Strategies Targeting Hemagglutinin as a Universal Influenza Vaccine
Abstract
1. Introduction
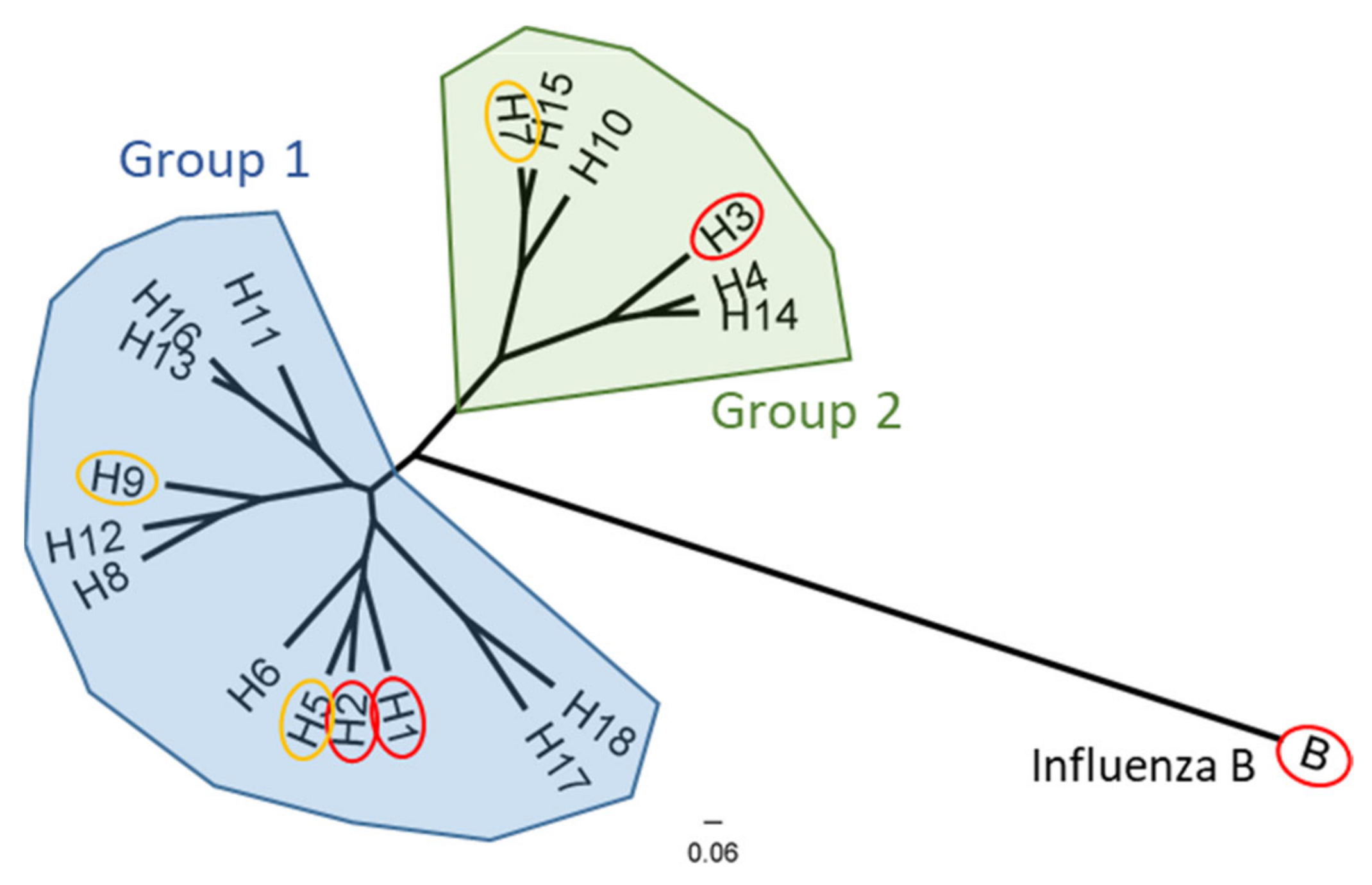
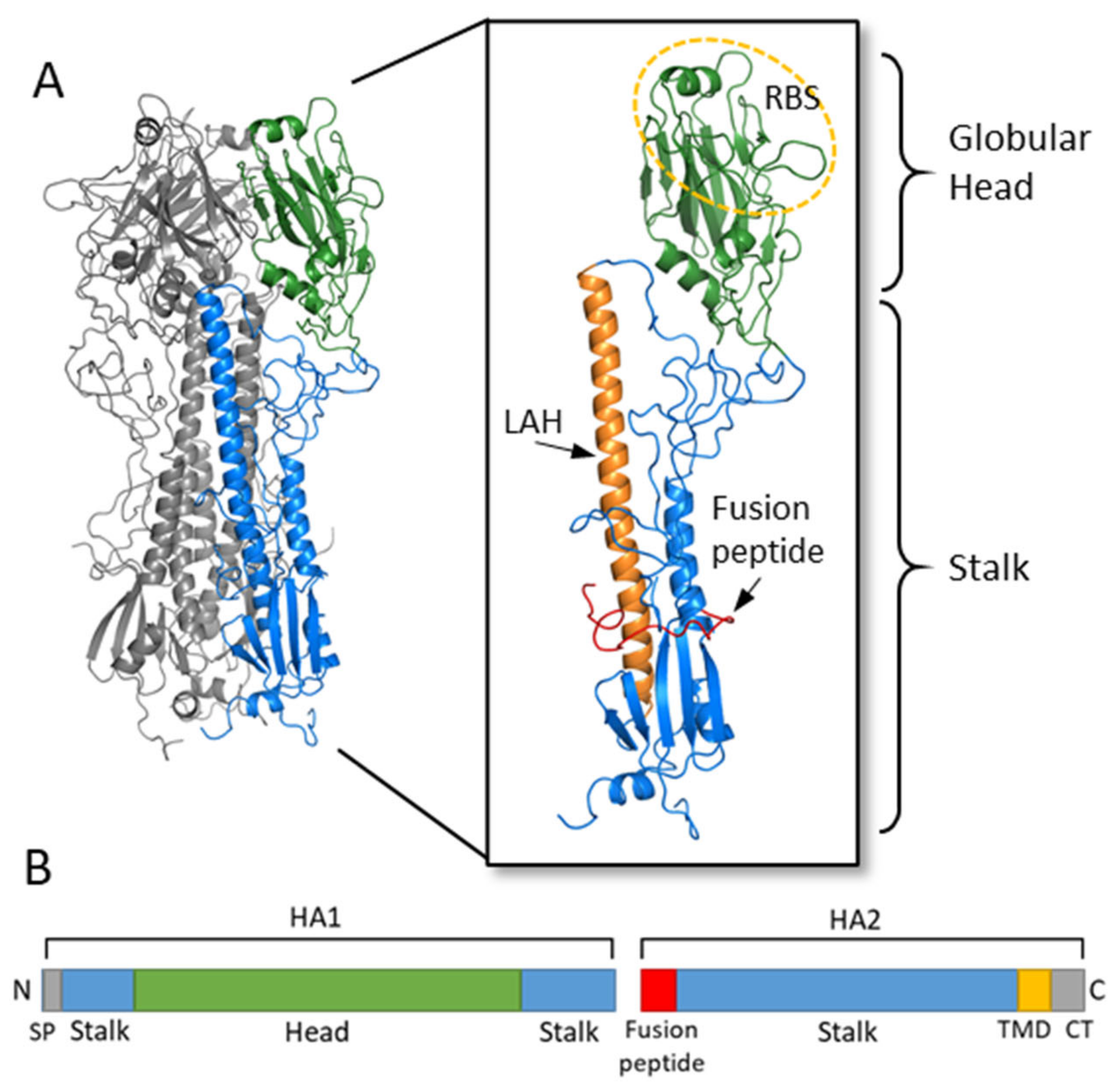
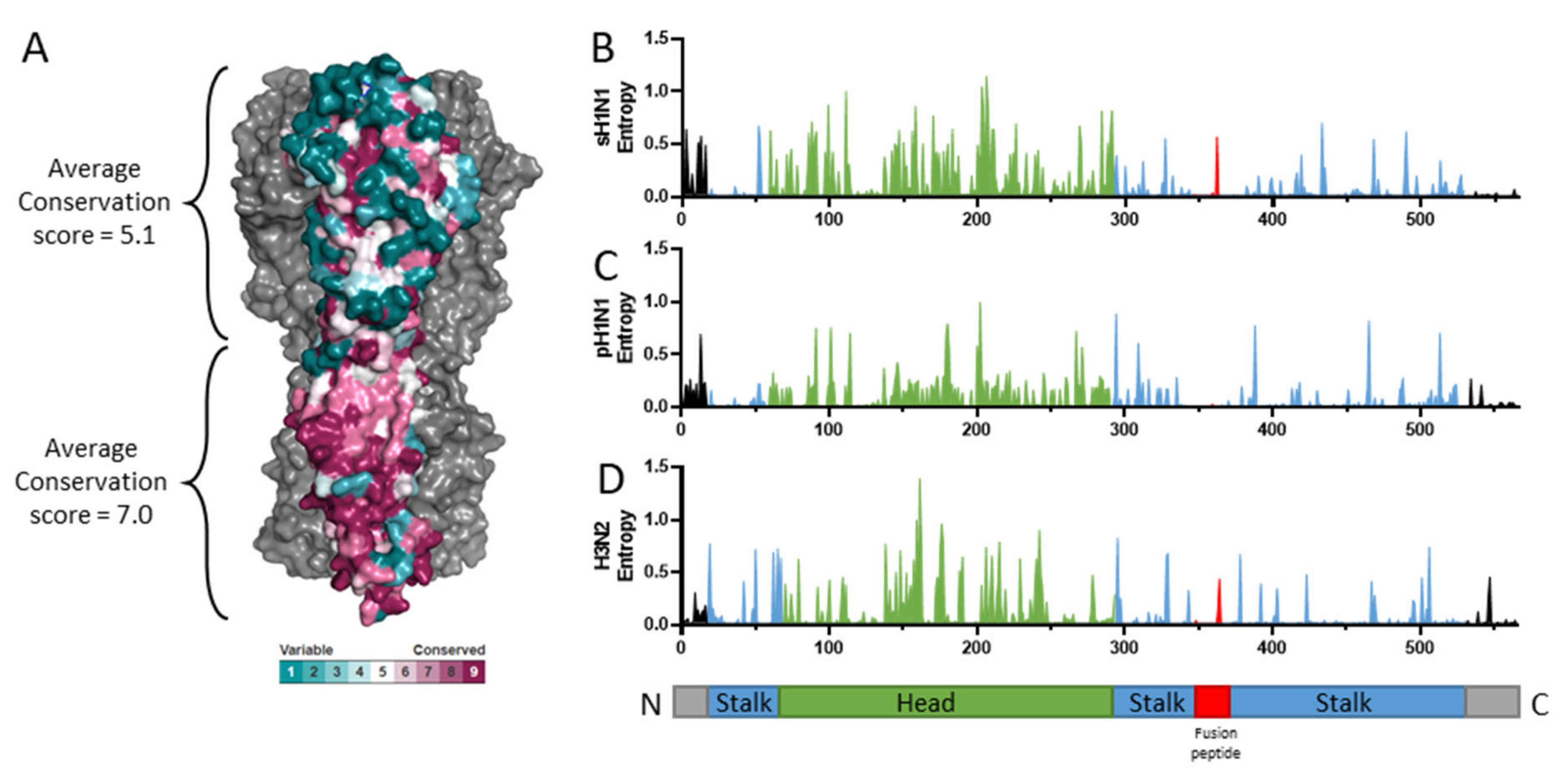
2. Hemagglutinin Structure and Function
3. Stalk-Directed Strategies
3.1. General Principles of Stalk-Directed Stratgies
3.2. Design of Stalk-Directed Vaccines
3.2.1. Headless HA Constructs
3.2.2. Chimeric HA
3.2.3. Hyperglycosylated HA
3.2.4. “Mosaic” HA
3.2.5. LAH Fragment
3.3. Challenges Facing Stalk-Directed Strategies
4. Consensus-Based Strategies
4.1. General Principles of Consensus-Based Stratgies
4.2. Design of Consensus-Based Vaccines
4.2.1. Consensus Hemagglutinins
4.2.2. Micro-Consensus Hemagglutinin
4.2.3. Centralized Hemagglutinins
4.2.4. COBRA Hemagglutinins
4.3. Challenges Facing Consensus-Based Approaches
5. Computational Algorithms for Immunogen Design
5.1. General Prinicples of Computational Algorithm Approaches
5.2. Design of Computional Algorithm Vaccines
5.2.1. Ancestral Hemagglutinin
5.2.2. Mosaic Algorithm
5.2.3. Epigraph Algorithm
5.3. Challenges Facing Computational Algorithm Approaches
6. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Erbelding, E.J.; Post, D.J.; Stemmy, E.J.; Roberts, P.C.; Augustine, A.D.; Ferguson, S.; Paules, C.I.; Graham, B.S.; Fauci, A.S. A Universal Influenza Vaccine: The Strategic Plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 2018, 218, 347–354. [Google Scholar] [CrossRef]
- WHO. Influenza (Seasonal). Available online: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (accessed on 9 February 2021).
