A Nanoscaffolded Spike-RBD Vaccine Provides Protection against SARS-CoV-2 with Minimal Anti-Scaffold Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Modeling of the Designed RBD-Scaffolded Protein Cages
2.2. Preparation of DNA Constructs
2.3. Cell Culture
2.4. Recombinant Viral Proteins
2.5. Production and Characterization of RBD Variants
2.6. ELISA Assay for Binding of Recombinant Proteins to Immobilized Human ACE2
2.7. Surface Plasmon Resonance (SPR) Experiments
2.8. Batch Dynamic Light Scattering Measurement
2.9. SEC-MALS Analysis
2.10. Western Blot
2.11. Plasmid Stability Assay
2.12. Mouse Immunization Studies
2.13. Analysis of Immune Response on Mice
2.14. VSVΔG* Pseudotyped Virus System and Pseudoviral Neutralization Assay
2.15. Neutralization Assay Based on Inhibition of ACE2 Spike Interaction
2.16. T-cell response on Mouse Splenocytes
2.17. Lymph Node Trafficking
2.18. Surrogate Assay of Protection of Viral Infection by Immunization
2.19. Reporter Virus Strain, Stock Preparation, Plaque Assay, and In Vitro Infection
2.20. Serum Virus Neutralization Assay (SARS-CoV-2-GFP)
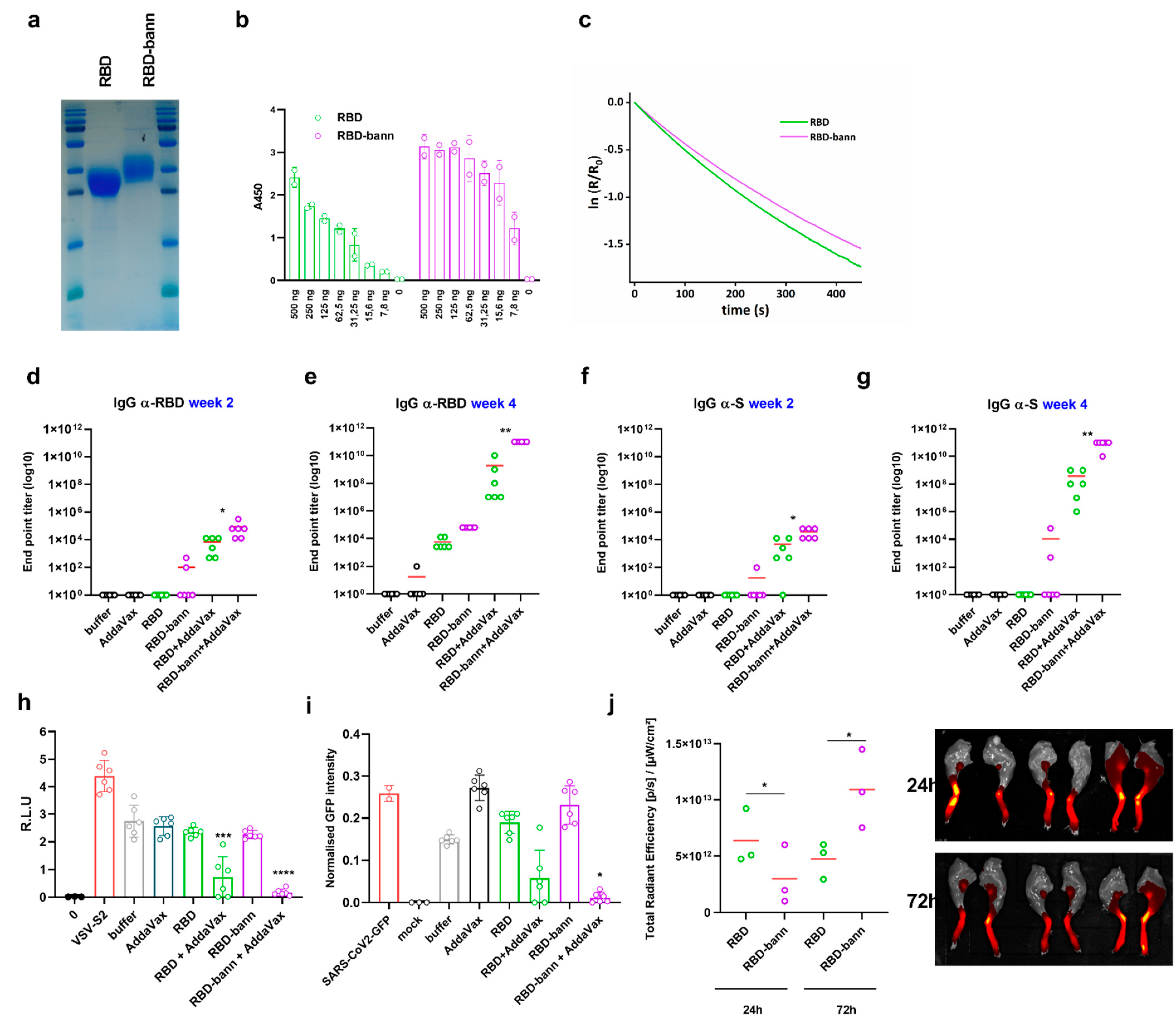
3. Results
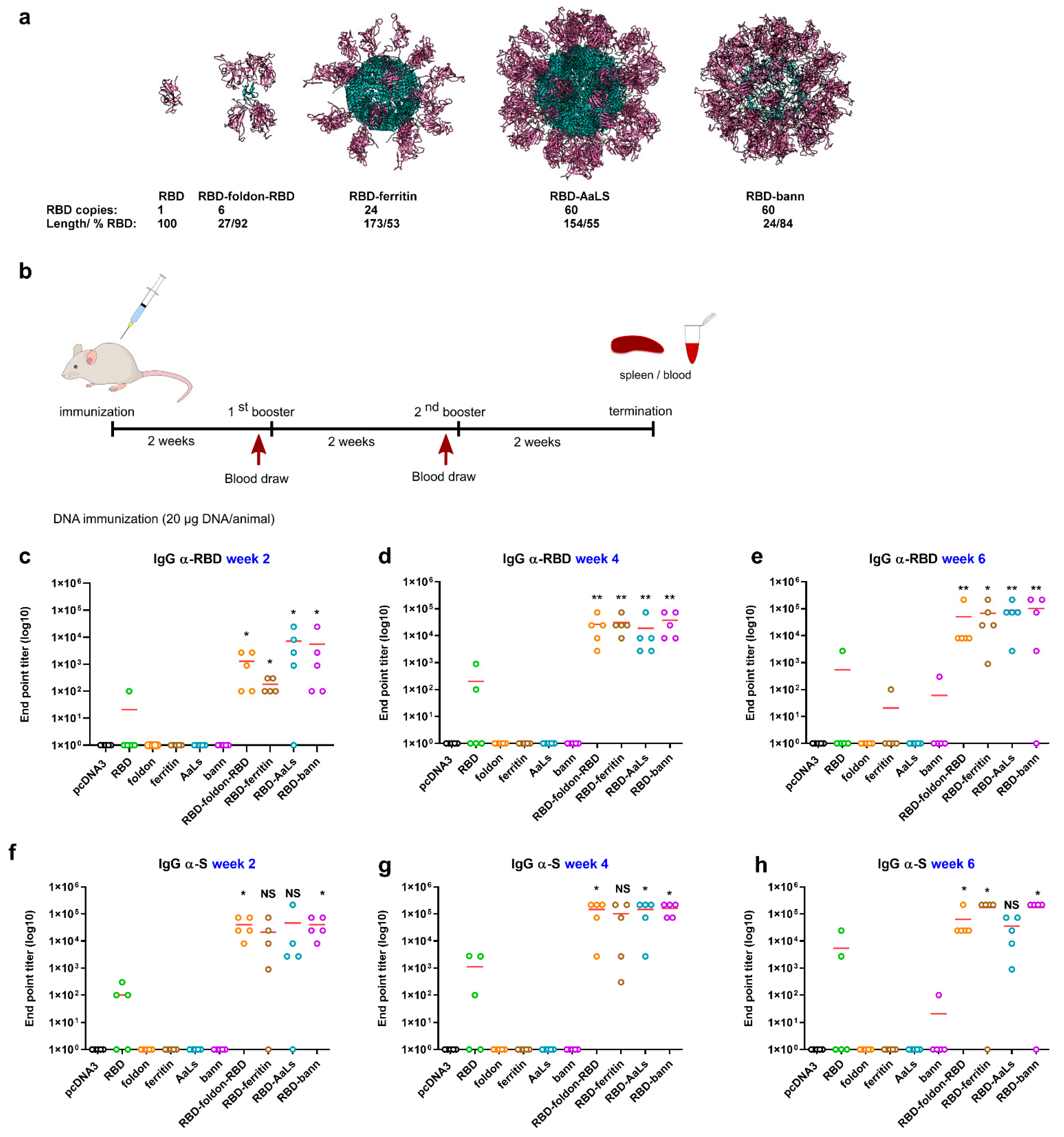
3.1. Design of RBD-Presenting Polypeptide Nanoparticle Scaffolds of Different Size and Oligomerization State
3.2. Scaffolded RBDs Induce a Robust Humoral Immune Response In Vivo
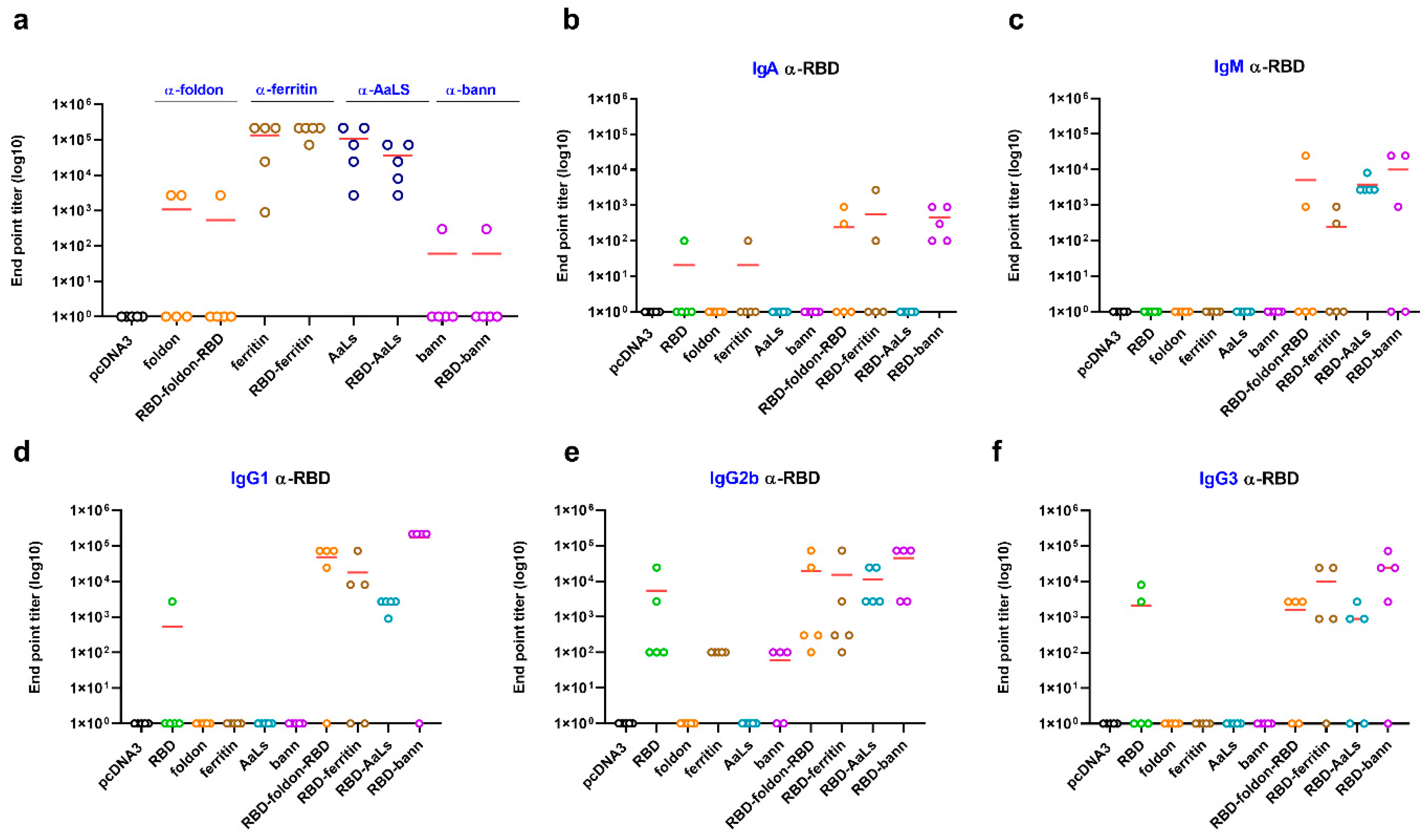
3.3. Immunogenic Properties of Scaffold Domains of the Nanoparticle Vaccines
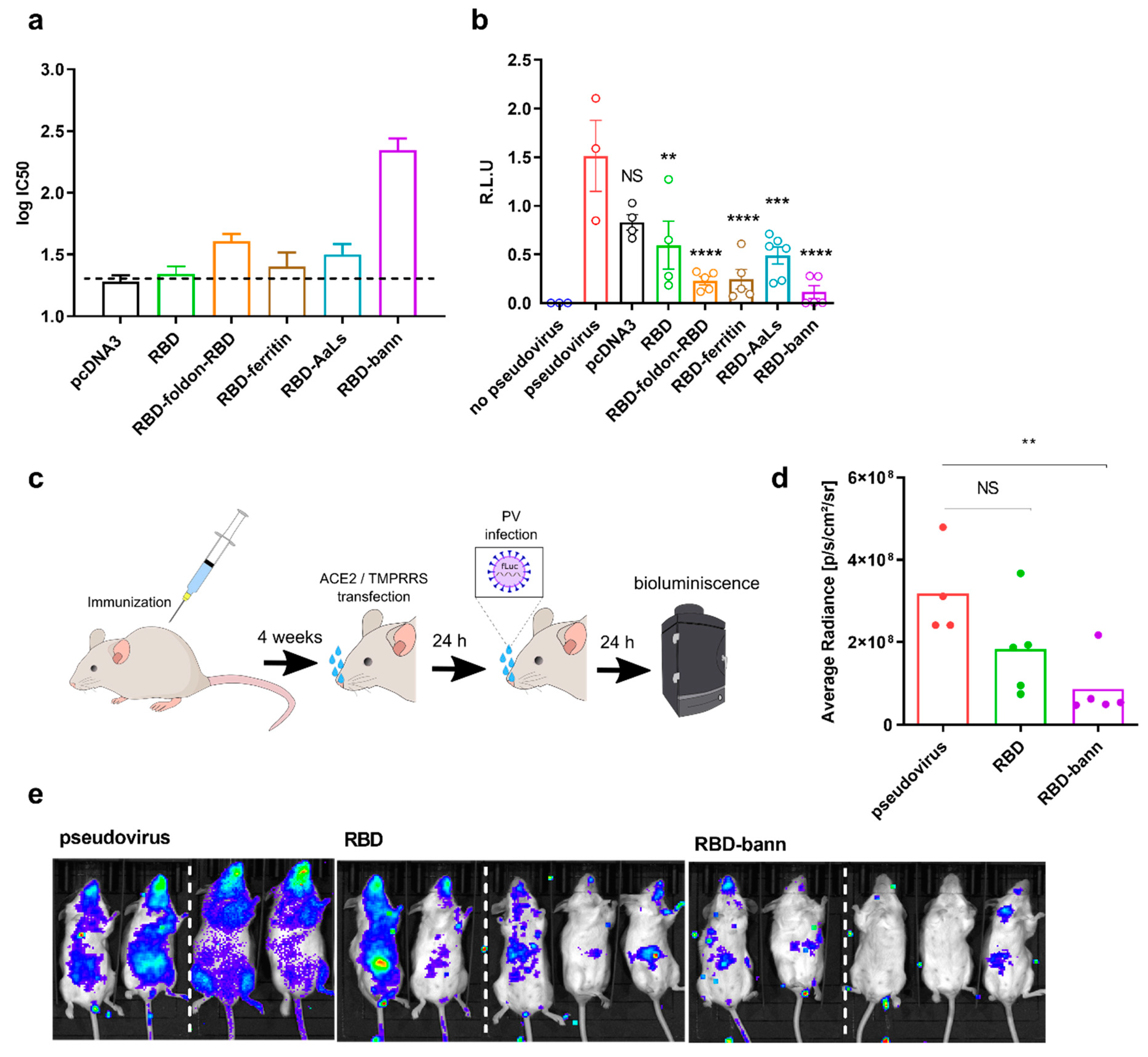
3.4. Sera of Mice Immunized with a RBD-Bann Encoding Vaccine Neutralize Spike-ACE2 Interaction In Vitro
3.5. Immunization with RBD-bann Is Protective In Vivo
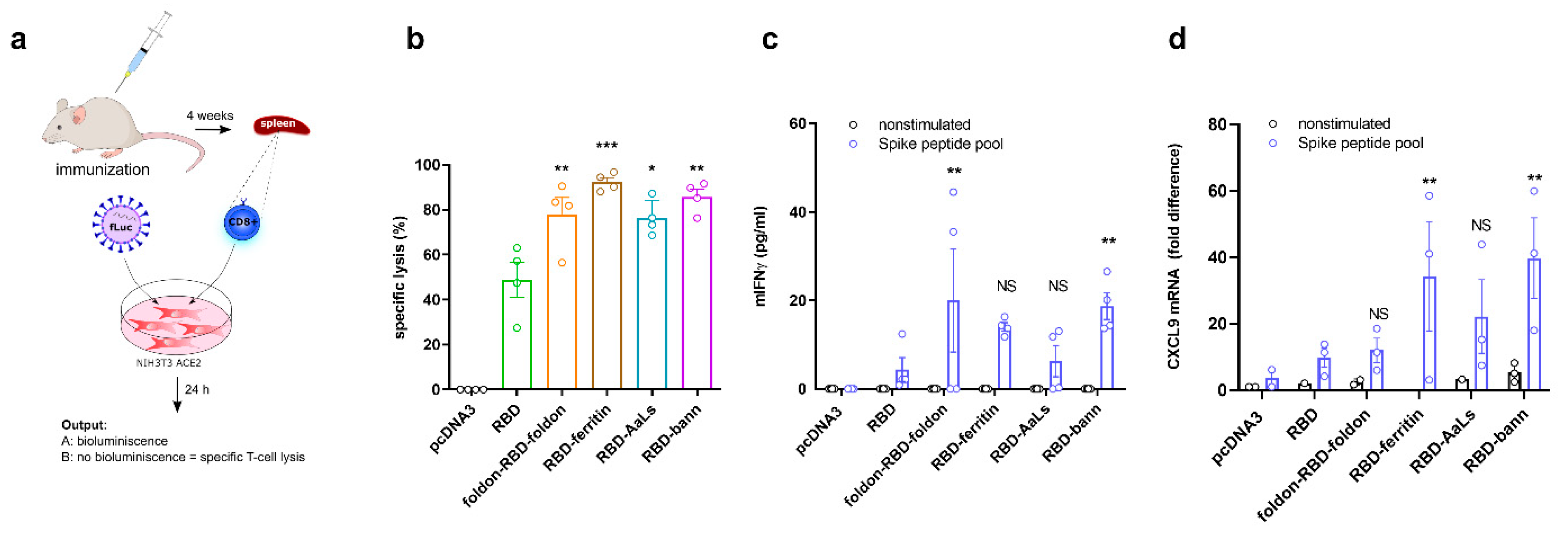
3.6. Immunization with RBD-Bann Plasmid Induces Spike Protein-Specific Cytotoxic T Cells
3.7. Immunization with RBD-Bann Protein Induces Potent Antibody Response
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Tostanoski, L.H.; Peter, L.; Mercado, N.B.; McMahan, K.; Mahrokhian, S.H.; Nkolola, J.P.; Liu, J.; Li, Z.; Chandrashekar, A.; et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020, 369, 806–811. [Google Scholar] [CrossRef]
