The HIV-1 Antisense Gene ASP: The New Kid on the Block
Abstract
1. Introduction: De Novo Creation of Genes
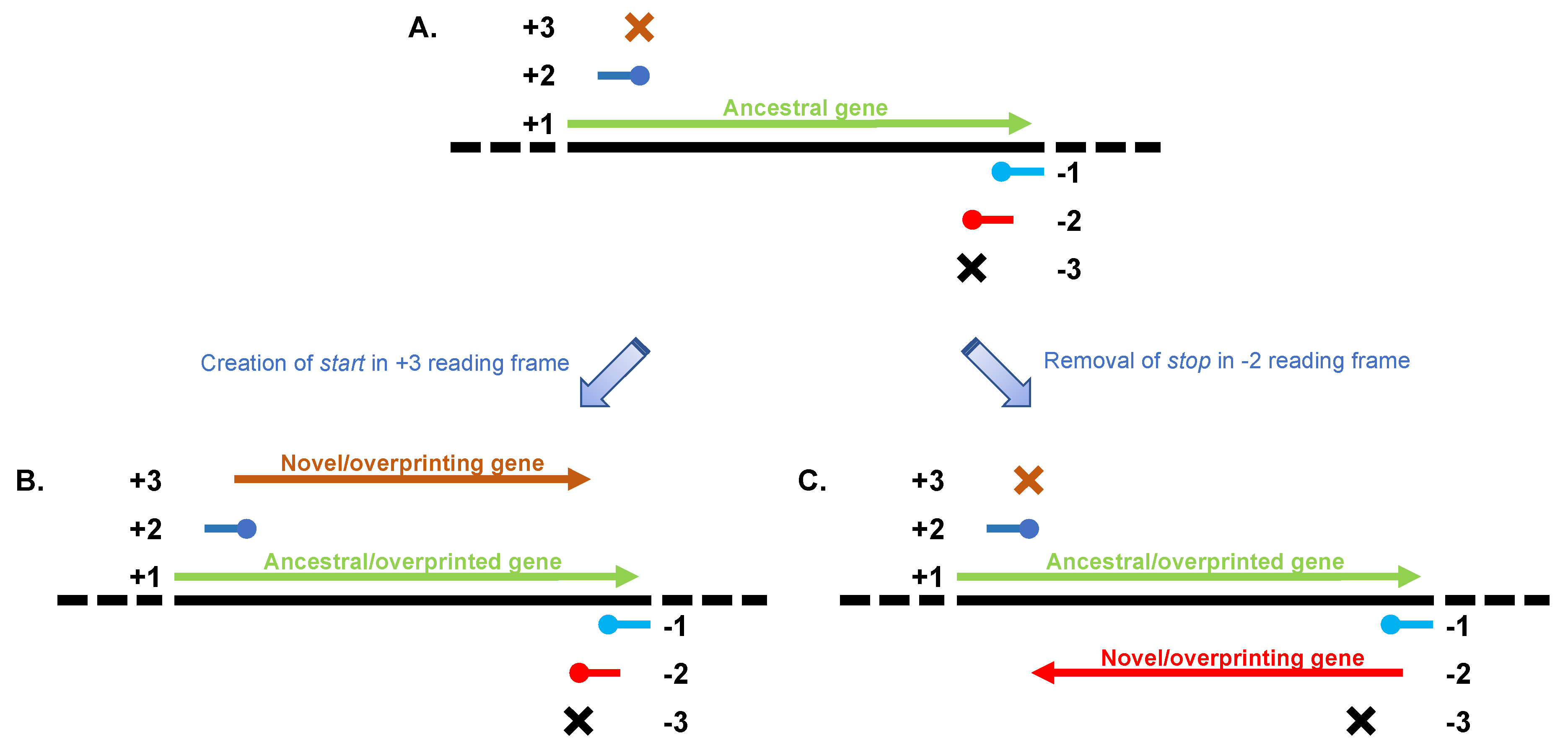
2. Overprinting in Viral Genomes
3. A Special Kind of Overprinting: Antisense Genes
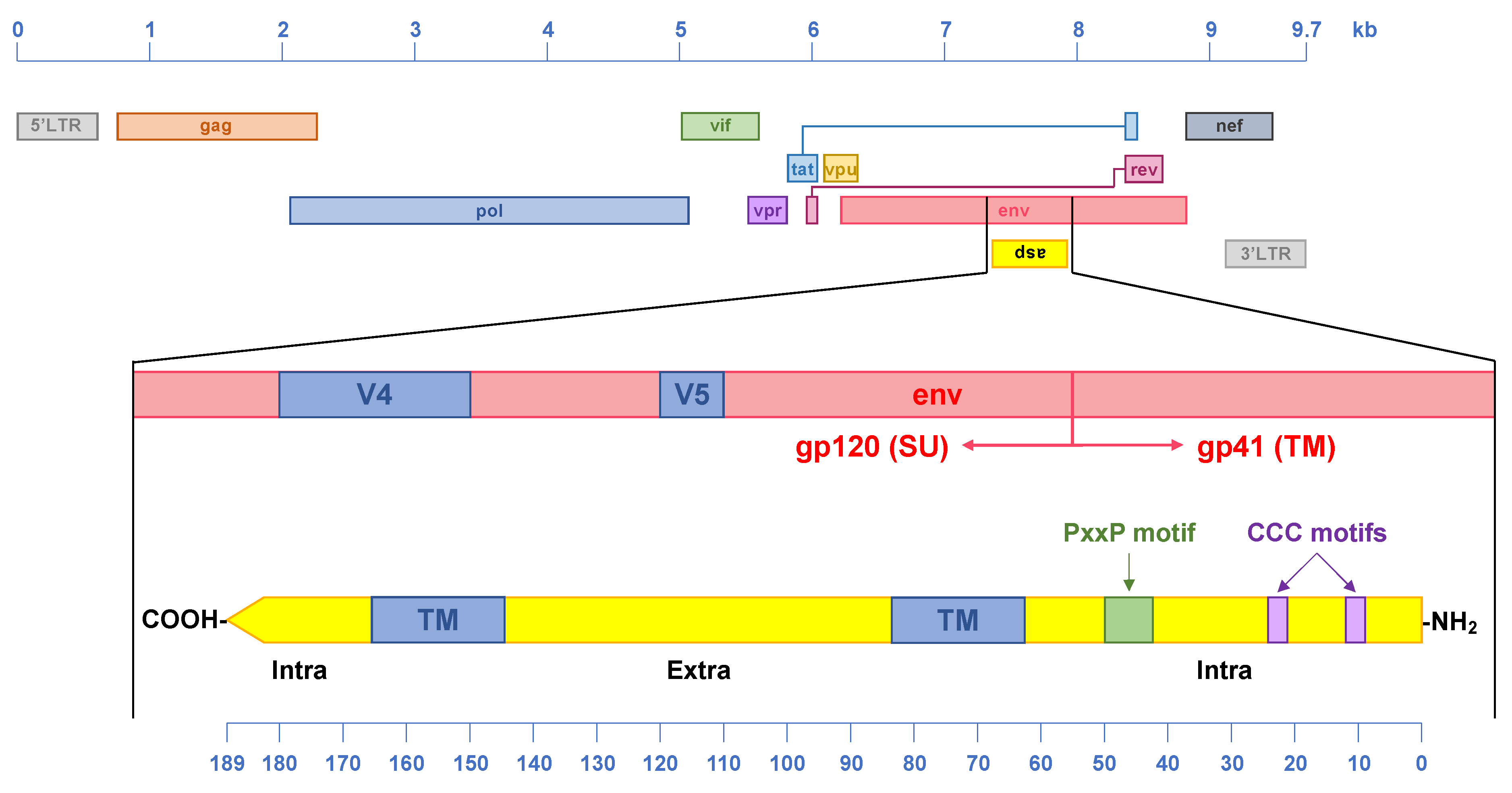
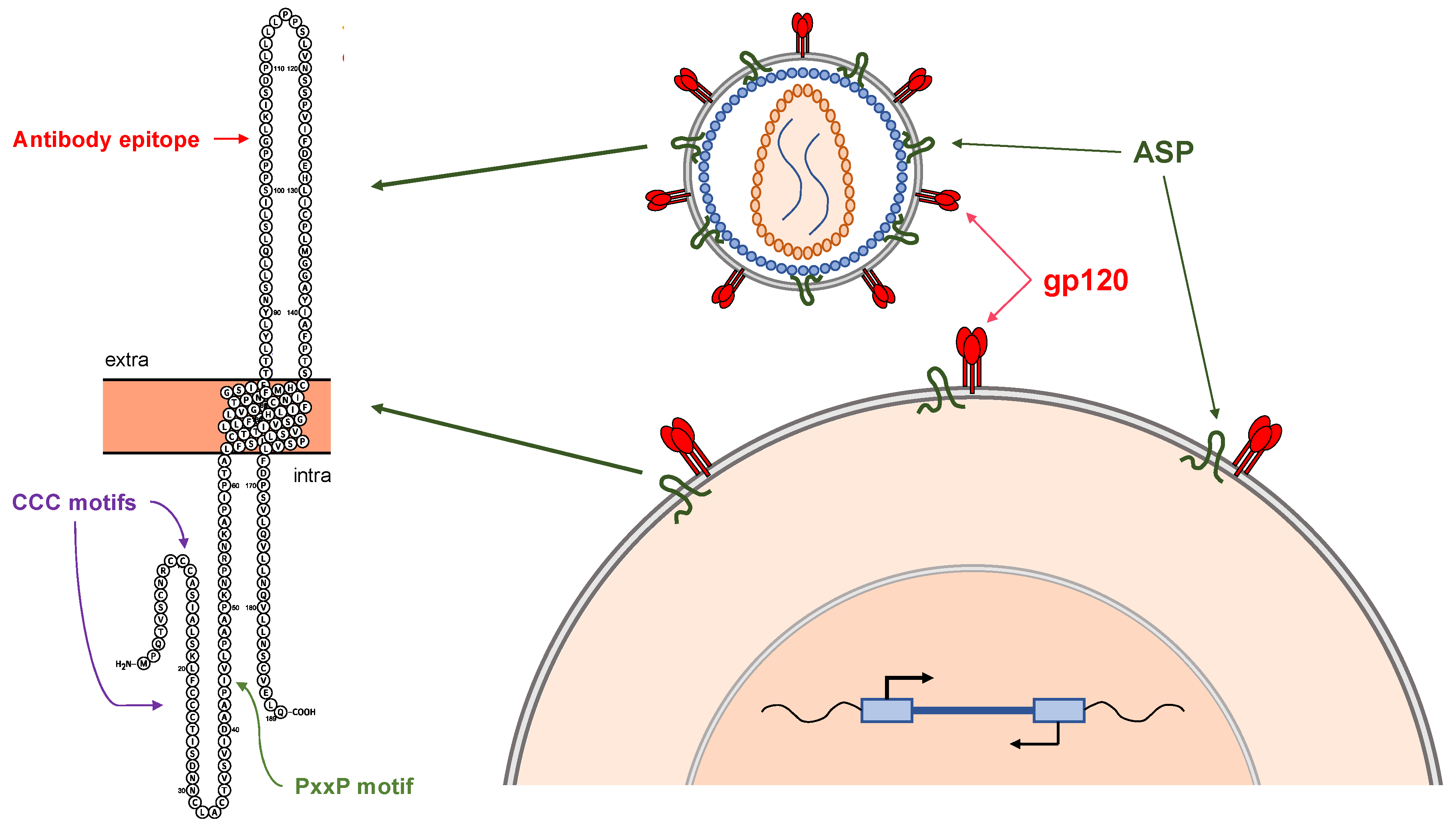
4. The HIV-1 Antisense Protein, ASP
5. ASP Expression in HIV-1 Infected Individuals
6. Expression and Functional Role of ASP in In Vitro Models
7. Origin, Conservation and Evolution of ASP
8. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Long, M.; Betran, E.; Thornton, K.; Wang, W. The origin of new genes: Glimpses from the young and old. Nat. Rev. Genet. 2003, 4, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F. Evolution and tinkering. Science 1977, 196, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Sorek, R. The birth of new exons: Mechanisms and evolutionary consequences. Rna 2007, 13, 1603–1608. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Zhang, Y.; Wang, Z.; Zhang, Y.; Cao, C.; Zhang, P.W.; Lu, S.J.; Li, X.M.; Yu, Q.; Zheng, X.; et al. A human-specific de novo protein-coding gene associated with human brain functions. PLoS Comput. Biol. 2010, 6, e1000734. [Google Scholar] [CrossRef]
- Rancurel, C.; Khosravi, M.; Dunker, A.K.; Romero, P.R.; Karlin, D. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J. Virol. 2009, 83, 10719–10736. [Google Scholar] [CrossRef]
- Sabath, N.; Wagner, A.; Karlin, D. Evolution of viral proteins originated de novo by overprinting. Mol. Biol. Evol. 2012, 29, 3767–3780. [Google Scholar] [CrossRef]
- Fischer, D.; Eisenberg, D. Finding families for genomic ORFans. Bioinformatics 1999, 15, 759–762. [Google Scholar] [CrossRef]
- Wilson, G.A.; Bertrand, N.; Patel, Y.; Hughes, J.B.; Feil, E.J.; Field, D. Orphans as taxonomically restricted and ecologically important genes. Microbiology 2005, 151, 2499–2501. [Google Scholar] [CrossRef]
- Yooseph, S.; Sutton, G.; Rusch, D.B.; Halpern, A.L.; Williamson, S.J.; Remington, K.; Eisen, J.A.; Heidelberg, K.B.; Manning, G.; Li, W.; et al. The Sorcerer II Global Ocean Sampling expedition: Expanding the universe of protein families. PLoS Biol. 2007, 5, e16. [Google Scholar] [CrossRef] [PubMed]
- Delaye, L.; Deluna, A.; Lazcano, A.; Becerra, A. The origin of a novel gene through overprinting in Escherichia coli. BMC Evol. Biol. 2008, 8, 31. [Google Scholar] [CrossRef]
- Ribrioux, S.; Brungger, A.; Baumgarten, B.; Seuwen, K.; John, M.R. Bioinformatics prediction of overlapping frameshifted translation products in mammalian transcripts. BMC Genom. 2008, 9, 122. [Google Scholar] [CrossRef]
