Vaccine Efficacy of Self-Assembled Multimeric Protein Scaffold Particles Displaying the Glycoprotein Gn Head Domain of Rift Valley Fever Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Gnhead Antigens Production
2.3. LS-SpyTag Production
2.4. Aldolase-SpyTag Production
2.5. E2-SpyTag Production
2.6. Transmission Electron Microscopy
2.7. Experiments with Mice
2.8. Experiments with Lambs
2.8.1. Lamb Trial 1
2.8.2. Lamb Trial 2
2.9. Preparation of Organ Suspensions
2.10. RNA Isolation and RT-qPCR
2.11. Serology
2.12. Statistical Analysis
3. Results
3.1. Production of SpyCatcher-RVFV-Gnhead and SpyTag-MPSPs
3.2. Coupling of the RVFV-Gnhead Subunit Antigen to the MPSPs
3.3. Coupling to MPSPs Results in Improved Efficacy in a Lethal RVFV Mouse Model
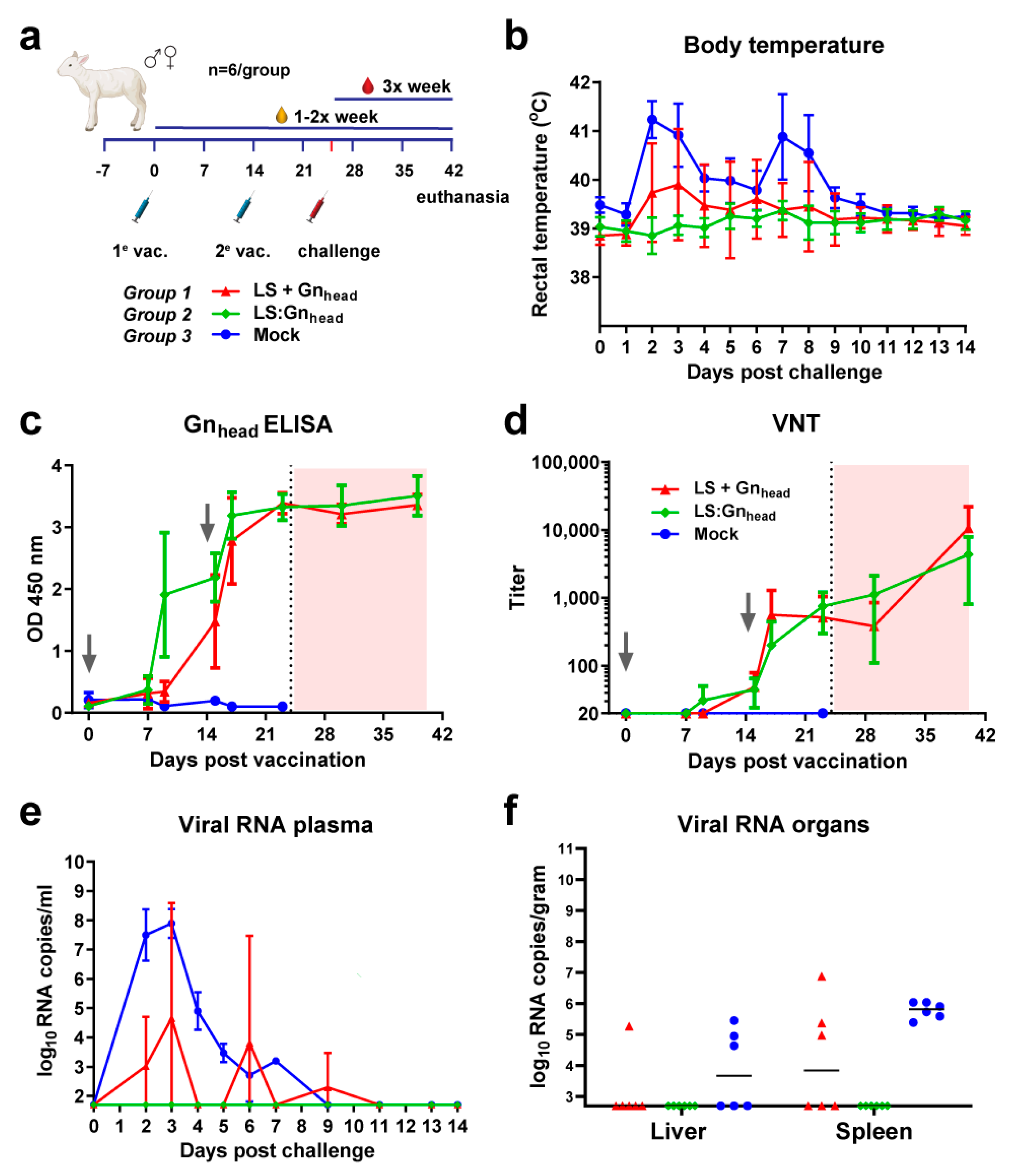
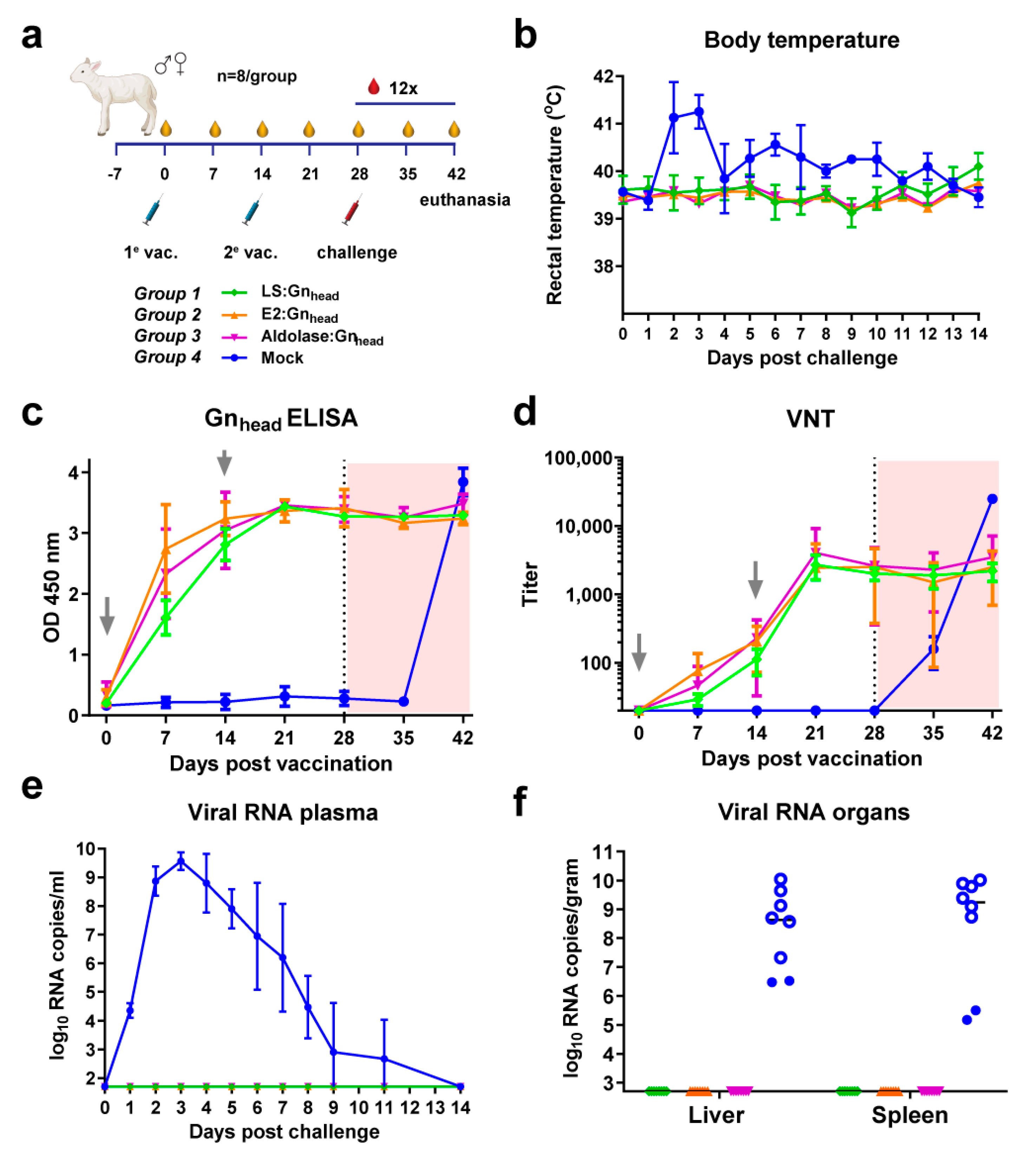
3.4. Coupled Particles Show Improved Efficacy in the RVFV Lamb Model
3.5. The Type of MPSP Evaluated Does Not Determine Immunogenicity and Vaccine Efficacy
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Pati, R.; Shevtsov, M.; Sonawane, A. Nanoparticle Vaccines Against Infectious Diseases. Front. Immunol. 2018, 9, 2224. [Google Scholar] [CrossRef] [PubMed]
- Butkovich, N.; Li, E.; Ramirez, A.; Burkhardt, A.M.; Wang, S.W. Advancements in protein nanoparticle vaccine platforms to combat infectious disease. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, e1681. [Google Scholar] [CrossRef]
- Kelly, H.G.; Tan, H.X.; Juno, J.A.; Esterbauer, R.; Ju, Y.; Jiang, W.; Wimmer, V.C.; Duckworth, B.C.; Groom, J.R.; Caruso, F.; et al. Self-assembling influenza nanoparticle vaccines drive extended germinal center activity and memory B cell maturation. JCI Insight 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Kanekiyo, M.; Wei, C.J.; Yassine, H.M.; McTamney, P.M.; Boyington, J.C.; Whittle, J.R.; Rao, S.S.; Kong, W.P.; Wang, L.; Nabel, G.J. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 2013, 499, 102–106. [Google Scholar] [CrossRef]
- Marcandalli, J.; Fiala, B.; Ols, S.; Perotti, M.; de van der Schueren, W.; Snijder, J.; Hodge, E.; Benhaim, M.; Ravichandran, R.; Carter, L.; et al. Induction of Potent Neutralizing Antibody Responses by a Designed Protein Nanoparticle Vaccine for Respiratory Syncytial Virus. Cell 2019, 176, 1420–1431.e1417. [Google Scholar] [CrossRef] [PubMed]