- Mercado, N.B.; Zahn, R.; Wegmann, F.; Loos, C.; Chandrashekar, A.; Yu, J.; Liu, J.; Peter, L.; McMahan, K.; Tostanoski, L.H.; et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 2020, 586, 583–588. [Google Scholar] [CrossRef]
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z.; et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020, 369, 77–81. [Google Scholar] [CrossRef]
- Yang, J.; Wang, W.; Chen, Z.; Lu, S.; Yang, F.; Bi, Z.; Bao, L.; Mo, F.; Li, X.; Huang, Y.; et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nat. Cell Biol. 2020, 586, 572–577. [Google Scholar] [CrossRef]
- Zhang, N.-N.; Li, X.-F.; Deng, Y.-Q.; Zhao, H.; Huang, Y.-J.; Yang, G.; Huang, W.-J.; Gao, P.; Zhou, C.; Zhang, R.-R.; et al. A thermostable mRNA vaccine against COVID-19. Cell 2020, 182, 1271–1283.e16. [Google Scholar] [CrossRef]
- Smith, T.R.F.; Patel, A.; Ramos, S.; Elwood, D.; Zhu, X.; Yan, J.; Gary, E.N.; Walker, S.N.; Schultheis, K.; Purwar, M.; et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020, 11, 2601. [Google Scholar] [CrossRef]
- Prather, K.J.; Sagar, S.; Murphy, J.; Chartrain, M. Industrial scale production of plasmid DNA for vaccine and gene therapy: Plasmid design, production, and purification. Enzym. Microb. Technol. 2003, 33, 865–883. [Google Scholar] [CrossRef]
- Racz, R.; Li, X.; Patel, M.; Xiang, Z.; He, Y. DNAVaxDB: The first web-based DNA vaccine database and its data analysis. BMC Bioinform. 2014, 15, 1–11. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Fu, Y.; Cheng, Y.; Wu, Y. Understanding SARS-CoV-2-Mediated Inflammatory Responses: From Mechanisms to Potential Therapeutic Tools. Virol. Sin. 2020, 35, 266–271. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Sun, S.; Tai, W.; Chen, J.; Geng, Q.; He, L.; Chen, Y.; Wu, J.; Shi, Z.; et al. Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry. J. Virol. 2019, 94, 94. [Google Scholar] [CrossRef]
- Liu, L.; Wei, Q.; Lin, Q.; Fang, J.; Wang, H.; Kwok, H.; Tang, H.; Nishiura, K.; Peng, J.; Tan, Z.; et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight 2019, 4, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, W.; Drabek, D.; Okba, N.M.A.; van Haperen, R.; Osterhaus, A.D.M.E.; van Kuppeveld, F.J.M.; Haagmans, B.L.; Grosveld, F.; Bosch, B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020, 11, 1–6. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, F.; Shen, C.; Peng, W.; Li, D.; Zhao, C.; Li, Z.; Li, S.; Bi, Y.; Yang, Y.; et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 2020, 368, 1274–1278. [Google Scholar] [CrossRef] [PubMed]
