F.; Huyop, F. Universal Oral Vaccine for Influenza Infections. J. Sains Kesihat. Malays. 2018, 16, 51–64. [Google Scholar] [CrossRef]
- Paules, C.I.; Sullivan, S.G.; Subbarao, K.; Fauci, A.S. Chasing seasonal influenza—the need for a universal influenza vaccine. N. Engl. J. Med. 2018, 378, 7–9. [Google Scholar] [CrossRef]
- Romeli, S.; Hassan, S.S.; Yap, W.B. Multi-Epitope Peptide-Based and Vaccinia-Based Universal Influenza Vaccine Candidates Subjected to Clinical Trials. Malays. J. Med.L Sci. Mjms 2020, 27, 10. [Google Scholar]
- Fan, J.; Liang, X.; Horton, M.S.; Perry, H.C.; Citron, M.P.; Heidecker, G.J.; Fu, T.-M.; Joyce, J.; Przysiecki, C.T.; Keller, P.M.; et al. Preclinical study of influenza virus A M2 peptide conjugate vaccines in mice, ferrets, and rhesus monkeys. Vaccine 2004, 22, 2993–3003. [Google Scholar] [CrossRef] [PubMed]
- Schotsaert, M.; De Filette, M.; Fiers, W.; Saelens, X. Universal M2 ectodomain-based influenza A vaccines: Preclinical and clinical developments. Expert Rev. Vaccines 2009, 8, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Kitikoon, P.; Vincent, A.L.; Janke, B.H.; Erickson, B.; Strait, E.L.; Yu, S.; Gramer, M.R.; Thacker, E.L. Swine influenza matrix 2 (M2) protein contributes to protection against infection with different H1 swine influenza virus (SIV) isolates. Vaccine 2009, 28, 523–531. [Google Scholar] [CrossRef]
- Saelens, X. The Role of Matrix Protein 2 Ectodomain in the Development of Universal Influenza Vaccines. J. Infect. Dis. 2019, 219, S68–S74. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, L.; Compans, R.W.; Wang, B.-Z. Universal Influenza Vaccines, a Dream to Be Realized Soon. Viruses 2014, 6, 1974–1991. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, R.; Igarashi, M.; Takada, A. Influenza A Virus M2 Protein: Roles from Ingress to Egress. Int. J. Mol. Sci. 2017, 18, 2649. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.A.; Zebedee, S.L.; Richardson, C.D. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 1985, 40, 627–633. [Google Scholar] [CrossRef]
- Skehel, J.J.; Wiley, D.C. Receptor Binding and Membrane Fusion in Virus Entry: The Influenza Hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.K.; Gupta, D.; Lal, S.K. Host Lipid Rafts Play a Major Role in Binding and Endocytosis of Influenza A Virus. Viruses 2018, 10, 650. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Matsumae, H.; Katoh, M.; Eisfeld, A.J.; Neumann, G.; Hase, T.; Ghosh, S.; Shoemaker, J.E.; Lopes, T.J.S.; Watanabe, T.; et al. A comprehensive map of the influenza A virus replication cycle. BMC Syst. Biol. 2013, 7, 97. [Google Scholar] [CrossRef] [PubMed]
- Lukarska, M. Structural and Functional Characterization of the Interaction between Influenza Polymerase and the Cellular Transcription Machinery. Doctoral Dissertation, Université Grenoble Alpes, Grenoble, France, 2018. [Google Scholar]
- Kapoor, S.; Dhama, K. Replication Cycle of Influenza Viruses. In Insight into Influenza Viruses of Animals and Humans; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2014; pp. 15–30. [Google Scholar]
- Gerhard, W.; Mozdzanowska, K.; Zharikova, D. Prospects for Universal Influenza Virus Vaccine. Emerg. Infect. Dis. 2006, 12, 569–574. [Google Scholar] [CrossRef]