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.J.; et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet 2018, 391, 1285–1300. [Google Scholar] [CrossRef]
- Lafond, K.E.; Nair, H.; Rasooly, M.H.; Valente, F.; Booy, R.; Rahman, M.; Kitsutani, P.; Yu, H.; Guzman, G.; Coulibaly, D.; et al. Global Role and Burden of Influenza in Pediatric Respiratory Hospitalizations, 1982–2012: A Systematic Analysis. Plos Med. 2016, 13, e1001977. [Google Scholar] [CrossRef] [PubMed]
- Van Kerkhove, M.D.; Hirve, S.; Koukounari, A.; Mounts, A.W. Estimating age-specific cumulative incidence for the 2009 influenza pandemic: A meta-analysis of A(H1N1)pdm09 serological studies from 19 countries. Influenza Other Respir. Viruses 2013, 7, 872–886. [Google Scholar] [CrossRef] [PubMed]
- Fields, B.N.; Knipe, D.M.; Howley, P.M. Fields Virology; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013. [Google Scholar]
- Bouvier, N.M.; Palese, P. The biology of influenza viruses. Vaccine 2008, 26 (Suppl. 4). [Google Scholar] [CrossRef] [PubMed]
- Reneer, Z.B.; Ross, T.M. H2 influenza viruses: Designing vaccines against future H2 pandemics. Biochem. Soc. Trans. 2019, 47, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.C.; Baranovich, T.; Marathe, B.M.; Danner, A.F.; Seiler, J.P.; Franks, J.; Govorkova, E.A.; Krauss, S.; Webster, R.G. Risk assessment of H2N2 influenza viruses from the avian reservoir. J. Virol. 2014, 88, 1175–1188. [Google Scholar] [CrossRef] [PubMed]
- Malik Peiris, J.S. Avian influenza viruses in humans. Rev. Sci. Tech. 2009, 28, 161–173. [Google Scholar] [CrossRef]
- Lai, S.; Qin, Y.; Cowling, B.J.; Ren, X.; Wardrop, N.A.; Gilbert, M.; Tsang, T.K.; Wu, P.; Feng, L.; Jiang, H.; et al. Global epidemiology of avian influenza A H5N1 virus infection in humans, 1997–2015: A systematic review of individual case data. Lancet Infect. Dis. 2016, 16, e108. [Google Scholar] [CrossRef]
- Yoon, S.-W.; Webby, R.J.; Webster, R.G. Evolution and Ecology of Influenza A Viruses. In Influenza Pathogenesis and Control; Compans, R.W., Oldstone, M.B.A., Eds.; Springer International Publishing: Cham, Switzerland, 2014; Volume I, pp. 359–375. [Google Scholar] [CrossRef]
- Fiore, A.E.; Bridges, C.B.; Cox, N.J. Seasonal influenza vaccines. Curr. Top. Microbiol. Immunol. 2009, 333, 43–82. [Google Scholar] [CrossRef]
- Paules, C.; Subbarao, K. Influenza. Lancet 2017, 390, 697–708. [Google Scholar] [CrossRef]
- Heaton, N.S.; Sachs, D.; Chen, C.-J.; Hai, R.; Palese, P. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 20248–20253. [Google Scholar] [CrossRef] [PubMed]
- CDC. CDC Seasonal Flu Vaccine Effectiveness Studies. Available online: https://www.cdc.gov/flu/vaccines-work/effectiveness-studies.htm (accessed on 9 February 2021).