- Maity, P.C.; Blount, A.; Jumaa, H.; Ronneberger, O.; Lillemeier, B.F.; Reth, M. B cell antigen receptors of the IgM and IgD classes are clustered in different protein islands that are altered during B cell activation. Sci. Signal. 2015, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.F.; Jennings, G.T. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010, 10, 787–796. [Google Scholar] [CrossRef]
- López-Sagaseta, J.; Malito, E.; Rappuoli, R.; Bottomley, M.J. Self-assembling protein nanoparticles in the design of vaccines. Comput. Struct. Biotechnol. J. 2016, 14, 58–68. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Zinkernagel, R.M. Neutralizing antiviral B cell responses. Annu. Rev. Immunol. 1997, 15, 235–270. [Google Scholar] [CrossRef]
- Zhang, Y.-N.; Lazarovits, J.; Poon, W.; Ouyang, B.; Nguyen, L.N.M.; Kingston, B.R.; Chan, W.C.W. Nanoparticle Size Influences Antigen Retention and Presentation in Lymph Node Follicles for Humoral Immunity. Nano Lett. 2019, 19, 7226–7235. [Google Scholar] [CrossRef]
- Kanekiyo, M.; Buck, C.B. Virus-Like Particle and Nanoparticle Vaccines; Elsevier Inc.: Amsterdam, The Netherlands, 2017; ISBN 9780128023020. [Google Scholar]
- Choi, B.; Moon, H.; Hong, S.J.; Shin, C.; Do, Y.; Ryu, S.; Kang, S. Effective Delivery of Antigen-Encapsulin Nanoparticle Fusions to Dendritic Cells Leads to Antigen-Specific Cytotoxic T Cell Activation and Tumor Rejection. ACS Nano 2016, 10, 7339–7350. [Google Scholar] [CrossRef] [PubMed]
- Kamp, H.D.; Swanson, K.A.; Wei, R.R.; Dhal, P.K.; Dharanipragada, R.; Kern, A.; Sharma, B.; Sima, R.; Hajdusek, O.; Hu, L.T.; et al. Design of a broadly reactive Lyme disease vaccine. NPJ Vaccines 2020, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kanekiyo, M.; Wei, C.-J.; Yassine, H.M.; McTamney, P.M.; Boyington, J.C.; Whittle, J.R.R.; Rao, S.S.; Kong, W.-P.; Wang, L.; Nabel, G.J. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 2013, 499, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.A.; Rainho-Tomko, J.N.; Williams, Z.P.; Lanza, L.; Peredelchuk, M.; Kishko, M.; Pavot, V.; Alamares-Sapuay, J.; Adhikarla, H.; Gupta, S.; et al. A respiratory syncytial virus (RSV) F protein nanoparticle vaccine focuses antibody responses to a conserved neutralization domain. Sci. Immunol. 2020, 5, eaba6466. [Google Scholar] [CrossRef]