- Rts, S.C.T.P. Efficacy and safety of RTS,S/AS01 malaria vaccine with or without a booster dose in infants and children in Africa: Final results of a phase 3, individually randomised, controlled trial. Lancet 2015, 386, 31–45. [Google Scholar] [CrossRef]
- Ladenstein, R.; Morgunova, E. Second career of a biosynthetic enzyme: Lumazine synthase as a virus-like nanoparticle in vaccine development. Biotechnol. Rep. 2020, 27, e00494. [Google Scholar] [CrossRef] [PubMed]
- Caivano, A.; Doria-Rose, N.A.; Buelow, B.; Sartorius, R.; Trovato, M.; D’Apice, L.; Domingo, G.J.; Sutton, W.F.; Haigwood, N.L.; De Berardinis, P. HIV-1 Gag p17 presented as virus-like particles on the E2 scaffold from Geobacillus stearothermophilus induces sustained humoral and cellular immune responses in the absence of IFNgamma production by CD4+ T cells. Virology 2010, 407, 296–305. [Google Scholar] [CrossRef]
- Hsia, Y.; Bale, J.B.; Gonen, S.; Shi, D.; Sheffler, W.; Fong, K.K.; Nattermann, U.; Xu, C.; Huang, P.S.; Ravichandran, R.; et al. Design of a hyperstable 60-subunit protein dodecahedron. Nature 2016, 535, 136–139. [Google Scholar] [CrossRef]
- Veggiani, G.; Zakeri, B.; Howarth, M. Superglue from bacteria: Unbreakable bridges for protein nanotechnology. Trends Biotechnol. 2014, 32, 506–512. [Google Scholar] [CrossRef]
- Zakeri, B.; Fierer, J.O.; Celik, E.; Chittock, E.C.; Schwarz-Linek, U.; Moy, V.T.; Howarth, M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 2012, 109, E690–E697. [Google Scholar] [CrossRef] [PubMed]
- Brune, K.D.; Howarth, M. New Routes and Opportunities for Modular Construction of Particulate Vaccines: Stick, Click, and Glue. Front. Immunol. 2018, 9, 1432. [Google Scholar] [CrossRef] [PubMed]
- Wuerth, J.D.; Weber, F. Phleboviruses and the Type I Interferon Response. Viruses 2016, 8, 174. [Google Scholar] [CrossRef]
- Kreher, F.; Tamietti, C.; Gommet, C.; Guillemot, L.; Ermonval, M.; Failloux, A.B.; Panthier, J.J.; Bouloy, M.; Flamand, M. The Rift Valley fever accessory proteins NSm and P78/NSm-GN are distinct determinants of virus propagation in vertebrate and invertebrate hosts. Emerg. Microbes Infect. 2014, 3, e71. [Google Scholar] [CrossRef] [PubMed]
- Kading, R.C.; Crabtree, M.B.; Bird, B.H.; Nichol, S.T.; Erickson, B.R.; Horiuchi, K.; Biggerstaff, B.J.; Miller, B.R. Deletion of the NSm virulence gene of Rift Valley fever virus inhibits virus replication in and dissemination from the midgut of Aedes aegypti mosquitoes. PLoS Negl. Trop. Dis. 2014, 8, e2670. [Google Scholar] [CrossRef] [PubMed]