- Chung, W.Y.; Wadhawan, S.; Szklarczyk, R.; Pond, S.K.; Nekrutenko, A. A first look at ARFome: Dual-coding genes in mammalian genomes. PLoS Comput. Biol. 2007, 3, e91. [Google Scholar] [CrossRef]
- McVeigh, A.; Fasano, A.; Scott, D.A.; Jelacic, S.; Moseley, S.L.; Robertson, D.C.; Savarino, S.J. IS1414, an Escherichia coli insertion sequence with a heat-stable enterotoxin gene embedded in a transposase-like gene. Infect. Immun. 2000, 68, 5710–5715. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Michel, A.M.; Choudhury, K.R.; Firth, A.E.; Ingolia, N.T.; Atkins, J.F.; Baranov, P.V. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome Res. 2012, 22, 2219–2229. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, D.; Lapointe, C.; Bissonnette, C.; Tremblay, G.; Motard, J.; Roucou, X. An out-of-frame overlapping reading frame in the ataxin-1 coding sequence encodes a novel ataxin-1 interacting protein. J. Biol. Chem. 2013, 288, 21824–21835. [Google Scholar] [CrossRef] [PubMed]
- Vanderperre, B.; Lucier, J.F.; Bissonnette, C.; Motard, J.; Tremblay, G.; Vanderperre, S.; Wisztorski, M.; Salzet, M.; Boisvert, F.M.; Roucou, X. Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLoS ONE 2013, 8, e70698. [Google Scholar] [CrossRef] [PubMed]
- Fellner, L.; Simon, S.; Scherling, C.; Witting, M.; Schober, S.; Polte, C.; Schmitt-Kopplin, P.; Keim, D.A.; Scherer, S.; Neuhaus, K. Evidence for the recent origin of a bacterial protein-coding, overlapping orphan gene by evolutionary overprinting. BMC Evol. Biol. 2015, 15, 283. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, G.; Zhang, Y.; Xu, S.; Zhao, R.; Zhan, Z.; Li, X.; Ding, Y.; Yang, S.; Wang, W. On the origin of new genes in Drosophila. Genome Res. 2008, 18, 1446–1455. [Google Scholar] [CrossRef]
- Toll-Riera, M.; Bosch, N.; Bellora, N.; Castelo, R.; Armengol, L.; Estivill, X.; Alba, M.M. Origin of primate orphan genes: A comparative genomics approach. Mol. Biol. Evol. 2009, 26, 603–612. [Google Scholar] [CrossRef]
- Ekman, D.; Elofsson, A. Identifying and quantifying orphan protein sequences in fungi. J. Mol. Biol. 2010, 396, 396–405. [Google Scholar] [CrossRef]
- Belshaw, R.; Pybus, O.G.; Rambaut, A. The evolution of genome compression and genomic novelty in RNA viruses. Genome Res. 2007, 17, 1496–1504. [Google Scholar] [CrossRef]
- Chirico, N.; Vianelli, A.; Belshaw, R. Why genes overlap in viruses. Proc. Biol. Sci. 2010, 277, 3809–3817. [Google Scholar] [CrossRef] [PubMed]