- King, N.P.; Sheffler, W.; Sawaya, M.R.; Vollmar, B.S.; Sumida, J.P.; Andre, I.; Gonen, T.; Yeates, T.O.; Baker, D. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science 2012, 336, 1171–1174. [Google Scholar] [CrossRef]
- Xu, C.; Huang, P.-S.; Ravichandran, R.; Yi, S.; Davis, T.N.; Gonen, T.; Neil, P.; Hsia, Y.; Bale, J.B.; Gonen, S.; et al. Design of a hyperstable 60-subunit protein icosahedron. Nature 2016, 535, 136–139. [Google Scholar] [CrossRef]
- Ljubetič, A.; Lapenta, F.; Gradišar, H.; Drobnak, I.; Aupič, J.; Strmšek, Ž.; Lainšček, D.; Hafner-Bratkovič, I.; Majerle, A.; Krivec, N.; et al. Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo. Nat. Biotechnol. 2017, 35, 1094–1101. [Google Scholar] [CrossRef]
- Lienert, F.; Lohmueller, J.J.; Garg, A.; Silver, P.A. Synthetic biology in mammalian cells: Next generation research tools and therapeutics. Nat. Rev. Mol. Cell Biol. 2014, 15, 95–107. [Google Scholar] [CrossRef]
- Veneziano, R.; Moyer, T.J.; Stone, M.B.; Wamhoff, E.-C.; Read, B.J.; Mukherjee, S.; Shepherd, T.R.; Das, J.; Schief, W.R.; Irvine, D.J.; et al. Role of nanoscale antigen organization on B-cell activation probed using DNA origami. Nat. Nanotechnol. 2020, 15, 716–723. [Google Scholar] [CrossRef]
- Ueda, G.; Antanasijevic, A.; Fallas, J.; Sheffler, W.; Copps, J.; Ellis, D.; Hutchinson, G.; Moyer, A.; Yasmeen, A.; Tsybovsky, Y.; et al. Tailored Design of Protein Nanoparticle Scaffolds for Multivalent Presentation of Viral Glycoprotein Antigens. eLife 2020, 9, e57659. [Google Scholar] [CrossRef]
- Matsuura, K.; Watanabe, K.; Matsuzaki, T.; Sakurai, K.; Kimizuka, N. Self-assembled synthetic viral capsids from a 24-mer viral peptide fragment. Angew. Chem. Int. Ed. 2010, 49, 9662–9665. [Google Scholar] [CrossRef] [PubMed]
- McCluskie, M.J.; Evans, D.M.; Zhang, N.; Benoit, M.; McElhiney, S.P.; Unnithan, M.; DeMarco, S.C.; Clay, B.; Huber, C.; Deora, A.; et al. The effect of preexisting anti-carrier immunity on subsequent responses to CRM197 or Qb-VLP conjugate vaccines. Immunopharmacol. Immunotoxicol. 2016, 38, 184–196. [Google Scholar] [CrossRef]