- Hutchinson, E.C.; Charles, P.D.; Hester, S.S.; Thomas, B.; Trudgian, D.C.; Martínez-Alonso, M.; Fodor, E.J.B. Conserved and host-specific features of influenza virion architecture. Nat. Commun. 2014, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Vahey, M.D.; Fletcher, D.A. Influenza A virus surface proteins are organized to help penetrate host mucus. eLife 2019, 8, 43764. [Google Scholar] [CrossRef]
- Eliasson, D.G.; Omokanye, A.; Schön, K.; Wenzel, U.A.; Bernasconi, V.; Bemark, M.; Kolpe, A.; El Bakkouri, K.; Ysenbaert, T.; Deng, L.; et al. M2e-tetramer-specific memory CD4 T cells are broadly protective against influenza infection. Mucosal Immunol. 2018, 11, 273–289. [Google Scholar] [CrossRef]
- Liu, W.; Zou, P.; Ding, J.; Lu, Y.; Chen, Y.-H. Sequence comparison between the extracellular domain of M2 protein human and avian influenza A virus provides new information for bivalent influenza vaccine design. Microbes Infect. 2005, 7, 171–177. [Google Scholar] [CrossRef]
- Rappazzo, C.G.; Watkins, H.C.; Guarino, C.M.; Chau, A.; Lopez, J.L.; DeLisa, M.; Leifer, C.A.; Whittaker, G.R.; Putnam, D. Recombinant M2e outer membrane vesicle vaccines protect against lethal influenza A challenge in BALB/c mice. Vaccine 2016, 34, 1252–1258. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, A.; Ngunjiri, J.M.; Xia, M.; Elaish, M.; Jang, H.; Mahesh, K.; Abundo, M.C.; Jiang, X.; Lee, C.-W. Heterosubtypic protection against avian influenza virus by live attenuated and chimeric norovirus P-particle-M2e vaccines in chickens. Vaccine 2019, 37, 1356–1364. [Google Scholar] [CrossRef]
- Zhu, W.; Pewin, W.; Wang, C.; Luo, Y.; Gonzalez, G.X.; Mohan, T.; Prausnitz, M.; Wang, B.-Z. A boosting skin vaccination with dissolving microneedle patch encapsulating M2e vaccine broadens the protective efficacy of conventional influenza vaccines. J. Control. Release 2017, 261, 1–9. [Google Scholar] [CrossRef]
- Simhadri, V.R.; Dimitrova, M.; Mariano, J.L.; Zenarruzabeitia, O.; Zhong, W.; Ozawa, T.; Muraguchi, A.; Kishi, H.; Eichelberger, M.C.; Borrego, F. A Human Anti-M2 Antibody Mediates Antibody-Dependent Cell-Mediated Cytotoxicity (ADCC) and Cytokine Secretion by Resting and Cytokine-Preactivated Natural Killer (NK) Cells. PLoS ONE 2015, 10, e0124677. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-H.; Kwon, Y.-M.; Lee, Y.-T.; Kim, M.-C.; Hwang, H.S.; Ko, E.-J.; Lee, Y.; Choi, H.-J.; Kang, S.-M. Virus-Like Particles Are a Superior Platform for Presenting M2e Epitopes to Prime Humoral and Cellular Immunity against Influenza Virus. Vaccines 2018, 6, 66. [Google Scholar] [CrossRef]
- Huleatt, J.W.; Nakaar, V.; Desai, P.; Huang, Y.; Hewitt, D.; Jacobs, A.; Tang, J.; McDonald, W.; Song, L.; Evans, R.K. Potent immunogenicity and efficacy of a universal influenza vaccine candidate comprising a recombinant fusion protein linking influenza M2e to the TLR5 ligand flagellin. Vaccine 2008, 26, 201–214. [Google Scholar] [CrossRef]
- Stepanova, L.A.; Kotlyarov, R.Y.; Kovaleva, A.A.; Potapchuk, M.V.; Korotkov, A.V.; Sergeeva, M.V.; Kasianenko, M.A.; Kuprianov, V.V.; Ravin, N.V.; Tsybalova, L.M. Protection against multiple influenza A virus strains induced by candidate recombinant vaccine based on heterologous M2e peptides linked to flagellin. PLoS ONE 2015, 10, e0119520. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-C.; Kim, K.-H.; Lee, J.W.; Lee, Y.-N.; Choi, H.-J.; Jung, Y.-J.; Kim, Y.-J.; Compans, R.W.; Prausnitz, M.R.; Kang, S.-M. Co-Delivery of M2e Virus-Like Particles with Influenza Split Vaccine to the Skin Using Microneedles Enhances the Efficacy of Cross Protection. Pharmaceutics 2019, 11, 188. [Google Scholar] [CrossRef]