- Barrell, B.G.; Air, G.M.; Hutchison, C.A., 3rd. Overlapping genes in bacteriophage phiX174. Nature 1976, 264, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Pavesi, A. New insights into the evolutionary features of viral overlapping genes by discriminant analysis. Virology 2020, 546, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Pavesi, A.; De Iaco, B.; Granero, M.I.; Porati, A. On the informational content of overlapping genes in prokaryotic and eukaryotic viruses. J. Mol. Evol. 1997, 44, 625–631. [Google Scholar] [CrossRef]
- Pavesi, A.; Magiorkinis, G.; Karlin, D.G. Viral proteins originated de novo by overprinting can be identified by codon usage: Application to the “gene nursery” of Deltaretroviruses. PLoS Comput. Biol. 2013, 9, e1003162. [Google Scholar] [CrossRef]
- Pavesi, A.; Vianelli, A.; Chirico, N.; Bao, Y.; Blinkova, O.; Belshaw, R.; Firth, A.; Karlin, D. Overlapping genes and the proteins they encode differ significantly in their sequence composition from non-overlapping genes. PLoS ONE 2018, 13, e0202513. [Google Scholar] [CrossRef]
- Pavesi, A. Asymmetric evolution in viral overlapping genes is a source of selective protein adaptation. Virology 2019, 532, 39–47. [Google Scholar] [CrossRef]
- Carter, C.W., Jr. Simultaneous codon usage, the origin of the proteome, and the emergence of de-novo proteins. Curr. Opin. Struct. Biol. 2021, 68, 142–148. [Google Scholar] [CrossRef]
- Karlin, D.; Ferron, F.; Canard, B.; Longhi, S. Structural disorder and modular organization in Paramyxovirinae N and P. J. Gen. Virol. 2003, 84, 3239–3252. [Google Scholar] [CrossRef]
- Willis, S.; Masel, J. Gene Birth Contributes to Structural Disorder Encoded by Overlapping Genes. Genetics 2018, 210, 303–313. [Google Scholar] [CrossRef]
- Wensman, J.J.; Munir, M.; Thaduri, S.; Hornaeus, K.; Rizwan, M.; Blomstrom, A.L.; Briese, T.; Lipkin, W.I.; Berg, M. The X proteins of bornaviruses interfere with type I interferon signalling. J. Gen. Virol. 2013, 94, 263–269. [Google Scholar] [CrossRef]
- Van Knippenberg, I.; Carlton-Smith, C.; Elliott, R.M. The N-terminus of Bunyamwera orthobunyavirus NSs protein is essential for interferon antagonism. J. Gen. Virol. 2010, 91, 2002–2006. [Google Scholar] [CrossRef]
- Li, F.; Ding, S.W. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 2006, 60, 503–531. [Google Scholar] [CrossRef]
- Chellappan, P.; Vanitharani, R.; Fauquet, C.M. MicroRNA-binding viral protein interferes with Arabidopsis development. Proc. Natl. Acad. Sci. USA 2005, 102, 10381–10386. [Google Scholar] [CrossRef]
- Chen, W.; Calvo, P.A.; Malide, D.; Gibbs, J.; Schubert, U.; Bacik, I.; Basta, S.; O’Neill, R.; Schickli, J.; Palese, P.; et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 2001, 7, 1306–1312. [Google Scholar] [CrossRef]