- Foppa, I.M.; Cheng, P.-Y.; Reynolds, S.B.; Shay, D.K.; Carias, C.; Bresee, J.S.; Kim, I.K.; Gambhir, M.; Fry, A.M. Deaths averted by influenza vaccination in the U.S. during the seasons 2005/06 through 2013/14. Vaccine 2015, 33, 3003–3009. [Google Scholar] [CrossRef] [PubMed]
- Coudeville, L.; Bailleux, F.; Riche, B.; Megas, F.; Andre, P.; Ecochard, R. Relationship between haemagglutination-inhibiting antibody titres and clinical protection against influenza: Development and application of a bayesian random-effects model. BMC Med. Res. Methodol. 2010, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Black, S.; Nicolay, U.; Vesikari, T.; Knuf, M.; Del Giudice, G.; Della Cioppa, G.; Tsai, T.; Clemens, R.; Rappuoli, R. Hemagglutination inhibition antibody titers as a correlate of protection for inactivated influenza vaccines in children. Pediatr. Infect Dis. J. 2011, 30, 1081–1085. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Holmes, E.C.; Pybus, O.G. The Genomic Rate of Molecular Adaptation of the Human Influenza A Virus. Mol. Biol. Evol. 2011, 28, 2443–2451. [Google Scholar] [CrossRef]
- Nobusawa, E.; Aoyama, T.; Kato, H.; Suzuki, Y.; Tateno, Y.; Nakajima, K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology 1991, 182, 475–485. [Google Scholar] [CrossRef]
- Steinhauer, D.A. Role of Hemagglutinin Cleavage for the Pathogenicity of Influenza Virus. Virology 1999, 258, 1–20. [Google Scholar] [CrossRef]
- Worch, R. Structural biology of the influenza virus fusion peptide. Acta Biochim. Pol. 2014, 61. [Google Scholar] [CrossRef]
- Knossow, M.; Skehel, J.J. Variation and infectivity neutralization in influenza. Immunology 2006, 119, 1–7. [Google Scholar] [CrossRef]
- Kirkpatrick, E.; Qiu, X.; Wilson, P.C.; Bahl, J.; Krammer, F. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci. Rep. 2018, 8, 10432. [Google Scholar] [CrossRef]
- Krammer, F.; Palese, P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr. Opin. Virol. 2013, 3, 521–530. [Google Scholar] [CrossRef] [PubMed]
- DiLillo, D.J.; Palese, P.; Wilson, P.C.; Ravetch, J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Investig. 2016, 126, 605–610. [Google Scholar] [CrossRef] [PubMed]
- DiLillo, D.J.; Tan, G.S.; Palese, P.; Ravetch, J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 2014, 20, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Nath Neerukonda, S.; Vassell, R.; Weiss, C.D. Neutralizing Antibodies Targeting the Conserved Stem Region of Influenza Hemagglutinin. Vaccines 2020, 8, 382. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-C.; Lin, Y.-F.; Chong, P.; Wu, S.-C. Broader Neutralizing Antibodies against H5N1 Viruses Using Prime-Boost Immunization of Hyperglycosylated Hemagglutinin DNA and Virus-Like Particles. PLoS ONE 2012, 7, e39075. [Google Scholar] [CrossRef]
- Lin, S.-C.; Liu, W.-C.; Jan, J.-T.; Wu, S.-C. Glycan Masking of Hemagglutinin for Adenovirus Vector and Recombinant Protein Immunizations Elicits Broadly Neutralizing Antibodies against H5N1 Avian Influenza Viruses. PLoS ONE 2014, 9, e92822. [Google Scholar] [CrossRef] [PubMed]
- Eggink, D.; Goff, P.H.; Palese, P. Guiding the Immune Response against Influenza Virus Hemagglutinin toward the Conserved Stalk Domain by Hyperglycosylation of the Globular Head Domain. J. Virol. 2014, 88, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Graves, P.N.; Schulman, J.L.; Young, J.F.; Palese, P. Preparation of influenza virus subviral particles lacking the HA1 subunit of hemagglutinin: Unmasking of cross-reactive HA2 determinants. Virology 1983, 126, 106–116. [Google Scholar] [CrossRef]
- Wohlbold, T.J.; Nachbagauer, R.; Margine, I.; Tan, G.S.; Hirsh, A.; Krammer, F. Vaccination with soluble headless hemagglutinin protects mice from challenge with divergent influenza viruses. Vaccine 2015, 33, 3314–3321. [Google Scholar] [CrossRef]
- Chen, J.; Wharton, S.A.; Weissenhorn, W.; Calder, L.J.; Hughson, F.M.; Skehel, J.J.; Wiley, D.C. A soluble domain of the membrane-anchoring chain of influenza virus hemagglutinin (HA2) folds in Escherichia coli into the low-pH-induced conformation. Proc. Natl. Acad. Sci. USA 1995, 92, 12205–12209. [Google Scholar] [CrossRef] [PubMed]
- Swalley, S.E.; Baker, B.M.; Calder, L.J.; Harrison, S.C.; Skehel, J.J.; Wiley, D.C. Full-Length Influenza Hemagglutinin HA2 Refolds into the Trimeric Low-pH-Induced Conformation. Biochemistry 2004, 43, 5902–5911. [Google Scholar] [CrossRef]
- Bommakanti, G.; Citron, M.P.; Hepler, R.W.; Callahan, C.; Heidecker, G.J.; Najar, T.A.; Lu, X.; Joyce, J.G.; Shiver, J.W.; Casimiro, D.R.; et al. Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc. Natl. Acad. Sci. USA 2010, 107, 13701–13706. [Google Scholar] [CrossRef]
- Bommakanti, G.; Lu, X.; Citron, M.P.; Najar, T.A.; Heidecker, G.J.; ter Meulen, J.; Varadarajan, R.; Liang, X. Design of Escherichia coli-Expressed Stalk Domain Immunogens of H1N1 Hemagglutinin That Protect Mice from Lethal Challenge. J. Virol. 2012, 86, 13434–13444. [Google Scholar] [CrossRef]