- Shim, B.-S.; Choi, Y.K.; Yun, C.-H.; Lee, E.-G.; Jeon, Y.S.; Park, S.-M.; Cheon, I.S.; Joo, D.-H.; Cho, C.H.; Song, M.-S.; et al. Sublingual Immunization with M2-Based Vaccine Induces Broad Protective Immunity against Influenza. PLoS ONE 2011, 6, e27953. [Google Scholar] [CrossRef] [PubMed]
- Andersson, A.-M.C.; Håkansson, K.O.; Jensen, B.A.H.; Christensen, D.; Andersen, P.; Thomsen, A.R.; Christensen, J.P. Increased immunogenicity and protective efficacy of influenza M2e fused to a tetramerizing protein. PLoS ONE 2012, 7, e46395. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Y.-C.; Feng, H.; Ahmed, T.; Compans, R.W.; Wang, B.-Z. Virus-like particles containing the tetrameric ectodomain of influenza matrix protein 2 and flagellin induce heterosubtypic protection in mice. BioMed Res. Int. 2013, 2013, 686549. [Google Scholar] [CrossRef]
- Mozdzanowska, K.; Zharikova, D.; Cudic, M.; Otvos, L.; Gerhard, W. Roles of adjuvant and route of vaccination in antibody response and protection engendered by a synthetic matrix protein 2-based influenza A virus vaccine in the mouse. Virology J. 2007, 4, 1–14. [Google Scholar] [CrossRef]
- Mazanec, M.B.; Coudret, C.L.; Fletcher, D.R. Intracellular neutralization of influenza virus by immunoglobulin A anti-hemagglutinin monoclonal antibodies. J. Virol. 1995, 69, 1339–1343. [Google Scholar] [CrossRef] [PubMed]
- Heinen, P.P.; De Boer-Luijtze, E.A.; Bianchi, A.T.J. Respiratory and systemic humoral and cellular immune responses of pigs to a heterosubtypic influenza A virus infection. J. Gen. Virol. 2001, 82, 2697–2707. [Google Scholar] [CrossRef] [PubMed]
- Kotomina, T.; Isakova-Sivak, I.; Kim, K.-H.; Park, B.R.; Jung, Y.-J.; Lee, Y.; Mezhenskaya, D.; Matyushenko, V.; Kang, S.-M.; Rudenko, L. Generation and Characterization of Universal Live-Attenuated Influenza Vaccine Candidates Containing Multiple M2e Epitopes. Vaccines 2020, 8, 648. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-C.; Lee, J.-S.; Kwon, Y.-M.; Eunju, O.; Lee, Y.-J.; Choi, J.-G.; Wang, B.-Z.; Compans, R.W.; Kang, S.-M. Multiple heterologous M2 extracellular domains presented on virus-like particles confer broader and stronger M2 immunity than live influenza A virus infection. Antivir. Res. 2013, 99, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Turley, C.B.; Rupp, R.E.; Johnson, C.; Taylor, D.N.; Wolfson, J.; Tussey, L.; Kavita, U.; Stanberry, L.; Shaw, A. Safety and immunogenicity of a recombinant M2e–flagellin influenza vaccine (STF2.4xM2e) in healthy adults. Vaccine 2011, 29, 5145–5152. [Google Scholar] [CrossRef] [PubMed]
- Ramos, E.L.; Mitcham, J.L.; Koller, T.D.; Bonavia, A.; Usner, D.W.; Balaratnam, G.; Fredlund, P.; Swiderek, K.M. Efficacy and Safety of Treatment With an Anti-M2e Monoclonal Antibody in Experimental Human Influenza. J. Infect. Dis. 2014, 211, 1038–1044. [Google Scholar] [CrossRef]
- Kolpe, A.; Schepens, B.; Fiers, W.; Saelens, X. M2-based influenza vaccines: Recent advances and clinical potential. Expert Rev. Vaccines 2017, 16, 123–136. [Google Scholar] [CrossRef]
- Beerli, R.R.; Bauer, M.; Schmitz, N.; Buser, R.B.; Gwerder, M.; Muntwiler, S.; Renner, W.A.; Saudan, P.; Bachmann, M.F. Prophylactic and therapeutic activity of fully human monoclonal antibodies directed against Influenza A M2 protein. Virol. J. 2009, 6, 224. [Google Scholar] [CrossRef]