- Boehme, K.W.; Hammer, K.; Tollefson, W.C.; Konopka-Anstadt, J.L.; Kobayashi, T.; Dermody, T.S. Nonstructural protein sigma1s mediates reovirus-induced cell cycle arrest and apoptosis. J. Virol. 2013, 87, 12967–12979. [Google Scholar] [CrossRef]
- Taliansky, M.; Roberts, I.M.; Kalinina, N.; Ryabov, E.V.; Raj, S.K.; Robinson, D.J.; Oparka, K.J. An umbraviral protein, involved in long-distance RNA movement, binds viral RNA and forms unique, protective ribonucleoprotein complexes. J. Virol. 2003, 77, 3031–3040. [Google Scholar] [CrossRef]
- Singh, M. No vaccine against HIV yet--are we not perfectly equipped? Virol. J. 2006, 3, 60. [Google Scholar] [CrossRef][Green Version]
- Pluta, A.; Jaworski, J.P.; Douville, R.N. Regulation of Expression and Latency in BLV and HTLV. Viruses 2020, 12, 1079. [Google Scholar] [CrossRef]
- Matsuoka, M.; Mesnard, J.M. HTLV-1 bZIP factor: The key viral gene for pathogenesis. Retrovirology 2020, 17, 2. [Google Scholar] [CrossRef] [PubMed]
- Satou, Y.; Yasunaga, J.; Yoshida, M.; Matsuoka, M. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc. Natl. Acad. Sci. USA 2006, 103, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, M.; Satou, Y.; Yasunaga, J.; Fujisawa, J.; Matsuoka, M. Transcriptional control of spliced and unspliced human T-cell leukemia virus type 1 bZIP factor (HBZ) gene. J. Virol. 2008, 82, 9359–9368. [Google Scholar] [CrossRef] [PubMed]
- Laverdure, S.; Polakowski, N.; Hoang, K.; Lemasson, I. Permissive Sense and Antisense Transcription from the 5′ and 3′ Long Terminal Repeats of Human T-Cell Leukemia Virus Type 1. J. Virol. 2016, 90, 3600–3610. [Google Scholar] [CrossRef] [PubMed]
- Clerc, I.; Polakowski, N.; Andre-Arpin, C.; Cook, P.; Barbeau, B.; Mesnard, J.M.; Lemasson, I. An interaction between the human T cell leukemia virus type 1 basic leucine zipper factor (HBZ) and the KIX domain of p300/CBP contributes to the down-regulation of tax-dependent viral transcription by HBZ. J. Biol. Chem. 2008, 283, 23903–23913. [Google Scholar] [CrossRef]
- Miyazaki, M.; Yasunaga, J.; Taniguchi, Y.; Tamiya, S.; Nakahata, T.; Matsuoka, M. Preferential selection of human T-cell leukemia virus type 1 provirus lacking the 5′ long terminal repeat during oncogenesis. J. Virol. 2007, 81, 5714–5723. [Google Scholar] [CrossRef]
- Taniguchi, Y.; Nosaka, K.; Yasunaga, J.; Maeda, M.; Mueller, N.; Okayama, A.; Matsuoka, M. Silencing of human T-cell leukemia virus type I gene transcription by epigenetic mechanisms. Retrovirology 2005, 2, 64. [Google Scholar] [CrossRef]