- Won, S.; Ikegami, T.; Peters, C.J.; Makino, S. NSm protein of Rift Valley fever virus suppresses virus-induced apoptosis. J. Virol. 2007, 81, 13335–13345. [Google Scholar] [CrossRef] [PubMed]
- Terasaki, K.; Won, S.; Makino, S. The C-terminal region of Rift Valley fever virus NSm protein targets the protein to the mitochondrial outer membrane and exerts antiapoptotic function. J. Virol. 2013, 87, 676–682. [Google Scholar] [CrossRef]
- Warimwe, G.M.; Gesharisha, J.; Carr, B.V.; Otieno, S.; Otingah, K.; Wright, D.; Charleston, B.; Okoth, E.; Elena, L.G.; Lorenzo, G.; et al. Chimpanzee Adenovirus Vaccine Provides Multispecies Protection against Rift Valley Fever. Sci. Rep. 2016, 6, 20617. [Google Scholar] [CrossRef] [PubMed]
- Kortekaas, J.; de Boer, S.M.; Kant, J.; Vloet, R.P.; Antonis, A.F.; Moormann, R.J. Rift Valley fever virus immunity provided by a paramyxovirus vaccine vector. Vaccine 2010, 28, 4394–4401. [Google Scholar] [CrossRef] [PubMed]
- Halldorsson, S.; Li, S.; Li, M.; Harlos, K.; Bowden, T.A.; Huiskonen, J.T. Shielding and activation of a viral membrane fusion protein. Nat. Commun. 2018, 9, 349. [Google Scholar] [CrossRef]
- Kortekaas, J.; Antonis, A.F.; Kant, J.; Vloet, R.P.; Vogel, A.; Oreshkova, N.; de Boer, S.M.; Bosch, B.J.; Moormann, R.J. Efficacy of three candidate Rift Valley fever vaccines in sheep. Vaccine 2012, 30, 3423–3429. [Google Scholar] [CrossRef]
- de Boer, S.M.; Kortekaas, J.; Antonis, A.F.; Kant, J.; van Oploo, J.L.; Rottier, P.J.; Moormann, R.J.; Bosch, B.J. Rift Valley fever virus subunit vaccines confer complete protection against a lethal virus challenge. Vaccine 2010, 28, 2330–2339. [Google Scholar] [CrossRef] [PubMed]
- Antonis, A.F.; Kortekaas, J.; Kant, J.; Vloet, R.P.; Vogel-Brink, A.; Stockhofe, N.; Moormann, R.J. Vertical transmission of Rift Valley fever virus without detectable maternal viremia. Vector Borne Zoonotic Dis. 2013, 13, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Kortekaas, J.; Oreshkova, N.; Cobos-Jimenez, V.; Vloet, R.P.; Potgieter, C.A.; Moormann, R.J. Creation of a nonspreading Rift Valley fever virus. J. Virol. 2011, 85, 12622–12630. [Google Scholar] [CrossRef] [PubMed]