- Grandea, A.G.; Olsen, O.A.; Cox, T.C.; Renshaw, M.; Hammond, P.W.; Chan-Hui, P.-Y.; Mitcham, J.L.; Cieplak, W.; Stewart, S.M.; Grantham, M.L.; et al. Human antibodies reveal a protective epitope that is highly conserved among human and nonhuman influenza A viruses. Proc. Natl. Acad. Sci. USA 2010, 107, 12658–12663. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Song, A.; Levin, J.; Dennis, D.; Zhang, N.J.; Yoshida, H.; Koriazova, L.; Madura, L.; Shapiro, L.; Matsumoto, A. Therapeutic potential of a fully human monoclonal antibody against influenza A virus M2 protein. Antivir. Res. 2008, 80, 168–177. [Google Scholar] [CrossRef]
- Liu, X.; Guo, J.; Han, S.; Yao, L.; Chen, A.; Yang, Q.; Bo, H.; Xu, P.; Yin, J.; Zhang, Z. Enhanced immune response induced by a potential influenza A vaccine based on branched M2e polypeptides linked to tuftsin. Vaccine 2012, 30, 6527–6533. [Google Scholar] [CrossRef]
- Blokhina, E.A.; Kuprianov, V.V.; Stepanova, L.A.; Tsybalova, L.M.; Kiselev, O.I.; Ravin, N.V.; Skryabin, K.G. A molecular assembly system for presentation of antigens on the surface of HBc virus-like particles. Virology 2013, 435, 293–300. [Google Scholar] [CrossRef]
- Ebrahimi, S.M.; Tebianian, M. Influenza A viruses: Why focusing on M2e-based universal vaccines. Virus Genes 2010, 42, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Łęga, T.; Weiher, P.; Obuchowski, M.; Nidzworski, D. Presenting Influenza A M2e Antigen on Recombinant Spores of Bacillus subtilis. PLoS ONE 2016, 11, e0167225. [Google Scholar] [CrossRef] [PubMed]
- Yap, W.B.; Tey, B.T.; Alitheen, N.B.M.; Tan, W.S. Display of the antigenic region of Nipah virus nucleocapsid protein on hepatitis B virus capsid. J. Biosci. Bioeng. 2012, 113, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Yap, W.B.; Tey, B.T.; Ng, M.; Ong, S.T.; Tan, W.S. N-terminally His-tagged hepatitis B core antigens: Construction, expression, purification and antigenicity. J. Virol. Methods 2009, 160, 125–131. [Google Scholar] [CrossRef]
- Westcott, M.M.; Clemens, E.A.; Holbrook, B.C.; King, S.B.; Alexander-Miller, M.A. The choice of linker for conjugating R848 to inactivated influenza virus determines the stimulatory capacity for innate immune cells. Vaccine 2018, 36, 1174–1182. [Google Scholar] [CrossRef]
- Ravin, N.V.; Blokhina, E.A.; Kuprianov, V.V.; Stepanova, L.A.; Shaldjan, A.A.; Kovaleva, A.A.; Tsybalova, L.M.; Skryabin, K.G. Development of a candidate influenza vaccine based on virus-like particles displaying influenza M2e peptide into the immunodominant loop region of hepatitis B core antigen: Insertion of multiple copies of M2e increases immunogenicity and protective efficiency. Vaccine 2015, 33, 3392–3397. [Google Scholar] [CrossRef]
- Neirynck, S.; DeRoo, T.; Saelens, X.; Vanlandschoot, P.; Jou, W.M.; Fiers, W. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 1999, 5, 1157–1163. [Google Scholar] [CrossRef]
- De Filette, M.; Jou, W.M.; Birkett, A.; Lyons, K.; Schultz, B.; Tonkyro, A.; Resch, S.; Fiers, W. Universal influenza A vaccine: Optimization of M2-based constructs. Virology 2005, 337, 149–161. [Google Scholar] [CrossRef]