- Da Silva, D.M.; Pastrana, D.V.; Schiller, J.T.; Kast, W.M. Effect of Preexisting Neutralizing Antibodies on the Anti-tumor Immune Response Induced by Chimeric Human Papillomavirus Virus-like Particle Vaccines. Virology 2001, 290, 350–360. [Google Scholar] [CrossRef]
- Eswar, N.; Webb, B.; Marti-Renom, M.A.; Madhusudhan, M.S.S.; Eramian, D.; Shen, M.M.-Y.; Pieper, U.; Sali, A.; Eswar, N.; Webb, B.; et al. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 2007, 15, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Batra, J.; Hultquist, J.F.; Liu, D.; Shtanko, O.; Von Dollen, J.; Satkamp, L.; Jang, G.M.; Luthra, P.; Schwarz, T.M.; Small, G.I.; et al. Protein Interaction Mapping Identifies RBBP6 as a Negative Regulator of Ebola Virus Replication. Cell 2018, 175, 1917–1930.e13. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Kanekiyo, M.; Bu, W.; Joyce, M.G.; Meng, G.; Whittle, J.R.R.R.; Baxa, U.; Yamamoto, T.; Narpala, S.; Todd, J.P.; Rao, S.S.; et al. Rational Design of an Epstein-Barr Virus Vaccine Targeting the Receptor-Binding Site. Cell 2015, 162, 1090–1100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Meining, W.; Fischer, M.; Bacher, A.; Ladenstein, R. X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 Å resolution: Determinants of thermostability revealed from structural comparisons. J. Mol. Biol. 2001, 306, 1099–1114. [Google Scholar] [CrossRef] [PubMed]
- Sliepen, K.; van Montfort, T.; Melchers, M.; Isik, G.; Sanders, R.W. Immunosilencing a highly immunogenic protein trimerization domain. J. Biol. Chem. 2015, 290, 7436–7442. [Google Scholar] [CrossRef] [PubMed]
- Frey, A.; Di Canzio, J.; Zurakowski, D. A statistically defined endpoint titer determination method for immunoassays. J. Immunol. Methods 1998, 221, 35–41. [Google Scholar] [CrossRef]
- Rentsch, M.B.; Zimmer, G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS ONE 2011, 6, e25858. [Google Scholar] [CrossRef]
- Dai, L.; Zheng, T.; Xu, K.; Han, Y.; Xu, L.; Huang, E.; An, Y.; Cheng, Y.; Li, S.; Liu, M.; et al. A universal design of betacoronavirus vaccines against COVID-19, MERS and SARS. Cell 2020, 182, 722–733. [Google Scholar] [CrossRef]