- Wichgers Schreur, P.J.; Oreshkova, N.; van Keulen, L.; Kant, J.; van de Water, S.; Soos, P.; Dehon, Y.; Kollar, A.; Penzes, Z.; Kortekaas, J. Safety and efficacy of four-segmented Rift Valley fever virus in young sheep, goats and cattle. NPJ Vaccines 2020, 5, 65. [Google Scholar] [CrossRef] [PubMed]
- Wichgers Schreur, P.J.; Kant, J.; van Keulen, L.; Moormann, R.J.; Kortekaas, J. Four-segmented Rift Valley fever virus induces sterile immunity in sheep after a single vaccination. Vaccine 2015, 33, 1459–1464. [Google Scholar] [CrossRef]
- Wichgers Schreur, P.J.; Oreshkova, N.; Moormann, R.J.; Kortekaas, J. Creation of Rift Valley fever viruses with four-segmented genomes reveals flexibility in bunyavirus genome packaging. J. Virol. 2014, 88, 10883–10893. [Google Scholar] [CrossRef]
- Kortekaas, J.; Dekker, A.; de Boer, S.M.; Weerdmeester, K.; Vloet, R.P.; de Wit, A.A.; Peeters, B.P.; Moormann, R.J. Intramuscular inoculation of calves with an experimental Newcastle disease virus-based vector vaccine elicits neutralizing antibodies against Rift Valley fever virus. Vaccine 2010, 28, 2271–2276. [Google Scholar] [CrossRef]
- Wichgers Schreur, P.J.; van de Water, S.; Harmsen, M.; Bermudez-Mendez, E.; Drabek, D.; Grosveld, F.; Wernike, K.; Beer, M.; Aebischer, A.; Daramola, O.; et al. Multimeric single-domain antibody complexes protect against bunyavirus infections. Elife 2020, 9, e52716. [Google Scholar] [CrossRef]
- Wu, Y.; Zhu, Y.; Gao, F.; Jiao, Y.; Oladejo, B.O.; Chai, Y.; Bi, Y.; Lu, S.; Dong, M.; Zhang, C.; et al. Structures of phlebovirus glycoprotein Gn and identification of a neutralizing antibody epitope. Proc. Natl. Acad. Sci. USA 2017, 114, E7564–E7573. [Google Scholar] [CrossRef]
- Allen, E.R.; Krumm, S.A.; Raghwani, J.; Halldorsson, S.; Elliott, A.; Graham, V.A.; Koudriakova, E.; Harlos, K.; Wright, D.; Warimwe, G.M.; et al. A Protective Monoclonal Antibody Targets a Site of Vulnerability on the Surface of Rift Valley Fever Virus. Cell Rep. 2018, 25, 3750–3758.e3754. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ma, T.; Wu, Y.; Chen, Z.; Zeng, H.; Tong, Z.; Gao, F.; Qi, J.; Zhao, Z.; Chai, Y.; et al. Neutralization mechanism of human monoclonal antibodies against Rift Valley fever virus. Nat. Microbiol. 2019, 4, 1231–1241. [Google Scholar] [CrossRef]