- Impagliazzo, A.; Milder, F.; Kuipers, H.; Wagner, M.V.; Zhu, X.; Hoffman, R.M.; van Meersbergen, R.; Huizingh, J.; Wanningen, P.; Verspuij, J.; et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 2015, 349, 1301–1306. [Google Scholar] [CrossRef] [PubMed]
- Van der Lubbe, J.E.M.; Verspuij, J.W.A.; Huizingh, J.; Schmit-Tillemans, S.P.R.; Tolboom, J.; Dekking, L.; Kwaks, T.; Brandenburg, B.; Meijberg, W.; Zahn, R.C.; et al. Mini-HA Is Superior to Full Length Hemagglutinin Immunization in Inducing Stem-Specific Antibodies and Protection Against Group 1 Influenza Virus Challenges in Mice. Front Immunol. 2018, 9, 2350. [Google Scholar] [CrossRef] [PubMed]
- Valkenburg, S.A.; Mallajosyula, V.V.A.; Li, O.T.W.; Chin, A.W.H.; Carnell, G.; Temperton, N.; Varadarajan, R.; Poon, L.L.M. Stalking influenza by vaccination with pre-fusion headless HA mini-stem. Sci. Rep. 2016, 6, 22666. [Google Scholar] [CrossRef]
- Mallajosyula, V.V.; Citron, M.; Ferrara, F.; Lu, X.; Callahan, C.; Heidecker, G.J.; Sarma, S.P.; Flynn, J.A.; Temperton, N.J.; Liang, X.; et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc. Natl. Acad. Sci. USA 2014, 111, E2514–E2523. [Google Scholar] [CrossRef]
- Lu, Y.; Welsh, J.P.; Swartz, J.R. Production and stabilization of the trimeric influenza hemagglutinin stem domain for potentially broadly protective influenza vaccines. Proc. Natl. Acad. Sci. USA 2014, 111, 125–130. [Google Scholar] [CrossRef]
- Thrane, S.; Aves, K.L.; Uddbäck, I.E.M.; Janitzek, C.M.; Han, J.; Yang, Y.R.; Ward, A.B.; Theander, T.G.; Nielsen, M.A.; Salanti, A.; et al. A Vaccine Displaying a Trimeric Influenza-A HA Stem Protein on Capsid-Like Particles Elicits Potent and Long-Lasting Protection in Mice. Vaccines 2020, 8, 389. [Google Scholar] [CrossRef]
- Yassine, H.M.; Boyington, J.C.; McTamney, P.M.; Wei, C.J.; Kanekiyo, M.; Kong, W.P.; Gallagher, J.R.; Wang, L.; Zhang, Y.; Joyce, M.G.; et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015, 21, 1065–1070. [Google Scholar] [CrossRef]
- Deng, L.; Mohan, T.; Chang, T.Z.; Gonzalez, G.X.; Wang, Y.; Kwon, Y.M.; Kang, S.M.; Compans, R.W.; Champion, J.A.; Wang, B.Z. Double-layered protein nanoparticles induce broad protection against divergent influenza A viruses. Nat. Commun. 2018, 9, 359. [Google Scholar] [CrossRef]
- Corbett, K.S.; Moin, S.M.; Yassine, H.M.; Cagigi, A.; Kanekiyo, M.; Boyoglu-Barnum, S.; Myers, S.I.; Tsybovsky, Y.; Wheatley, A.K.; Schramm, C.A.; et al. Design of Nanoparticulate Group 2 Influenza Virus Hemagglutinin Stem Antigens That Activate Unmutated Ancestor B Cell Receptors of Broadly Neutralizing Antibody Lineages. mBio 2019, 10. [Google Scholar] [CrossRef]
- Wang, T.T.; Tan, G.S.; Hai, R.; Pica, N.; Ngai, L.; Ekiert, D.C.; Wilson, I.A.; García-Sastre, A.; Moran, T.M.; Palese, P. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc. Natl. Acad. Sci. USA 2010, 107, 18979–18984. [Google Scholar] [CrossRef]
- Chen, S.; Zheng, D.; Li, C.; Zhang, W.; Xu, W.; Liu, X.; Fang, F.; Chen, Z. Protection against Multiple Subtypes of Influenza Viruses by Virus-Like Particle Vaccines Based on a Hemagglutinin Conserved Epitope. BioMed Res. Int. 2015, 2015, 901817. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-S.; Chowdhury, M.Y.E.; Moon, H.-J.; Choi, Y.-K.; Talactac, M.R.; Kim, J.-H.; Park, M.-E.; Son, H.-Y.; Shin, K.-S.; Kim, C.-J. The highly conserved HA2 protein of the influenza a virus induces a cross protective immune response. J. Virol. Methods 2013, 194, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Hai, R.; Yondola, M.; Tan, G.S.; Leyva-Grado, V.H.; Ryder, A.B.; Miller, M.S.; Rose, J.K.; Palese, P.; García-Sastre, A.; et al. Assessment of influenza virus hemagglutinin stalk-based immunity in ferrets. J. Virol. 2014, 88, 3432–3442. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Pica, N.; Hai, R.; Margine, I.; Palese, P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol. 2013, 87, 6542–6550. [Google Scholar] [CrossRef] [PubMed]
- Ryder, A.B.; Nachbagauer, R.; Buonocore, L.; Palese, P.; Krammer, F.; Rose, J.K. Vaccination with Vesicular Stomatitis Virus-Vectored Chimeric Hemagglutinins Protects Mice against Divergent Influenza Virus Challenge Strains. J. Virol. 2015, 90, 2544–2550. [Google Scholar] [CrossRef]
- Isakova-Sivak, I.; Korenkov, D.; Smolonogina, T.; Kotomina, T.; Donina, S.; Matyushenko, V.; Mezhenskaya, D.; Krammer, F.; Rudenko, L. Broadly protective anti-hemagglutinin stalk antibodies induced by live attenuated influenza vaccine expressing chimeric hemagglutinin. Virology 2018, 518, 313–323. [Google Scholar] [CrossRef]