- Sliepen, K.; Ozorowski, G.; Burger, J.A.; van Montfort, T.; Stunnenberg, M.; LaBranche, C.; Montefiori, D.C.; Moore, J.P.; Ward, A.B.; Sanders, R.W. Presenting native-like HIV-1 envelope trimers on ferritin nanoparticles improves their immunogenicity. Retrovirology 2015, 12, 1–5. [Google Scholar] [CrossRef]
- Güthe, S.; Kapinos, L.; Möglich, A.; Meier, S.; Grzesiek, S.; Kiefhaber, T. Very Fast Folding and Association of a Trimerization Domain from Bacteriophage T4 Fibritin. J. Mol. Biol. 2004, 337, 905–915. [Google Scholar] [CrossRef]
- Ra, J.-S.; Shin, H.-H.; Kang, S.; Do, Y. Lumazine synthase protein cage nanoparticles as antigen delivery nanoplatforms for dendritic cell-based vaccine development. Clin. Exp. Vaccine Res. 2014, 3, 227. [Google Scholar] [CrossRef]
- Nakamura, Y.; Yamada, S.; Nishikawa, S.; Matsuura, K. DNA-modified artificial viral capsids self-assembled from DNA-conjugated β-annulus peptide. J. Pept. Sci. 2017, 23, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; Watanabe, K.; Matsushita, Y.; Kimizuka, N. Guest-binding behavior of peptide nanocapsules self-assembled from viral peptide fragments. Polym. J. 2013, 45, 529–534. [Google Scholar] [CrossRef]
- Matsuura, K.; Ota, J.; Fujita, S.; Shiomi, Y.; Inaba, H. Construction of Ribonuclease-Decorated Artificial Virus-like Capsid by Peptide Self-assembly. J. Org. Chem. 2020, 85, 1668–1673. [Google Scholar] [CrossRef]
- Boyle, J.; Koniaras, C.; Lew, A.M. Influence of cellular location of expressed antigen on the efficacy of DNA vaccination: Cytotoxic T lymphocyte and antibody responses are suboptimal when antigen is cytoplasmic after intramuscular DNA immunization. Int. Immunol. 1997, 9, 1897–1906. [Google Scholar] [CrossRef]
- Li, L.; Petrovsky, N. Molecular mechanisms for enhanced DNA vaccine immunogenicity. Expert Rev. Vaccines 2016, 15, 313–329. [Google Scholar] [CrossRef]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.C.; Tiu, C.; Hu, Z.; Chen, V.C.W.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef]
- McMahan, K.; Yu, J.; Mercado, N.B.; Loos, C.; Tostanoski, L.H.; Chandrashekar, A.; Liu, J.; Peter, L.; Atyeo, C.; Zhu, A.; et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 2021, 590, 630–634. [Google Scholar] [CrossRef]
