mRNA Vaccines against Flaviviruses
Abstract
:1. Introduction
2. RNA Vaccine Overview
3. Flavivirus Molecular Biology
4. Antibody-Dependent Enhancement
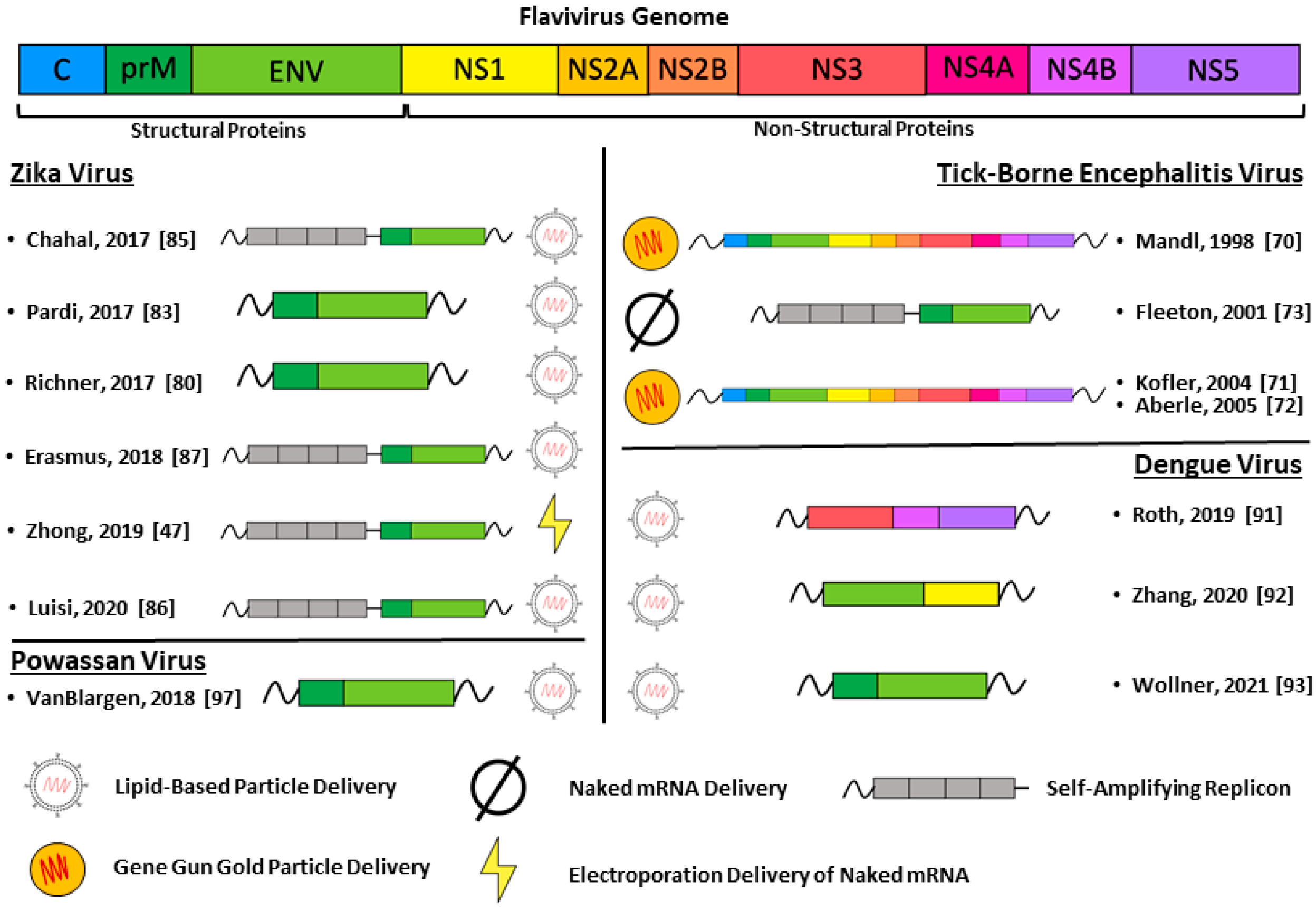
5. Flavivirus mRNA Vaccines
5.1. Tick-Borne Encephalitis Virus
5.2. Zika Virus
5.3. Dengue Virus
5.4. Powassan Virus
6. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lindenbach, B.D.; Murray, C.L.; Thiel, H.J.; Rice, C.M. Flaviviridae. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume One, pp. 712–746. [Google Scholar]
- Holbrook, M.R. Historical Perspectives on Flavivirus Research. Viruses 2017, 9, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faria, N.R.; Azevedo, R.; Kraemer, M.U.G.; Souza, R.; Cunha, M.S.; Hill, S.C.; Theze, J.; Bonsall, M.B.; Bowden, T.A.; Rissanen, I.; et al. Zika virus in the Americas: Early epidemiological and genetic findings. Science 2016, 352, 345–349. [Google Scholar] [CrossRef] [Green Version]
- Barrett, A.D.T. Yellow fever live attenuated vaccine: A very successful live attenuated vaccine but still we have problems controlling the disease. Vaccine 2017, 35, 5951–5955. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.H.; Metz, S.W. Progress and Works in Progress: Update on Flavivirus Vaccine Development. Clin. Ther. 2017, 39, 1519–1536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theiler, M.; Smith, H.H. The Use of Yellow Fever Virus Modified by in Vitro Cultivation for Human Immunization. J. Exp. Med. 1937, 65, 787–800. [Google Scholar] [CrossRef] [Green Version]
- Hoke, C.H.; Nisalak, A.; Sangawhipa, N.; Jatanasen, S.; Laorakapongse, T.; Innis, B.L.; Kotchasenee, S.; Gingrich, J.B.; Latendresse, J.; Fukai, K.; et al. Protection against Japanese encephalitis by inactivated vaccines. N. Engl. J. Med. 1988, 319, 608–614. [Google Scholar] [CrossRef]
- Smorodintseff, A.A.; Kagan, N.W.; Levkovitsch, E.N.; Dankovskij, N.L. Experimenteller und epidemiologischer Beitrag zur aktiven Immunisierung gegen die Frühling-Sommer-Zeckenencephalitis. Arch. Für die Gesamte Virusforsch. 1941, 2, 1–25. [Google Scholar] [CrossRef]
- Yun, S.I.; Lee, Y.M. Japanese encephalitis: The virus and vaccines. Hum. Vaccin Immunother 2014, 10, 263–279. [Google Scholar] [CrossRef] [Green Version]
- Lindquist, L.; Vapalahti, O. Tick-borne encephalitis. Lancet 2008, 371, 1861–1871. [Google Scholar] [CrossRef]
- Kaiser, J.A.; Barrett, A.D.T. Twenty Years of Progress Toward West Nile Virus Vaccine Development. Viruses 2019, 11, 823. [Google Scholar] [CrossRef] [Green Version]
- De Silva, A.M.; Harris, E. Which Dengue Vaccine Approach Is the Most Promising, and Should We Be Concerned about Enhanced Disease after Vaccination? The Path to a Dengue Vaccine: Learning from Human Natural Dengue Infection Studies and Vaccine Trials. Cold Spring Harb. Perspect Biol. 2018, 10, a029371. [Google Scholar] [CrossRef]
- Thomas, S.J.; Rothman, A.L. Trials and Tribulations on the Path to Developing a Dengue Vaccine. Am. J. Prev. Med. 2015, 49, S334–344. [Google Scholar] [CrossRef]