- Nachbagauer, R.; Krammer, F.; Albrecht, R.A. A Live-Attenuated Prime, Inactivated Boost Vaccination Strategy with Chimeric Hemagglutinin-Based Universal Influenza Virus Vaccines Provides Protection in Ferrets: A Confirmatory Study. Vaccines 2018, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Isakova-Sivak, I.; Matyushenko, V.; Kotomina, T.; Kiseleva, I.; Krutikova, E.; Donina, S.; Rekstin, A.; Larionova, N.; Mezhenskaya, D.; Sivak, K.; et al. Sequential Immunization with Universal Live Attenuated Influenza Vaccine Candidates Protects Ferrets against a High-Dose Heterologous Virus Challenge. Vaccines 2019, 7, 61. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.C.; Nachbagauer, R.; Stadlbauer, D.; Solórzano, A.; Berlanda-Scorza, F.; García-Sastre, A.; Palese, P.; Krammer, F.; Albrecht, R.A. Sequential Immunization With Live-Attenuated Chimeric Hemagglutinin-Based Vaccines Confers Heterosubtypic Immunity Against Influenza A Viruses in a Preclinical Ferret Model. Front Immunol. 2019, 10, 756. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.C.; Nachbagauer, R.; Stadlbauer, D.; Strohmeier, S.; Solórzano, A.; Berlanda-Scorza, F.; Innis, B.L.; García-Sastre, A.; Palese, P.; Krammer, F.; et al. Chimeric Hemagglutinin-Based Live-Attenuated Vaccines Confer Durable Protective Immunity against Influenza A Viruses in a Preclinical Ferret Model. Vaccines 2021, 9, 40. [Google Scholar] [CrossRef]
- Margine, I.; Krammer, F.; Hai, R.; Heaton, N.S.; Tan, G.S.; Andrews, S.A.; Runstadler, J.A.; Wilson, P.C.; Albrecht, R.A.; García-Sastre, A.; et al. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J. Virol. 2013, 87, 10435–10446. [Google Scholar] [CrossRef]
- Ermler, M.E.; Kirkpatrick, E.; Sun, W.; Hai, R.; Amanat, F.; Chromikova, V.; Palese, P.; Krammer, F. Chimeric Hemagglutinin Constructs Induce Broad Protection against Influenza B Virus Challenge in the Mouse Model. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Nachbagauer, R.; Feser, J.; Naficy, A.; Bernstein, D.I.; Guptill, J.; Walter, E.B.; Berlanda-Scorza, F.; Stadlbauer, D.; Wilson, P.C.; Aydillo, T.; et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 2021, 27, 106–114. [Google Scholar] [CrossRef]
- Bernstein, D.I.; Guptill, J.; Naficy, A.; Nachbagauer, R.; Berlanda-Scorza, F.; Feser, J.; Wilson, P.C.; Solórzano, A.; Van der Wielen, M.; Walter, E.B.; et al. Immunogenicity of chimeric haemagglutinin-based, universal influenza virus vaccine candidates: Interim results of a randomised, placebo-controlled, phase 1 clinical trial. Lancet Infect. Dis. 2020, 20, 80–91. [Google Scholar] [CrossRef]
- Broecker, F.; Liu, S.T.H.; Suntronwong, N.; Sun, W.; Bailey, M.J.; Nachbagauer, R.; Krammer, F.; Palese, P. A mosaic hemagglutinin-based influenza virus vaccine candidate protects mice from challenge with divergent H3N2 strains. NPJ Vaccines 2019, 4, 31. [Google Scholar] [CrossRef]
- Sun, W.; Kirkpatrick, E.; Ermler, M.; Nachbagauer, R.; Broecker, F.; Krammer, F.; Palese, P. Development of Influenza B Universal Vaccine Candidates Using the “Mosaic” Hemagglutinin Approach. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Steel, J.; Lowen, A.C.; Wang, T.T.; Yondola, M.; Gao, Q.; Haye, K.; García-Sastre, A.; Palese, P. Influenza Virus Vaccine Based on the Conserved Hemagglutinin Stalk Domain. mBio 2010, 1, e00018-10. [Google Scholar] [CrossRef]
- Jang, Y.H.; Seong, B.L. The Quest for a Truly Universal Influenza Vaccine. Front. Cell. Infect. Microbiol. 2019, 9. [Google Scholar] [CrossRef]
- Park, J.K.; Han, A.; Czajkowski, L.; Reed, S.; Athota, R.; Bristol, T.; Rosas, L.A.; Cervantes-Medina, A.; Taubenberger, J.K.; Memoli, M.J. Evaluation of Preexisting Anti-Hemagglutinin Stalk Antibody as a Correlate of Protection in a Healthy Volunteer Challenge with Influenza A/H1N1pdm Virus. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; Nachbagauer, R.; Balmaseda, A.; Stadlbauer, D.; Ojeda, S.; Patel, M.; Rajabhathor, A.; Lopez, R.; Guglia, A.F.; Sanchez, N.; et al. Novel correlates of protection against pandemic H1N1 influenza A virus infection. Nat. Med. 2019, 25, 962–967. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.R.; Toulmin, S.A.; Griesman, T.; Lamerato, L.E.; Petrie, J.G.; Martin, E.T.; Monto, A.S.; Hensley, S.E. Assessing the Protective Potential of H1N1 Influenza Virus Hemagglutinin Head and Stalk Antibodies in Humans. J. Virol. 2019, 93, e02134-18. [Google Scholar] [CrossRef] [PubMed]
- Han, A.; Czajkowski, L.; Rosas, L.A.; Cervantes-Medina, A.; Xiao, Y.; Gouzoulis, M.; Lumbard, K.; Hunsberger, S.; Reed, S.; Athota, R.; et al. Safety and Efficacy of CR6261 in an Influenza A H1N1 Healthy Human Challenge Model. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ellebedy, A.H.; Webby, R.J. Influenza vaccines. Vaccine 2009, 27, D65–D68. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-W.; Cheng, T.-J.R.; Huang, Y.; Jan, J.-T.; Ma, S.-H.; Yu, A.L.; Wong, C.-H.; Ho, D.D. A consensus-hemagglutinin-based DNA vaccine that protects mice against divergent H5N1 influenza viruses. Proc. Natl. Acad. Sci. USA 2008, 105, 13538–13543. [Google Scholar] [CrossRef]
- Laddy, D.J.; Yan, J.; Kutzler, M.; Kobasa, D.; Kobinger, G.P.; Khan, A.S.; Greenhouse, J.; Sardesai, N.Y.; Draghia-Akli, R.; Weiner, D.B. Heterosubtypic protection against pathogenic human and avian influenza viruses via in vivo electroporation of synthetic consensus DNA antigens. PLoS ONE 2008, 3, e2517. [Google Scholar] [CrossRef]
- Wu, P.; Lu, J.; Zhang, X.; Mei, M.; Feng, L.; Peng, D.; Hou, J.; Kang, S.-M.; Liu, X.; Tang, Y. Single Dose of Consensus Hemagglutinin-Based Virus-Like Particles Vaccine Protects Chickens against Divergent H5 Subtype Influenza Viruses. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef]