- Fan, J.; Ma, G.; Nosaka, K.; Tanabe, J.; Satou, Y.; Koito, A.; Wain-Hobson, S.; Vartanian, J.P.; Matsuoka, M. APOBEC3G generates nonsense mutations in human T-cell leukemia virus type 1 proviral genomes in vivo. J. Virol. 2010, 84, 7278–7287. [Google Scholar] [CrossRef]
- Arnold, J.; Zimmerman, B.; Li, M.; Lairmore, M.D.; Green, P.L. Human T-cell leukemia virus type-1 antisense-encoded gene, Hbz, promotes T-lymphocyte proliferation. Blood 2008, 112, 3788–3797. [Google Scholar] [CrossRef]
- Satou, Y.; Yasunaga, J.; Zhao, T.; Yoshida, M.; Miyazato, P.; Takai, K.; Shimizu, K.; Ohshima, K.; Green, P.L.; Ohkura, N.; et al. HTLV-1 bZIP factor induces T-cell lymphoma and systemic inflammation in vivo. PLoS Pathog. 2011, 7, e1001274. [Google Scholar] [CrossRef]
- Zhao, T.; Satou, Y.; Matsuoka, M. Development of T cell lymphoma in HTLV-1 bZIP factor and Tax double transgenic mice. Arch. Virol. 2014, 159, 1849–1856. [Google Scholar] [CrossRef]
- Savoret, J.; Mesnard, J.M.; Gross, A.; Chazal, N. Antisense Transcripts and Antisense Protein: A New Perspective on Human Immunodeficiency Virus Type 1. Front. Microbiol. 2020, 11, 625941. [Google Scholar] [CrossRef]
- Miura, M.; Yasunaga, J.; Tanabe, J.; Sugata, K.; Zhao, T.; Ma, G.; Miyazato, P.; Ohshima, K.; Kaneko, A.; Watanabe, A.; et al. Characterization of simian T-cell leukemia virus type 1 in naturally infected Japanese macaques as a model of HTLV-1 infection. Retrovirology 2013, 10, 118. [Google Scholar] [CrossRef]
- Briquet, S.; Richardson, J.; Vanhee-Brossollet, C.; Vaquero, C. Natural antisense transcripts are detected in different cell lines and tissues of cats infected with feline immunodeficiency virus. Gene 2001, 267, 157–164. [Google Scholar] [CrossRef]
- Miller, R.H. Human immunodeficiency virus may encode a novel protein on the genomic DNA plus strand. Science 1988, 239, 1420–1422. [Google Scholar] [CrossRef]
- Casino, A.; Cipollaro, M.; Guerrini, A.M.; Mastrocinque, G.; Spena, A.; Scarlato, V. Coding capacity of complementary DNA strands. Nucleic Acids Res. 1981, 9, 1499–1518. [Google Scholar] [CrossRef][Green Version]
- Affram, Y.; Zapata, J.C.; Gholizadeh, Z.; Tolbert, W.D.; Zhou, W.; Iglesias-Ussel, M.D.; Pazgier, M.; Ray, K.; Latinovic, O.S.; Romerio, F. The HIV-1 Antisense Protein ASP Is a Transmembrane Protein of the Cell Surface and an Integral Protein of the Viral Envelope. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Clerc, I.; Laverdure, S.; Torresilla, C.; Landry, S.; Borel, S.; Vargas, A.; Arpin-Andre, C.; Gay, B.; Briant, L.; Gross, A.; et al. Polarized expression of the membrane ASP protein derived from HIV-1 antisense transcription in T cells. Retrovirology 2011, 8, 74. [Google Scholar] [CrossRef] [PubMed]