- Kim, M.-C.; Song, J.-M.; Eunju, O.; Kwon, Y.-M.; Lee, Y.-J.; Compans, R.; Kang, S.-M. Virus-like Particles Containing Multiple M2 Extracellular Domains Confer Improved Cross-protection Against Various Subtypes of Influenza Virus. Mol. Ther. 2013, 21, 485–492. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Lee, Y.-T.; Kim, M.-C.; Lee, Y.-N.; Kim, K.-H.; Ko, E.-J.; Song, J.-M.; Kang, S.-M. Cross-protective efficacy of influenza virus M2e containing virus-like particles is superior to hemagglutinin vaccines and variable depending on the genetic backgrounds of mice. Front. Immunol. 2017, 8, 1730. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, J.R.; Torian, U.; McCraw, D.M.; Harris, A.K. Structural studies of influenza virus RNPs by electron microscopy indicate molecular contortions within NP supra-structures. J. Struct. Biol. 2017, 197, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Portela, A.; Digard, P. The influenza virus nucleoprotein: A multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 2002, 83, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Babar, M.M.; Zaidi, N.-U.-S.S. Protein sequence conservation and stable molecular evolution reveals influenza virus nucleoprotein as a universal druggable target. Infect. Genet. Evol. 2015, 34, 200–210. [Google Scholar] [CrossRef]
- Yewdell, J.W.; Bennink, J.R.; Smith, G.L.; Moss, B. Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. USA 1985, 82, 1785–1789. [Google Scholar] [CrossRef]
- Zheng, M.; Liu, F.; Shen, Y.; Wang, S.; Xu, W.; Fang, F.; Sun, B.; Xie, Z.; Chen, Z. Cross-protection against influenza virus infection by intranasal administration of nucleoprotein-based vaccine with compound 48/80 adjuvant. Hum. Vaccines Immunother. 2015, 11, 397–406. [Google Scholar] [CrossRef]
- Stevens, M.P.; Barclay, W.S. The N-terminal extension of the influenza B virus nucleoprotein is not required for nuclear accumulation or the expression and replication of a model RNA. J. Virol. 1998, 72, 5307–5312. [Google Scholar] [CrossRef]
- Fernando, G.J.; Zhang, J.; Ng, H.-I.; Haigh, O.L.; Yukiko, S.R.; Kendall, M.A. Influenza nucleoprotein DNA vaccination by a skin targeted, dry coated, densely packed microprojection array (Nanopatch) induces potent antibody and CD8+ T cell responses. J. Contr. Rel. 2016, 237, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.K.; Zhang, H.; Tan, K.; Li, Z.; Liu, J.; Chan, P.; Li, S.; Chan, W.Y.; Au, S.W.; Joachimiak, A.; et al. Structure of the influenza virus A H5N1 nucleoprotein: Implications for RNA binding, oligomerization, and vaccine design. Faseb J. 2008, 22, 3638–3647. [Google Scholar] [CrossRef]
- Gao, X.M.; Liew, F.Y.; Tite, J.P. Identification and characterization of T helper epitopes in the nucleoprotein of influenza A virus. J. Immunol. 1989, 143, 3007–3014. [Google Scholar]
- Taylor, P.M.; Davey, J.; Howland, K.; Rothbard, J.B.; Askonas, B.A. Class I MHC molecules rather than other mouse genes dictate influenza epitope recognition by cytotoxic T cells. Immunogenetics 1987, 26, 267–272. [Google Scholar] [CrossRef]
- Rothbard, J.; Pemberton, R.; Bodmer, H.; Askonas, B.; Taylor, W. Identification of residues necessary for clonally specific recognition of a cytotoxic T cell determinant. Embo J. 1989, 8, 2321–2328. [Google Scholar] [CrossRef] [PubMed]
- Levi, R.; Arnon, R. Synthetic recombinant influenza vaccine induces efficient long-term immunity and cross-strain protection. Vaccine 1996, 14, 85–92. [Google Scholar] [CrossRef]
- Sharma, P.; Kumar, P.; Sharma, R. THE MAJOR HISTOCOMPATIBILITY COMPLEX: A REVIEW. Asian J. Pharm. Clin. Res. 2017, 10, 33–36. [Google Scholar] [CrossRef][Green Version]