- Bao, L.; Deng, W.; Huang, B.; Gao, H.; Liu, J.; Ren, L.; Wei, Q.; Yu, P.; Xu, Y.; Qi, F.; et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020, 583, 830–833. [Google Scholar] [CrossRef]
- Israelow, B.; Song, E.; Mao, T.; Lu, P.; Meir, A.; Liu, F.; Alfajaro, M.M.; Wei, J.; Dong, H.; Homer, R.J.; et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med. 2020, 217, e20201241. [Google Scholar] [CrossRef]
- Ma, X.; Zou, F.; Yu, F.; Li, R.; Yuan, Y.; Zhang, Y.; Zhang, X.; Deng, J.; Chen, T.; Song, Z.; et al. Nanoparticle Vaccines Based on the Receptor Binding Domain (RBD) and Heptad Repeat (HR) of SARS-CoV-2 Elicit Robust Protective Immune Responses. Immunity 2020, 53, 1315–1330. [Google Scholar] [CrossRef]
- Wu, Y.; Kelly, S.H.; Sanchez-Perez, L.; Sampson, J.H.; Collier, J.H. Comparative study of α-helical and β-sheet self-assembled peptide nanofiber vaccine platforms: Influence of integrated T-cell epitopes. Biomater. Sci. 2020, 8, 3522–3535. [Google Scholar] [CrossRef]
- Hudalla, G.A.; Sun, T.; Gasiorowski, J.Z.; Han, H.; Tian, Y.F.; Chong, A.S.; Collier, J.H. Gradated assembly of multiple proteins into supramolecular nanomaterials. Nat. Mater. 2014, 13, 829–836. [Google Scholar] [CrossRef]
- Reddy, S.T.; van der Vlies, A.J.; Simeoni, E.; Angeli, V.; Randolph, G.J.; O’Neil, C.P.; Lee, L.K.; Swartz, M.A.; Hubbell, J.A. Exploiting lymphatic transport and complement activation in nanoparticle vaccines. Nat. Biotechnol. 2007, 25, 1159–1164. [Google Scholar] [CrossRef]
- Lainšček, D.; Fink, T.; Forstnerič, V.; Hafner-Bratkovič, I.; Orehek, S.; Strmšek, Ž.; Manček-Keber, M.; Pečan, P.; Esih, H.; Malenšek, Š.; et al. Immune response to vaccine candidates based on different types of nanoscaffolded RBD domain of the SARS-CoV-2 spike protein. bioRxiv 2020. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2020, 397, 99–111. [Google Scholar] [CrossRef]
- Bernelin-Cottet, C.; Urien, C.; Fretaud, M.; Langevin, C.; Trus, I.; Jouneau, L.; Blanc, F.; Leplat, J.J.; Barc, C.; Boulesteix, O.; et al. A DNA prime immuno-potentiates a modified live vaccine against the porcine reproductive and respiratory syndrome virus but does not improve heterologous protection. Viruses 2019, 11, 576. [Google Scholar] [CrossRef]