- Shan, C.; Xie, X.; Shi, P.Y. Zika Virus Vaccine: Progress and Challenges. Cell Host Microbe 2018, 24, 12–17. [Google Scholar] [CrossRef] [Green Version]
- Jackson, N.A.C.; Kester, K.E.; Casimiro, D.; Gurunathan, S.; DeRosa, F. The promise of mRNA vaccines: A biotech and industrial perspective. NPJ Vaccines 2020, 5, 11. [Google Scholar] [CrossRef]
- Kis, Z.; Kontoravdi, C.; Shattock, R.; Shah, N. Resources, Production Scales and Time Required for Producing RNA Vaccines for the Global Pandemic Demand. Vaccines 2020, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardi, N.; Parkhouse, K.; Kirkpatrick, E.; McMahon, M.; Zost, S.J.; Mui, B.L.; Tam, Y.K.; Kariko, K.; Barbosa, C.J.; Madden, T.D.; et al. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018, 9, 3361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahl, K.; Senn, J.J.; Yuzhakov, O.; Bulychev, A.; Brito, L.A.; Hassett, K.J.; Laska, M.E.; Smith, M.; Almarsson, O.; Thompson, J.; et al. Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses. Mol. Ther. 2017, 25, 1316–1327. [Google Scholar] [CrossRef] [Green Version]
- Brazzoli, M.; Magini, D.; Bonci, A.; Buccato, S.; Giovani, C.; Kratzer, R.; Zurli, V.; Mangiavacchi, S.; Casini, D.; Brito, L.M.; et al. Induction of Broad-Based Immunity and Protective Efficacy by Self-amplifying mRNA Vaccines Encoding Influenza Virus Hemagglutinin. J. Virol. 2016, 90, 332–344. [Google Scholar] [CrossRef] [Green Version]
- Vogel, A.B.; Lambert, L.; Kinnear, E.; Busse, D.; Erbar, S.; Reuter, K.C.; Wicke, L.; Perkovic, M.; Beissert, T.; Haas, H.; et al. Self-Amplifying RNA Vaccines Give Equivalent Protection against Influenza to mRNA Vaccines but at Much Lower Doses. Mol. Ther. 2018, 26, 446–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardi, N.; LaBranche, C.C.; Ferrari, G.; Cain, D.W.; Tombacz, I.; Parks, R.J.; Muramatsu, H.; Mui, B.L.; Tam, Y.K.; Kariko, K.; et al. Characterization of HIV-1 Nucleoside-Modified mRNA Vaccines in Rabbits and Rhesus Macaques. Mol. Ther. Nucleic Acids 2019, 15, 36–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnee, M.; Vogel, A.B.; Voss, D.; Petsch, B.; Baumhof, P.; Kramps, T.; Stitz, L. An mRNA Vaccine Encoding Rabies Virus Glycoprotein Induces Protection against Lethal Infection in Mice and Correlates of Protection in Adult and Newborn Pigs. PLoS Negl. Trop Dis. 2016, 10, e0004746. [Google Scholar] [CrossRef]
- Alberer, M.; Gnad-Vogt, U.; Hong, H.S.; Mehr, K.T.; Backert, L.; Finak, G.; Gottardo, R.; Bica, M.A.; Garofano, A.; Koch, S.D.; et al. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: An open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet 2017, 390, 1511–1520. [Google Scholar] [CrossRef]