- Thomas, P.G.; Keating, R.; Hulse-Post, D.J.; Doherty, P.C. Cell-mediated Protection in Influenza Infection. Emerg. Infect. Dis. 2006, 12, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-Y.; Kang, J.-O.; Chang, J. Nucleoprotein vaccine induces cross-protective cytotoxic T lymphocytes against both lineages of influenza B virus. Clin. Exp. Vaccine Res. 2019, 8, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Epstein, S.L.; Price, G.E. Cross-protective immunity to influenza A viruses. Expert Rev. Vaccines 2010, 9, 1325–1341. [Google Scholar] [CrossRef]
- Cookenham, T.; Lanzer, K.G.; Gage, E.; Lorenzo, E.C.; Carter, D.; Coler, R.N.; Baldwin, S.L.; Haynes, L.; Reiley, W.W.; Blackman, M.A. Vaccination of aged mice with adjuvanted recombinant influenza nucleoprotein enhances protective immunity. Vaccine 2020, 38, 5256–5267. [Google Scholar] [CrossRef]
- Jegaskanda, S.; Luke, C.; Hickman, H.D.; Sangster, M.Y.; Wieland-Alter, W.F.; McBride, J.M.; Yewdell, J.W.; Wright, P.F.; Treanor, J.; Rosenberger, C.M. Generation and protective ability of Influenza virus–specific antibody-dependent cellular cytotoxicity in humans elicited by vaccination, natural infection, and experimental challenge. J. Inf. Dis. 2016, 214, 945–952. [Google Scholar] [CrossRef]
- Gonzalez, S.F.; Jayasekera, J.P.; Carroll, M.C. Complement and natural antibody are required in the long-term memory response to influenza virus. Vaccine 2008, 26, I86–I93. [Google Scholar] [CrossRef]
- Terajima, M.; Cruz, J.; Co, M.D.T.; Lee, J.-H.; Kaur, K.; Wilson, P.C.; Ennis, F.A. Complement-Dependent Lysis of Influenza A Virus-Infected Cells by Broadly Cross-Reactive Human Monoclonal Antibodies. J. Virol. 2011, 85, 13463–13467. [Google Scholar] [CrossRef]
- Chenavas, S.; Estrozi, L.; Slama-Schwok, A.; Delmas, B.; Di Primo, C.; Baudin, F.; Li, X.; Crépin, T.; Ruigrok, R.W.H. Monomeric Nucleoprotein of Influenza A Virus. Plos Pathog. 2013, 9, e1003275. [Google Scholar] [CrossRef]
- Zepeda-Cervantes, J.; Ramírez-Jarquín, J.O.; Vaca, L. Interaction Between Virus-Like Particles (VLPs) and Pattern Recognition Receptors (PRRs) From Dendritic Cells (DCs): Toward Better Engineering of VLPs. Front. Immunol. 2020, 11, 1100. [Google Scholar] [CrossRef]
- Klimek, L.; Kündig, T.; Kramer, M.F.; Guethoff, S.; Jensen-Jarolim, E.; Schmidt-Weber, C.B.; Palomares, O.; Mohsen, M.O.; Jakob, T.; Bachmann, M. Virus-like particles (VLP) in prophylaxis and immunotherapy of allergic diseases. Allergo J. Int. 2018, 27, 245–255. [Google Scholar] [CrossRef]
- Yolshin, N.D.; Shaldzhyan, A.A.; Klotchenko, S.A. Efficient soluble expression and purification of influenza A and B nucleoproteins in E. coli. Mir J. 2021, 6, 43–48. [Google Scholar] [CrossRef]
- Tavakoli, S.; Bandehpour, M.; Soleimanifar, Z.; Goudarzi, M.; Kazemi, B. Cloning and Expression of Recombinant Nucleoprotein of Influenza H1N1. Nov. Biomed. 2015, 3, 53–56. [Google Scholar]
- Woo, E.J. Allergic Reactions After Egg-Free Recombinant Influenza Vaccine: Reports to the US Vaccine Adverse Event Reporting System. Clin. Infect. Dis. 2014, 60, 777–780. [Google Scholar] [CrossRef]