- Sasaki, E.; Bohringer, D.; van de Waterbeemd, M.; Leibundgut, M.; Zschoche, R.; Heck, A.J.; Ban, N.; Hilvert, D. Structure and assembly of scalable porous protein cages. Nat. Commun. 2017, 8, 14663. [Google Scholar] [CrossRef] [PubMed]
- Herzik, M.A., Jr.; Wu, M.; Lander, G.C. Achieving better-than-3-A resolution by single-particle cryo-EM at 200 keV. Nat. Methods 2017, 14, 1075–1078. [Google Scholar] [CrossRef]
- Dalmau, M.; Lim, S.; Chen, H.C.; Ruiz, C.; Wang, S.W. Thermostability and molecular encapsulation within an engineered caged protein scaffold. Biotechnol. Bioeng. 2008, 101, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Wichgers Schreur, P.J.; Paweska, J.T.; Kant, J.; Kortekaas, J. A novel highly sensitive, rapid and safe Rift Valley fever virus neutralization test. J. Virol. Methods 2017, 248, 26–30. [Google Scholar] [CrossRef]
- Janitzek, C.M.; Matondo, S.; Thrane, S.; Nielsen, M.A.; Kavishe, R.; Mwakalinga, S.B.; Theander, T.G.; Salanti, A.; Sander, A.F. Bacterial superglue generates a full-length circumsporozoite protein virus-like particle vaccine capable of inducing high and durable antibody responses. Malar. J. 2016, 15, 545. [Google Scholar] [CrossRef]
- Thrane, S.; Janitzek, C.M.; Matondo, S.; Resende, M.; Gustavsson, T.; de Jongh, W.A.; Clemmensen, S.; Roeffen, W.; van de Vegte-Bolmer, M.; van Gemert, G.J.; et al. Bacterial superglue enables easy development of efficient virus-like particle based vaccines. J. Nanobiotechnol. 2016, 14, 30. [Google Scholar] [CrossRef]
- Brune, K.D.; Leneghan, D.B.; Brian, I.J.; Ishizuka, A.S.; Bachmann, M.F.; Draper, S.J.; Biswas, S.; Howarth, M. Plug-and-Display: Decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci. Rep. 2016, 6, 19234. [Google Scholar] [CrossRef]
- Escolano, A.; Gristick, H.B.; Abernathy, M.E.; Merkenschlager, J.; Gautam, R.; Oliveira, T.Y.; Pai, J.; West, A.P., Jr.; Barnes, C.O.; Cohen, A.A.; et al. Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques. Nature 2019, 570, 468–473. [Google Scholar] [CrossRef]
- Wang, W.; Liu, Z.; Zhou, X.; Guo, Z.; Zhang, J.; Zhu, P.; Yao, S.; Zhu, M. Ferritin nanoparticle-based SpyTag/SpyCatcher-enabled click vaccine for tumor immunotherapy. Nanomed. Nanotechnol. Biol. Med. 2019, 16, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chao, C.W.; Tsybovsky, Y.; Abiona, O.M.; Hutchinson, G.B.; Moliva, J.I.; Olia, A.S.; Pegu, A.; Phung, E.; Stewart-Jones, G.B.E.; et al. A platform incorporating trimeric antigens into self-assembling nanoparticles reveals SARS-CoV-2-spike nanoparticles to elicit substantially higher neutralizing responses than spike alone. Sci. Rep. 2020, 10, 18149. [Google Scholar] [CrossRef] [PubMed]