- Saksela, K.; Cheng, G.; Baltimore, D. Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 1995, 14, 484–491. [Google Scholar] [CrossRef]
- Zapata, J.C.; Campilongo, F.; Barclay, R.A.; DeMarino, C.; Iglesias-Ussel, M.D.; Kashanchi, F.; Romerio, F. The Human Immunodeficiency Virus 1 ASP RNA promotes viral latency by recruiting the Polycomb Repressor Complex 2 and promoting nucleosome assembly. Virology 2017, 506, 34–44. [Google Scholar] [CrossRef]
- Mancarella, A.; Procopio, F.A.; Achsel, T.; De Crignis, E.; Foley, B.T.; Corradin, G.; Bagni, C.; Pantaleo, G.; Graziosi, C. Detection of antisense protein (ASP) RNA transcripts in individuals infected with human immunodeficiency virus type 1 (HIV-1). J. Gen. Virol. 2019, 100, 863–876. [Google Scholar] [CrossRef] [PubMed]
- Vanhee-Brossollet, C.; Thoreau, H.; Serpente, N.; D’Auriol, L.; Levy, J.P.; Vaquero, C. A natural antisense RNA derived from the HIV-1 env gene encodes a protein which is recognized by circulating antibodies of HIV+ individuals. Virology 1995, 206, 196–202. [Google Scholar] [CrossRef]
- Savoret, J.; Chazal, N.; Moles, J.P.; Tuaillon, E.; Boufassa, F.; Meyer, L.; Lecuroux, C.; Lambotte, O.; Van De Perre, P.; Mesnard, J.M.; et al. A Pilot Study of the Humoral Response against the AntiSense Protein (ASP) in HIV-1-Infected Patients. Front. Microbiol. 2020, 11, 20. [Google Scholar] [CrossRef]
- Champiat, S.; Raposo, R.A.; Maness, N.J.; Lehman, J.L.; Purtell, S.E.; Hasenkrug, A.M.; Miller, J.C.; Dean, H.; Koff, W.C.; Hong, M.A.; et al. Influence of HAART on alternative reading frame immune responses over the course of HIV-1 infection. PLoS ONE 2012, 7, e39311. [Google Scholar] [CrossRef][Green Version]
- Bet, A.; Maze, E.A.; Bansal, A.; Sterrett, S.; Gross, A.; Graff-Dubois, S.; Samri, A.; Guihot, A.; Katlama, C.; Theodorou, I.; et al. The HIV-1 antisense protein (ASP) induces CD8 T cell responses during chronic infection. Retrovirology 2015, 12, 15. [Google Scholar] [CrossRef]
- Berger, C.T.; Llano, A.; Carlson, J.M.; Brumme, Z.L.; Brockman, M.A.; Cedeno, S.; Harrigan, P.R.; Kaufmann, D.E.; Heckerman, D.; Meyerhans, A.; et al. Immune screening identifies novel T cell targets encoded by antisense reading frames of HIV-1. J. Virol. 2015, 89, 4015–4019. [Google Scholar] [CrossRef]
- Michael, N.L.; Vahey, M.T.; d’Arcy, L.; Ehrenberg, P.K.; Mosca, J.D.; Rappaport, J.; Redfield, R.R. Negative-strand RNA transcripts are produced in human immunodeficiency virus type 1-infected cells and patients by a novel promoter downregulated by Tat. J. Virol. 1994, 68, 979–987. [Google Scholar] [CrossRef]