- Chung, E.H. Vaccine allergies. Clin. Exp. Vaccine Res. 2014, 3, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.; Hassan, S.S.; Yap, W. Expression of surface-bound nonstructural 1 (NS1) protein of influenza virus A H5N1 on Lactobacillus casei strain C1. Lett. Appl. Microbiol. 2017, 64, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, S.; Robinson, J.M.; Cunningham, G.; Iqbal, R.; Larsen, S. The complexity and cost of vaccine manufacturing—An overview. Vaccine 2017, 35, 4064–4071. [Google Scholar] [CrossRef]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Gómez, A.; Mirón, A.S.; Garcia-Camacho, F.; Grima, E.M.; Chisti, Y. Protein production using the baculovirus-insect cell expression system. Biotechnol. Prog. 2013, 30, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-Y.; Yeh, Y.-C.; Yang, Y.-C.; Chou, C.; Liu, M.-T.; Wu, H.-S.; Chan, J.-T.; Hsiao, P.-W. Mammalian expression of virus-like particles for advanced mimicry of authentic influenza virus. PLoS ONE 2010, 5, e9784. [Google Scholar] [CrossRef] [PubMed]
- Griffith, D.A.; Delipala, C.; Leadsham, J.; Jarvis, S.M.; Oesterhelt, D. A novel yeast expression system for the overproduction of quality-controlled membrane proteins. FEBS Lett. 2003, 553, 45–50. [Google Scholar] [CrossRef]
- Gordon, E.; Horsefield, R.; Swarts, H.G.P.; Pont, J.J.H.H.M.d.; Neutze, R.; Snijder, A. Effective high-throughput overproduction of membrane proteins in Escherichia coli. Protein Expr. Purif. 2008, 62, 1–8. [Google Scholar] [CrossRef]
- Koehn, J.; Hunt, I. High-Throughput Protein Production (HTPP): A review of enabling technologies to expedite protein production. Methods Mol. Biol. 2009, 498, 1–18. [Google Scholar]
- Valkenburg, S.A.; Venturi, V.; Dang, T.H.Y.; Bird, N.L.; Doherty, P.C.; Turner, S.J.; Davenport, M.P.; Kedzierska, K. Early priming minimizes the age-related immune compromise of CD8+ T cell diversity and function. PLoS Pathog 2012, 8, e1002544. [Google Scholar] [CrossRef]
- Doherty, P.C.; Turner, S.J.; Webby, R.G.; Thomas, P.G. Influenza and the challenge for immunology. Nat. Immunol. 2006, 7, 449–455. [Google Scholar] [CrossRef]
- Pizzolla, A.; Nguyen, T.H.O.; Smith, J.M.; Brooks, A.G.; Kedzierska, K.; Heath, W.R.; Reading, P.; Wakim, L.M. Resident memory CD8+T cells in the upper respiratory tract prevent pulmonary influenza virus infection. Sci. Immunol. 2017, 2, eaam6970. [Google Scholar] [CrossRef]
- Kreijtz, J.H.C.M.; de Mutsert, G.; van Baalen, C.A.; Fouchier, R.A.M.; Osterhaus, A.D.M.E.; Rimmelzwaan, G.F. Cross-Recognition of Avian H5N1 Influenza Virus by Human Cytotoxic T-Lymphocyte Populations Directed to Human Influenza A Virus. J. Virol. 2008, 82, 5161–5166. [Google Scholar] [CrossRef]
- Lee, L.Y.-H.; Ha, D.L.A.; Simmons, C.P.; De Jong, M.D.; Chau, N.V.V.; Schumacher, R.; Peng, Y.C.; McMichael, A.J.; Farrar, J.; Smith, G.L.; et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Investig. 2008, 118, 3478–3490. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.; Izzard, L.; Hurt, A.C. A Review of DNA Vaccines Against Influenza. Front. Immunol. 2018, 9, 1568. [Google Scholar] [CrossRef]
- Heinen, P.P.; Rijsewijk, F.A.; de Boer-Luijtze, E.A.; Bianchi, A.T. Vaccination of pigs with a DNA construct expressing an influenza virus M2–nucleoprotein fusion protein exacerbates disease after challenge with influenza A virusThe GenBank accession numbers of the sequences reported in this paper are AF385293, AF385294, AF385295, AF385296 and AF385297. J. Gen. Virol. 2002, 83, 1851–1859. [Google Scholar] [PubMed]
- Yao, Y.; Wang, H.; Chen, J.; Shao, Z.; He, B.; Chen, J.; Lan, J.; Chen, Q.; Chen, Z. Protection against homo and hetero-subtypic inflfluenza A virus by optimized M2e DNA vaccine. Emerg. Microbes Infect. 2019, 8, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Wu, T.-L.; Lasaro, M.O.; Latimer, B.P.; Parzych, E.M.; Bian, A.; Li, Y.; Li, H.; Erikson, J.; Xiang, Z. A universal influenza A vaccine based on adenovirus expressing matrix-2 ectodomain and nucleoprotein protects mice from lethal challenge. Molec. Ther. 2010, 18, 2182–2188. [Google Scholar] [CrossRef]