- Chrun, T.; Lacote, S.; Urien, C.; Richard, C.A.; Tenbusch, M.; Aubrey, N.; Pulido, C.; Lakhdar, L.; Marianneau, P.; Schwartz-Cornil, I. A DNA Vaccine Encoding the Gn Ectodomain of Rift Valley Fever Virus Protects Mice via a Humoral Response Decreased by DEC205 Targeting. Front. Immunol. 2019, 10, 860. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, N.; Heise, M.T.; Ross, T.M. Vaccination with DNA plasmids expressing Gn coupled to C3d or alphavirus replicons expressing gn protects mice against Rift Valley fever virus. PLoS Negl. Trop. Dis. 2010, 4, e725. [Google Scholar] [CrossRef]
- Gomes, A.C.; Mohsen, M.; Bachmann, M.F. Harnessing Nanoparticles for Immunomodulation and Vaccines. Vaccines 2017, 5, 6. [Google Scholar] [CrossRef]
- Cruz, F.M.; Colbert, J.D.; Merino, E.; Kriegsman, B.A.; Rock, K.L. The Biology and Underlying Mechanisms of Cross-Presentation of Exogenous Antigens on MHC-I Molecules. Annu. Rev. Immunol. 2017, 35, 149–176. [Google Scholar] [CrossRef]
- Hao, M.; Zhang, G.; Zhang, S.; Chen, Z.; Chi, X.; Dong, Y.; Fan, P.; Liu, Y.; Chen, Y.; Song, X.; et al. Characterization of Two Neutralizing Antibodies against Rift Valley Fever Virus Gn Protein. Viruses 2020, 12, 259. [Google Scholar] [CrossRef]
- Singh, B. Myceliophthora thermophila syn. Sporotrichum thermophile: A thermophilic mould of biotechnological potential. Crit. Rev. Biotechnol. 2016, 36, 59–69. [Google Scholar] [CrossRef]
| Protein Domain | Amino Acid Sequence |
|---|---|
| GP64 signal sequence | MPMLSAIVLYVLLAAAAHSAFA |
| SpyCatcher | DIPTTENLYFQGAMVDTLSGLSSE QGQSGDMTIEEDSATHIKFSKRD EDGKELAGATMELRDSSGKTIS TWISDGQVKDFYLYPGKYTFVETAAPDGYEV ATAITFTVNEQGQVTVNGKATKGDA HIDGPQGIWGQLEWKK |
| 10 GlySer linker | GGGGSGGGGS |
| RVFV-Gnhead | EDPHLRNRPGKGHNYIDGMTQEDATCK PVTYAGACSSFDVLLEKGKFPLFQSYAHHRTL LEAVHDTIIAKADPP SCDLQSAHGNPCMKEKLVMKTHCPNDYQSA HYLNNDGKMASVKCPPKYELT EDCNFCRQMTGASLKKGSYPLQDLFCQSSED DGSKLKTKMKGVCEVGVQAL KKCDGQLSTAHEVVPFAVFKNSKKVYLDKLD LKTEENLL PDSFVCFEHKGQYKGTMDSGQTKRELKSFDIS QCPKIGGHGSKKCTG DAAFCSAYECTAQYANAYCSHANGSGVVQI QVSGVWKKPLCVGYERVVVKRE |
| Entero Kinase site | DDDDK |
| Twin Strep-tag | GSAWSHPQFEKGGGSGGGSGGSAWSHPQFEK |