- Laddy, D.J.; Yan, J.; Corbitt, N.; Kobinger, G.P.; Weiner, D.B. Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine 2007, 25, 2984–2989. [Google Scholar] [CrossRef] [PubMed]
- Ducatez, M.F.; Bahl, J.; Griffin, Y.; Stigger-Rosser, E.; Franks, J.; Barman, S.; Vijaykrishna, D.; Webb, A.; Guan, Y.; Webster, R.G.; et al. Feasibility of reconstructed ancestral H5N1 influenza viruses for cross-clade protective vaccine development. Proc. Natl. Acad. Sci. USA 2011, 108, 349–354. [Google Scholar] [CrossRef]
- Elliott, S.T.C.; Keaton, A.A.; Chu, J.D.; Reed, C.C.; Garman, B.; Patel, A.; Yan, J.; Broderick, K.E.; Weiner, D.B. A Synthetic Micro-Consensus DNA Vaccine Generates Comprehensive Influenza A H3N2 Immunity and Protects Mice Against Lethal Challenge by Multiple H3N2 Viruses. Hum. Gene Ther. 2018, 29, 1044–1055. [Google Scholar] [CrossRef]
- Weaver, E.A.; Rubrum, A.M.; Webby, R.J.; Barry, M.A. Protection against Divergent Influenza H1N1 Virus by a Centralized Influenza Hemagglutinin. PLoS ONE 2011, 6, e18314. [Google Scholar] [CrossRef]
- Webby, R.J.; Weaver, E.A. Centralized Consensus Hemagglutinin Genes Induce Protective Immunity against H1, H3 and H5 Influenza Viruses. PLoS ONE 2015, 10, e0140702. [Google Scholar] [CrossRef]
- Lingel, A.; Bullard, B.L.; Weaver, E.A. Efficacy of an Adenoviral Vectored Multivalent Centralized Influenza Vaccine. Sci. Rep. 2017, 7, 14912. [Google Scholar] [CrossRef]
- Carter, D.M.; Darby, C.A.; Johnson, S.K.; Carlock, M.A.; Kirchenbaum, G.A.; Allen, J.D.; Vogel, T.U.; Delagrave, S.; DiNapoli, J.; Kleanthous, H.; et al. Elicitation of Protective Antibodies against a Broad Panel of H1N1 Viruses in Ferrets Preimmune to Historical H1N1 Influenza Viruses. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Carter, D.M.; Darby, C.A.; Lefoley, B.C.; Crevar, C.J.; Alefantis, T.; Oomen, R.; Anderson, S.F.; Strugnell, T.; Cortés-Garcia, G.; Vogel, T.U. Design and characterization of a computationally optimized broadly reactive hemagglutinin vaccine for H1N1 influenza viruses. J. Virol. 2016, 90, 4720–4734. [Google Scholar] [CrossRef] [PubMed]
- Sautto, G.A.; Kirchenbaum, G.A.; Ecker, J.W.; Bebin-Blackwell, A.G.; Pierce, S.R.; Ross, T.M. Elicitation of Broadly Protective Antibodies following Infection with Influenza Viruses Expressing H1N1 Computationally Optimized Broadly Reactive Hemagglutinin Antigens. ImmunoHorizons 2018, 2, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Sautto, G.A.; Kirchenbaum, G.A.; Abreu, R.B.; Ecker, J.W.; Pierce, S.R.; Kleanthous, H.; Ross, T.M. A Computationally Optimized Broadly Reactive Antigen Subtype-Specific Influenza Vaccine Strategy Elicits Unique Potent Broadly Neutralizing Antibodies against Hemagglutinin. J. Immunol. 2020, 204, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Darricarrère, N.; Pougatcheva, S.; Duan, X.; Rudicell, R.S.; Chou, T.H.; DiNapoli, J.; Ross, T.M.; Alefantis, T.; Vogel, T.U.; Kleanthous, H.; et al. Development of a Pan-H1 Influenza Vaccine. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Reneer, Z.B.; Jamieson, P.J.; Skarlupka, A.L.; Huang, Y.; Ross, T.M. Computationally Optimized Broadly Reactive H2 HA Influenza Vaccines Elicited Broadly Cross-Reactive Antibodies and Protected Mice from Viral Challenges. J. Virol. 2020, 95, e01526-20. [Google Scholar] [CrossRef]
- Wong, T.M.; Allen, J.D.; Bebin-Blackwell, A.G.; Carter, D.M.; Alefantis, T.; DiNapoli, J.; Kleanthous, H.; Ross, T.M. Computationally Optimized Broadly Reactive Hemagglutinin Elicits Hemagglutination Inhibition Antibodies against a Panel of H3N2 Influenza Virus Cocirculating Variants. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Allen, J.D.; Jang, H.; DiNapoli, J.; Kleanthous, H.; Ross, T.M. Elicitation of Protective Antibodies against 20 Years of Future H3N2 Cocirculating Influenza Virus Variants in Ferrets Preimmune to Historical H3N2 Influenza Viruses. J. Virol. 2019, 93, e00946-18. [Google Scholar] [CrossRef] [PubMed]
- Giles, B.M.; Ross, T.M. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 2011, 29, 3043–3054. [Google Scholar] [CrossRef] [PubMed]
- Giles, B.M.; Bissel, S.J.; Dealmeida, D.R.; Wiley, C.A.; Ross, T.M. Antibody breadth and protective efficacy are increased by vaccination with computationally optimized hemagglutinin but not with polyvalent hemagglutinin-based H5N1 virus-like particle vaccines. Clin. Vaccine Immunol. Cvi. 2012, 19, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Giles, B.M.; Crevar, C.J.; Carter, D.M.; Bissel, S.J.; Schultz-Cherry, S.; Wiley, C.A.; Ross, T.M. A Computationally Optimized Hemagglutinin Virus-Like Particle Vaccine Elicits Broadly Reactive Antibodies that Protect Nonhuman Primates from H5N1 Infection. J. Infect. Dis. 2012, 205, 1562–1570. [Google Scholar] [CrossRef]
- Crevar, C.J.; Carter, D.M.; Lee, K.Y.; Ross, T.M. Cocktail of H5N1 COBRA HA vaccines elicit protective antibodies against H5N1 viruses from multiple clades. Hum. Vaccin Immunother. 2015, 11, 572–583. [Google Scholar] [CrossRef]
- Allen, J.D.; Owino, S.O.; Carter, D.M.; Crevar, C.J.; Reese, V.A.; Fox, C.B.; Coler, R.N.; Reed, S.G.; Baldwin, S.L.; Ross, T.M. Broadened immunity and protective responses with emulsion-adjuvanted H5 COBRA-VLP vaccines. Vaccine 2017, 35, 5209–5216. [Google Scholar] [CrossRef] [PubMed]