- Bentley, K.; Deacon, N.; Sonza, S.; Zeichner, S.; Churchill, M. Mutational analysis of the HIV-1 LTR as a promoter of negative sense transcription. Arch. Virol. 2004, 149, 2277–2294. [Google Scholar] [CrossRef]
- Kobayashi-Ishihara, M.; Yamagishi, M.; Hara, T.; Matsuda, Y.; Takahashi, R.; Miyake, A.; Nakano, K.; Yamochi, T.; Ishida, T.; Watanabe, T. HIV-1-encoded antisense RNA suppresses viral replication for a prolonged period. Retrovirology 2012, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Torresilla, C.; Xiao, Y.; Nguyen, P.; Caté, C.; Barbosa, K.; Rassart, E.; Cen, S.; Bourgault, S.; Barbeaub, B. HIV-1 Antisense Protein of Different Clades Induces Autophagy and Associates with the Autophagy Factor p62. J. Virol. 2019, 93, e01757-18. [Google Scholar] [CrossRef]
- Cassan, E.; Arigon-Chifolleau, A.M.; Mesnard, J.M.; Gross, A.; Gascuel, O. Concomitant emergence of the antisense protein gene of HIV-1 and of the pandemic. Proc. Natl. Acad. Sci. USA 2016, 113, 11537–11542. [Google Scholar] [CrossRef] [PubMed]
- Briquet, S.; Vaquero, C. Immunolocalization studies of an antisense protein in HIV-1-infected cells and viral particles. Virology 2002, 292, 177–184. [Google Scholar] [CrossRef]
- Laverdure, S.; Gross, A.; Arpin-Andre, C.; Clerc, I.; Beaumelle, B.; Barbeau, B.; Mesnard, J.M. HIV-1 antisense transcription is preferentially activated in primary monocyte-derived cells. J. Virol. 2012, 86, 13785–13789. [Google Scholar] [CrossRef] [PubMed]
- Torresilla, C.; Larocque, E.; Landry, S.; Halin, M.; Coulombe, Y.; Masson, J.Y.; Mesnard, J.M.; Barbeau, B. Detection of the HIV-1 minus-strand-encoded antisense protein and its association with autophagy. J. Virol. 2013, 87, 5089–5105. [Google Scholar] [CrossRef] [PubMed]
- Dreux, M.; Chisari, F.V. Viruses and the autophagy machinery. Cell Cycle 2010, 9, 1295–1307. [Google Scholar] [CrossRef] [PubMed]
- Kyei, G.B.; Dinkins, C.; Davis, A.S.; Roberts, E.; Singh, S.B.; Dong, C.; Wu, L.; Kominami, E.; Ueno, T.; Yamamoto, A.; et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 2009, 186, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Pushker, R.; Jacque, J.M.; Shields, D.C. Meta-analysis to test the association of HIV-1 nef amino acid differences and deletions with disease progression. J. Virol. 2010, 84, 3644–3653. [Google Scholar] [CrossRef]
- Dimonte, S. Different HIV-1 env frames: gp120 and ASP (antisense protein) biosynthesis, and theirs co-variation tropic amino acid signatures in X4- and R5-viruses. J. Med. Virol. 2017, 89, 112–122. [Google Scholar] [CrossRef]