| LS-SpyTag 1 | MQIYEGKLTAEGLRFGIVASRFN HALVDRLVEGAIDAIVRHGGREEDITLVRVPG SWEIPVAAGELARKEDIDAVIAIGVLIRGATPH FDYIASEVSKGLANLSLELRKPITFGVIT ADTLEQAIERAGTKHGNKGWEAALSAIE MANLFKSLRGGGGSGGGGSGGGGS AHIVMVDAYKPTK |
| Aldolase-SpyTag 1 | MAHIVMVDAYKPTKLINGGSGGSGGSGGSM KMEELFKKHKIVAVLRANS VEEAKKKALAVFLGGVHLIEITFTVPDADTVIK ELSFLKEMGAIIGAGTV TSVEQCRKAVESGAEFIVSPHLDEE ISQFCKEKGVFYMPGVMTPTELVKAMKLGHTI LKLFPGEVVGPQFV KAMKGPFPNVKFVPTGGVNLDNVCEW FKAGVLAVGVG SALVKGTPVEVAEKAKAFVEKIRGCTE |
| E2-Spytag 1 | MAHIVMVDAYKPTKAAAEEKAAPAAAK PATTEGEFPETREKMSGIRRAIAKAMVH SKHTAPHVTLMDEADVTKLVAHRKKFKAIAA EKGIKLTFLPYVVKALVSALREYPVLNTSID DETEEIIQKHYYNIGIAADT DRGLLVPVIKHADRKPIFALAQEINELAEKAR DGKLTPGEMKGASCTIT NIGSAGGQWFTPVINHPEVAILGIGRIAEK PIVRDGEIVAAP MLALSLSFDHRMIDGATAQKALNHIKRLLSD PELLLMEA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wichgers Schreur, P.J.; Tacken, M.; Gutjahr, B.; Keller, M.; van Keulen, L.; Kant, J.; van de Water, S.; Lin, Y.; Eiden, M.; Rissmann, M.; et al. Vaccine Efficacy of Self-Assembled Multimeric Protein Scaffold Particles Displaying the Glycoprotein Gn Head Domain of Rift Valley Fever Virus. Vaccines 2021, 9, 301. https://doi.org/10.3390/vaccines9030301
Wichgers Schreur PJ, Tacken M, Gutjahr B, Keller M, van Keulen L, Kant J, van de Water S, Lin Y, Eiden M, Rissmann M, et al. Vaccine Efficacy of Self-Assembled Multimeric Protein Scaffold Particles Displaying the Glycoprotein Gn Head Domain of Rift Valley Fever Virus. Vaccines. 2021; 9(3):301. https://doi.org/10.3390/vaccines9030301
Chicago/Turabian StyleWichgers Schreur, Paul J., Mirriam Tacken, Benjamin Gutjahr, Markus Keller, Lucien van Keulen, Jet Kant, Sandra van de Water, Yanyin Lin, Martin Eiden, Melanie Rissmann, and et al. 2021. "Vaccine Efficacy of Self-Assembled Multimeric Protein Scaffold Particles Displaying the Glycoprotein Gn Head Domain of Rift Valley Fever Virus" Vaccines 9, no. 3: 301. https://doi.org/10.3390/vaccines9030301
APA StyleWichgers Schreur, P. J., Tacken, M., Gutjahr, B., Keller, M., van Keulen, L., Kant, J., van de Water, S., Lin, Y., Eiden, M., Rissmann, M., von Arnim, F., König, R., Brix, A., Charreyre, C., Audonnet, J.-C., Groschup, M. H., & Kortekaas, J. (2021). Vaccine Efficacy of Self-Assembled Multimeric Protein Scaffold Particles Displaying the Glycoprotein Gn Head Domain of Rift Valley Fever Virus. Vaccines, 9(3), 301. https://doi.org/10.3390/vaccines9030301