- Bar-Peled, Y.; Huang, J.; Nuñez, I.A.; Pierce, S.R.; Ecker, J.W.; Ross, T.M.; Mousa, J.J. Structural and antigenic characterization of a computationally-optimized H5 hemagglutinin influenza vaccine. Vaccine 2019, 37, 6022–6029. [Google Scholar] [CrossRef]
- Ross, T.M.; DiNapoli, J.; Giel-Moloney, M.; Bloom, C.E.; Bertran, K.; Balzli, C.; Strugnell, T.; Mariana, S.E.S.; Mebatsion, T.; Bublot, M.; et al. A computationally designed H5 antigen shows immunological breadth of coverage and protects against drifting avian strains. Vaccine 2019, 37, 2369–2376. [Google Scholar] [CrossRef]
- Nuñez, I.A.; Ross, T.M. Human COBRA 2 vaccine contains two major epitopes that are responsible for eliciting neutralizing antibody responses against heterologous clades of viruses. Vaccine 2020, 38, 830–839. [Google Scholar] [CrossRef]
- Skarlupka, A.L.; Owino, S.O.; Suzuki-Williams, L.P.; Crevar, C.J.; Carter, D.M.; Ross, T.M. Computationally optimized broadly reactive vaccine based upon swine H1N1 influenza hemagglutinin sequences protects against both swine and human isolated viruses. Hum. Vaccin Immunother. 2019, 15, 2013–2029. [Google Scholar] [CrossRef]
- Cai, W.; Pei, J.; Grishin, N.V. Reconstruction of ancestral protein sequences and its applications. BMC Evol. Biol. 2004, 4, 33. [Google Scholar] [CrossRef]
- Ducatez, M.F.; Becker, J.; Freudenstein, A.; Delverdier, M.; Delpont, M.; Sutter, G.; Guérin, J.-L.; Volz, A. Low pathogenic avian influenza (H9N2) in chicken: Evaluation of an ancestral H9-MVA vaccine. Vet. Microbiol. 2016, 189, 59–67. [Google Scholar] [CrossRef]
- Qiu, X.; Duvvuri, V.R.; Bahl, J. Computational Approaches and Challenges to Developing Universal Influenza Vaccines. Vaccines 2019, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Thurmond, J.; Yoon, H.; Kuiken, C.; Yusim, K.; Perkins, S.; Theiler, J.; Bhattacharya, T.; Korber, B.; Fischer, W. Web-based design and evaluation of T-cell vaccine candidates. Bioinformatics 2008, 24, 1639–1640. [Google Scholar] [CrossRef]
- Fischer, W.; Perkins, S.; Theiler, J.; Bhattacharya, T.; Yusim, K.; Funkhouser, R.; Kuiken, C.; Haynes, B.; Letvin, N.L.; Walker, B.D.; et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat. Med. 2007, 13, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Kamlangdee, A.; Kingstad-Bakke, B.; Anderson, T.K.; Goldberg, T.L.; Osorio, J.E. Broad protection against avian influenza virus by using a modified vaccinia Ankara virus expressing a mosaic hemagglutinin gene. J. Virol. 2014, 88, 13300–13309. [Google Scholar] [CrossRef] [PubMed]
- Kamlangdee, A.; Kingstad-Bakke, B.; Osorio, J.E. Mosaic H5 Hemagglutinin Provides Broad Humoral and Cellular Immune Responses against Influenza Viruses. J. Virol. 2016, 90, 6771–6783. [Google Scholar] [CrossRef] [PubMed]
- Florek, N.W.; Kamlangdee, A.; Mutschler, J.P.; Kingstad-Bakke, B.; Schultz-Darken, N.; Broman, K.W.; Osorio, J.E.; Friedrich, T.C. A modified vaccinia Ankara vaccine vector expressing a mosaic H5 hemagglutinin reduces viral shedding in rhesus macaques. PLoS ONE 2017, 12, e0181738. [Google Scholar] [CrossRef] [PubMed]
- Kingstad-Bakke, B.A.; Chandrasekar, S.S.; Phanse, Y.; Ross, K.A.; Hatta, M.; Suresh, M.; Kawaoka, Y.; Osorio, J.E.; Narasimhan, B.; Talaat, A.M. Effective mosaic-based nanovaccines against avian influenza in poultry. Vaccine 2019, 37, 5051–5058. [Google Scholar] [CrossRef] [PubMed]
- Corder, B.N.; Bullard, B.L.; DeBeauchamp, J.L.; Ilyushina, N.A.; Webby, R.J.; Weaver, E.A. Influenza H1 Mosaic Hemagglutinin Vaccine Induces Broad Immunity and Protection in Mice. Vaccines 2019, 7, 195. [Google Scholar] [CrossRef]
- Theiler, J.; Korber, B. Graph-based optimization of epitope coverage for vaccine antigen design. Stat. Med. 2018, 37, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Theiler, J.; Yoon, H.; Yusim, K.; Picker, L.J.; Fruh, K.; Korber, B. Epigraph: A Vaccine Design Tool Applied to an HIV Therapeutic Vaccine and a Pan-Filovirus Vaccine. Sci. Rep. 2016, 6, 33987. [Google Scholar] [CrossRef] [PubMed]
- Bullard, B.L.; Corder, B.N.; DeBeauchamp, J.; Rubrum, A.; Korber, B.; Webby, R.J.; Weaver, E.A. Epigraph hemagglutinin vaccine induces broad cross-reactive immunity against swine H3 influenza virus. Nat. Commun. 2021, 12, 1203. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bullard, B.L.; Weaver, E.A. Strategies Targeting Hemagglutinin as a Universal Influenza Vaccine. Vaccines 2021, 9, 257. https://doi.org/10.3390/vaccines9030257
Bullard BL, Weaver EA. Strategies Targeting Hemagglutinin as a Universal Influenza Vaccine. Vaccines. 2021; 9(3):257. https://doi.org/10.3390/vaccines9030257
Chicago/Turabian StyleBullard, Brianna L., and Eric A. Weaver. 2021. "Strategies Targeting Hemagglutinin as a Universal Influenza Vaccine" Vaccines 9, no. 3: 257. https://doi.org/10.3390/vaccines9030257
APA StyleBullard, B. L., & Weaver, E. A. (2021). Strategies Targeting Hemagglutinin as a Universal Influenza Vaccine. Vaccines, 9(3), 257. https://doi.org/10.3390/vaccines9030257