- Bowder, D.; Hollingsead, H.; Durst, K.; Hu, D.; Wei, W.; Wiggins, J.; Medjahed, H.; Finzi, A.; Sodroski, J.; Xiang, S.H. Contribution of the gp120 V3 loop to envelope glycoprotein trimer stability in primate immunodeficiency viruses. Virology 2018, 521, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.W.; Ardern, Z.; Wei, X. OLGenie: Estimating Natural Selection to Predict Functional Overlapping Genes. Mol. Biol. Evol. 2020, 37, 2440–2449. [Google Scholar] [CrossRef]

| Target | Samples | Assay | Findings | Refs. |
|---|---|---|---|---|
| Anti-ASP antibodies | Sera from HIV-1 patients | IP | Sera from 15 HIV-1 patients were able to immunoprecipitate in vitro translated ASP; patients were at stage I, III and IV of infection during pre-HAART era | [62] |
| LIPS | Antibodies to ASP are detectable in serum of untreated patients, but not in serum of treated patients or in serum of HIV-1 controllers | [63] | ||
| CTL responses against ASP | PBMC; acute and chronic; on and off HAART | ELISpot | PBMC from chronically infected patients both on and off HAART reacted to peptide pools spanning the ASP open reading frame | [64] |
| PBMC; on and off HAART | ELISpot ICS | Detection of CD8+ T cell responses against ASP in patients off HAART; CD8+ T cells responses included production of cytokines and chemokines | [65] | |
| PBMC; chronic patients | ELISpot | Detection of CTL responses against several peptides matching the ASP consensus sequence | [66] | |
| Antisense RNA | CD4+ T cells from treated patients | Strand-specific RT-qPCR | 10–30 copies of HIV-1 antisense RNA per 106 resting CD4+ T cells without in vitro activation | [67] |
| CD4+ T cells from treated and untreated patients | RT-qPCR with biotinylated RT primer | After anti-CD3/CD28 activation for 5 days: 5–10 copies of antisense RNA per 106 cells from treated patients; 20–2 × 106 copies of antisense RNA per 106 cells from untreated patients | [68] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gholizadeh, Z.; Iqbal, M.S.; Li, R.; Romerio, F. The HIV-1 Antisense Gene ASP: The New Kid on the Block. Vaccines 2021, 9, 513. https://doi.org/10.3390/vaccines9050513
Gholizadeh Z, Iqbal MS, Li R, Romerio F. The HIV-1 Antisense Gene ASP: The New Kid on the Block. Vaccines. 2021; 9(5):513. https://doi.org/10.3390/vaccines9050513
Chicago/Turabian StyleGholizadeh, Zahra, Mohd. Shameel Iqbal, Rui Li, and Fabio Romerio. 2021. "The HIV-1 Antisense Gene ASP: The New Kid on the Block" Vaccines 9, no. 5: 513. https://doi.org/10.3390/vaccines9050513
APA StyleGholizadeh, Z., Iqbal, M. S., Li, R., & Romerio, F. (2021). The HIV-1 Antisense Gene ASP: The New Kid on the Block. Vaccines, 9(5), 513. https://doi.org/10.3390/vaccines9050513