- Eo, S.K.; Gierynska, M.; Kamar, A.A.; Rouse, B.T. Prime-Boost Immunization with DNA Vaccine: Mucosal Route of Administration Changes the Rules. J. Immunol. 2001, 166, 5473–5479. [Google Scholar] [CrossRef]
- Du, L.; Zhao, G.; Lin, Y.; Chan, C.; He, Y.; Jiang, S.; Wu, C.; Jin, D.Y.; Yuen, K.Y.; Zhou, Y.; et al. Priming with rAAV encoding RBD of SARS-CoV S protein and boosting with RBD-specific peptides for T cell epitopes elevated humoral and cellular immune responses against SARS-CoV infection. Vaccine 2008, 26, 1644–1651. [Google Scholar] [CrossRef]
- Kardani, K.; Bolhassani, A.; Shahbazi, S. Prime-boost vaccine strategy against viral infections: Mechanisms and benefits. Vaccine 2016, 34, 413–423. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lainšček, D.; Fink, T.; Forstnerič, V.; Hafner-Bratkovič, I.; Orehek, S.; Strmšek, Ž.; Manček-Keber, M.; Pečan, P.; Esih, H.; Malenšek, Š.; et al. A Nanoscaffolded Spike-RBD Vaccine Provides Protection against SARS-CoV-2 with Minimal Anti-Scaffold Response. Vaccines 2021, 9, 431. https://doi.org/10.3390/vaccines9050431
Lainšček D, Fink T, Forstnerič V, Hafner-Bratkovič I, Orehek S, Strmšek Ž, Manček-Keber M, Pečan P, Esih H, Malenšek Š, et al. A Nanoscaffolded Spike-RBD Vaccine Provides Protection against SARS-CoV-2 with Minimal Anti-Scaffold Response. Vaccines. 2021; 9(5):431. https://doi.org/10.3390/vaccines9050431
Chicago/Turabian StyleLainšček, Duško, Tina Fink, Vida Forstnerič, Iva Hafner-Bratkovič, Sara Orehek, Žiga Strmšek, Mateja Manček-Keber, Peter Pečan, Hana Esih, Špela Malenšek, and et al. 2021. "A Nanoscaffolded Spike-RBD Vaccine Provides Protection against SARS-CoV-2 with Minimal Anti-Scaffold Response" Vaccines 9, no. 5: 431. https://doi.org/10.3390/vaccines9050431
APA StyleLainšček, D., Fink, T., Forstnerič, V., Hafner-Bratkovič, I., Orehek, S., Strmšek, Ž., Manček-Keber, M., Pečan, P., Esih, H., Malenšek, Š., Aupič, J., Dekleva, P., Plaper, T., Vidmar, S., Kadunc, L., Benčina, M., Omersa, N., Anderluh, G., Pojer, F., ... Jerala, R. (2021). A Nanoscaffolded Spike-RBD Vaccine Provides Protection against SARS-CoV-2 with Minimal Anti-Scaffold Response. Vaccines, 9(5), 431. https://doi.org/10.3390/vaccines9050431