- Lalor, P.A.; Webby, R.J.; Morrow, J.; Rusalov, D.; Kaslow, D.C.; Rolland, A.; Smith, L.R. Plasmid DNA-Based Vaccines Protect Mice and Ferrets against Lethal Challenge with A/Vietnam/1203/04 (H5N1) Influenza Virus. J. Inf. Dis. 2008, 197, 1643–1651. [Google Scholar] [CrossRef]
- Lo, C.-Y.; Wu, Z.; Misplon, J.A.; Price, G.E.; Pappas, C.; Kong, W.-P.; Tumpey, T.M.; Epstein, S.L. Comparison of vaccines for induction of heterosubtypic immunity to influenza A virus: Cold-adapted vaccine versus DNA prime-adenovirus boost strategies. Vaccine 2008, 26, 2062–2072. [Google Scholar] [CrossRef]
- Khan, K.H. DNA vaccines: Roles against diseases. Germs 2013, 3, 26–35. [Google Scholar] [CrossRef]
- Barouch, D.H.; Nabel, G.J. Adenovirus Vector-Based Vaccines for Human Immunodeficiency Virus Type 1. Hum. Gene 2005, 16, 149–156. [Google Scholar] [CrossRef]
- Corbett, K.S.; Edwards, D.K.; Leist, S.R.; Abiona, O.M.; Boyoglu-Barnum, S.; Gillespie, R.A.; Himansu, S.; Schäfer, A.; Ziwaaol, C.T.; DiPiazza, A.T.; et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 596, 567–587. [Google Scholar] [CrossRef]
- Graham, B.S. Rapid COVID-19 vaccine development. Science 2020, 368, 945–946. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zheng, D.; Zhang, W.; Fang, F.; Wang, H.; Sun, Y.; Ding, Y.; Xu, C.; Chen, Q.; Zhang, H.; et al. Induction of cross-protection against influenza A virus by DNA prime-intranasal protein boost strategy based on nucleoprotein. Virol. J. 2012, 9, 286. [Google Scholar] [CrossRef] [PubMed]
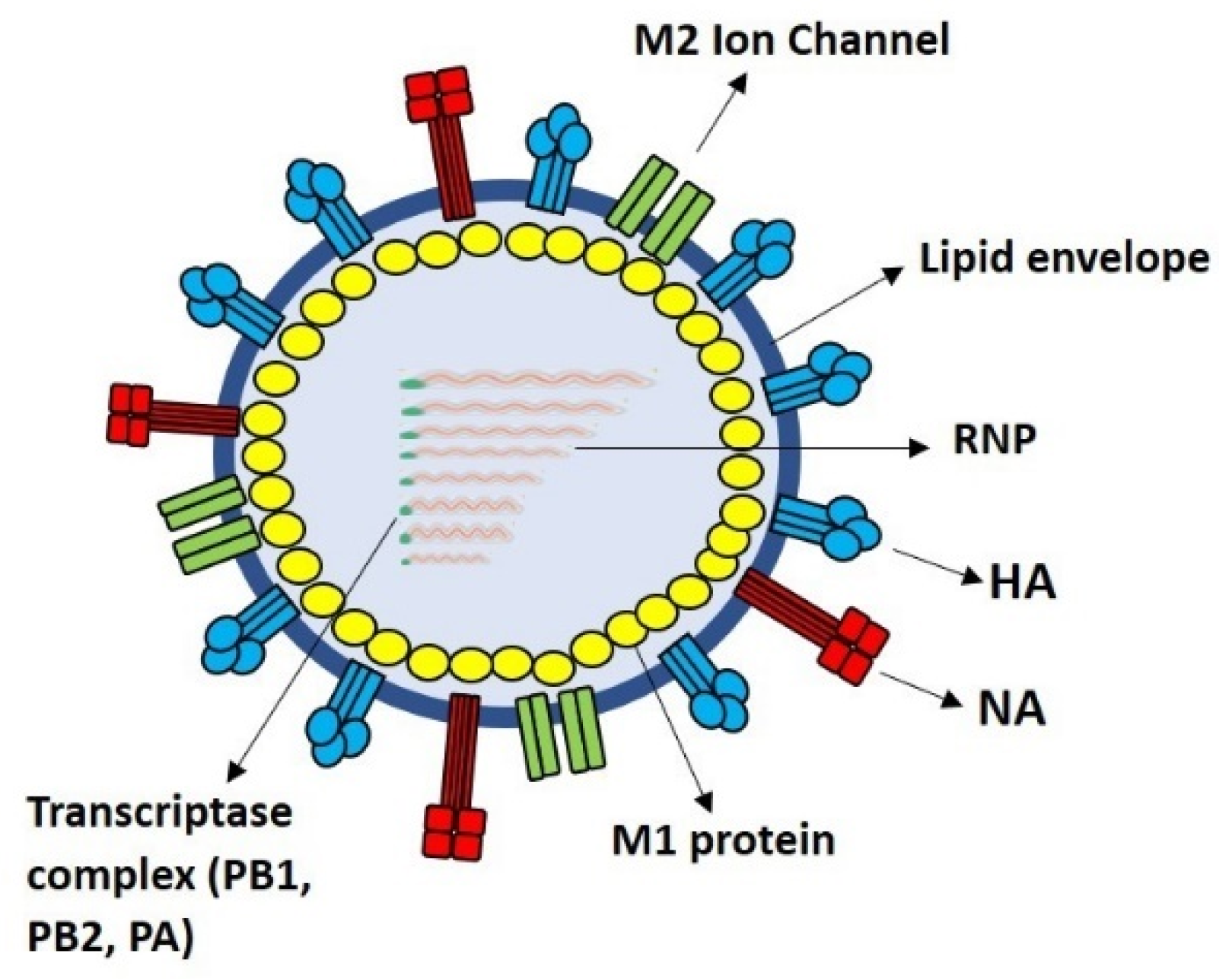
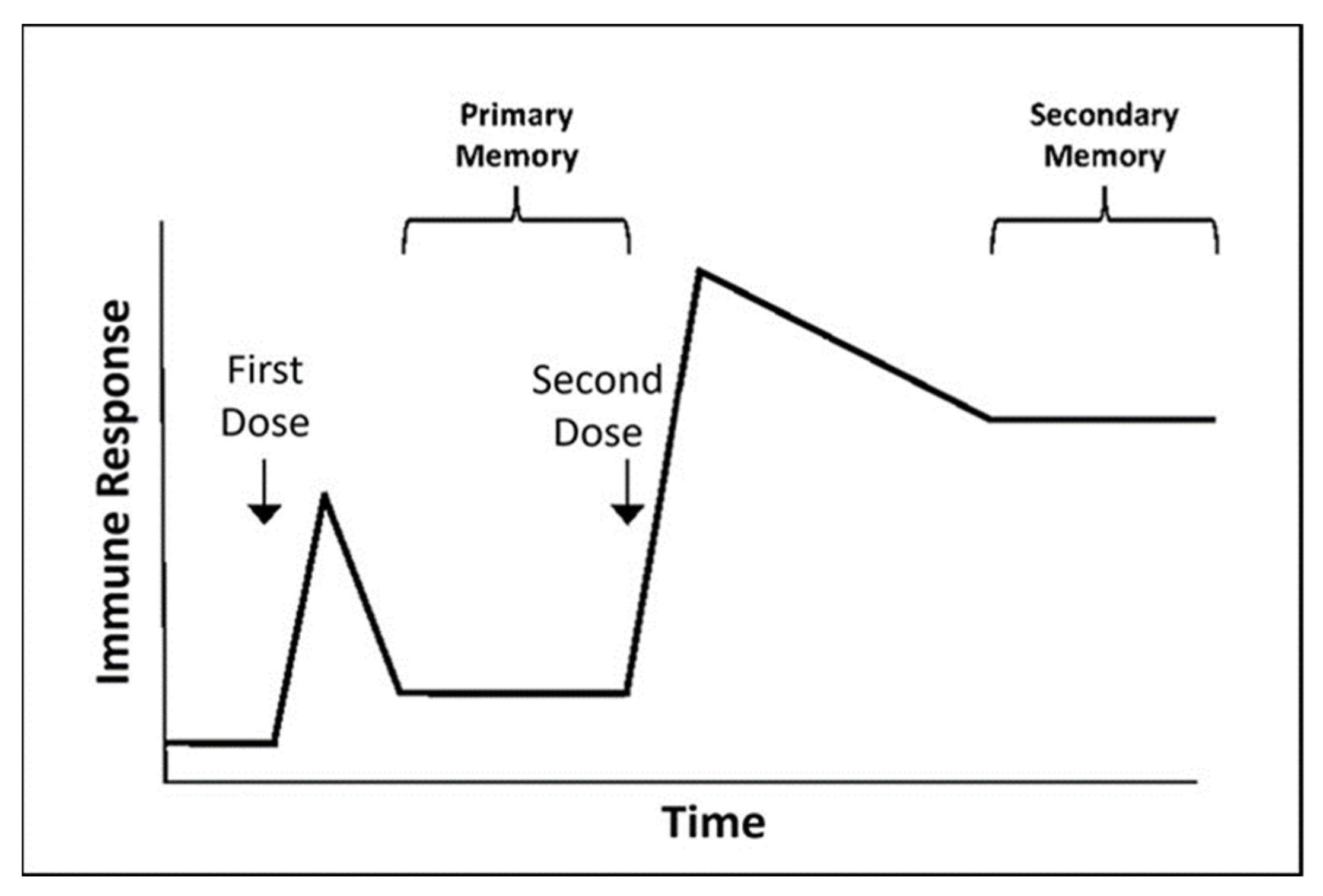

| LAIV | Inactivated Influenza Vaccine | Recombinant HA Quadrivalent Vaccine (e.g., Flublok Quadrivalent) |
|---|---|---|
| Induces multifaceted immune responses including humoral (antibody) and T-cell responses. | Inclusive of viral major surface glycoproteins, i.e., hemagglutinin and neuraminidase. Induces mostly antibody responses. | Contains three times more HA than the standard inactivated influenza vaccine. It induces the production of anti-HA antibodies to prevent influenza virus infections. |
| Licensed for use in recipients aged 2 years and older. | Licensed for use in recipients aged 6 months and older. | Licensed for use in adults aged 18 years old and older. |
| Administered intranasally (mimicking natural infection). | Administered intramuscularly. | Administered intramuscularly. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, M.P.; Tan, W.S.; Mohamed Alitheen, N.B.; Yap, W.B. M2e-Based Influenza Vaccines with Nucleoprotein: A Review. Vaccines 2021, 9, 739. https://doi.org/10.3390/vaccines9070739
Tan MP, Tan WS, Mohamed Alitheen NB, Yap WB. M2e-Based Influenza Vaccines with Nucleoprotein: A Review. Vaccines. 2021; 9(7):739. https://doi.org/10.3390/vaccines9070739
Chicago/Turabian StyleTan, Mei Peng, Wen Siang Tan, Noorjahan Banu Mohamed Alitheen, and Wei Boon Yap. 2021. "M2e-Based Influenza Vaccines with Nucleoprotein: A Review" Vaccines 9, no. 7: 739. https://doi.org/10.3390/vaccines9070739
APA StyleTan, M. P., Tan, W. S., Mohamed Alitheen, N. B., & Yap, W. B. (2021). M2e-Based Influenza Vaccines with Nucleoprotein: A Review. Vaccines, 9(7), 739. https://doi.org/10.3390/vaccines9070739