- Kose, N.; Fox, J.M.; Sapparapu, G.; Bombardi, R.; Tennekoon, R.N.; de Silva, A.D.; Elbashir, S.M.; Theisen, M.A.; Humphris-Narayanan, E.; Ciaramella, G.; et al. A lipid-encapsulated mRNA encoding a potently neutralizing human monoclonal antibody protects against chikungunya infection. Sci. Immunol. 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.S.; Jenks, J.A.; Pardi, N.; Goodwin, M.; Roark, H.; Edwards, W.; McLellan, J.S.; Pollara, J.; Weissman, D.; Permar, S.R. Human Cytomegalovirus Glycoprotein B Nucleoside-Modified mRNA Vaccine Elicits Antibody Responses with Greater Durability and Breadth than MF59-Adjuvanted gB Protein Immunization. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020. [CrossRef]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef]
- Mulligan, M.J.; Lyke, K.E.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Raabe, V.; Bailey, R.; Swanson, K.A.; et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 2020, 586, 589–593. [Google Scholar] [CrossRef]
- Ross, J.; Sullivan, T.D. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood 1985, 66, 1149–1154. [Google Scholar] [CrossRef] [Green Version]
- Holtkamp, S.; Kreiter, S.; Selmi, A.; Simon, P.; Koslowski, M.; Huber, C.; Tureci, O.; Sahin, U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 2006, 108, 4009–4017. [Google Scholar] [CrossRef]
- Gallie, D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991, 5, 2108–2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsui, N.B.; Ng, E.K.; Lo, Y.M. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin. Chem. 2002, 48, 1647–1653. [Google Scholar] [CrossRef] [PubMed]
- Kariko, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008, 16, 1833–1840. [Google Scholar] [CrossRef]
- Weissman, D.; Pardi, N.; Muramatsu, H.; Kariko, K. HPLC purification of in vitro transcribed long RNA. Methods Mol. Biol. 2013, 969, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Kariko, K.; Muramatsu, H.; Ludwig, J.; Weissman, D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011, 39, e142. [Google Scholar] [CrossRef] [Green Version]
- Baiersdorfer, M.; Boros, G.; Muramatsu, H.; Mahiny, A.; Vlatkovic, I.; Sahin, U.; Kariko, K. A Facile Method for the Removal of dsRNA Contaminant from In Vitro-Transcribed mRNA. Mol. Ther. Nucleic Acids 2019, 15, 26–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kariko, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Isaacs, A.; Cox, R.A.; Rotem, Z. Foreign nucleic acids as the stimulus to make interferon. Lancet 1963, 2, 113–116. [Google Scholar] [CrossRef]
- Anderson, B.R.; Muramatsu, H.; Jha, B.K.; Silverman, R.H.; Weissman, D.; Kariko, K. Nucleoside modifications in RNA limit activation of 2’-5’-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011, 39, 9329–9338. [Google Scholar] [CrossRef] [Green Version]
- Anderson, B.R.; Muramatsu, H.; Nallagatla, S.R.; Bevilacqua, P.C.; Sansing, L.H.; Weissman, D.; Kariko, K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010, 38, 5884–5892. [Google Scholar] [CrossRef] [Green Version]
- Pardi, N.; Tuyishime, S.; Muramatsu, H.; Kariko, K.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; Hope, M.J.; Weissman, D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release 2015, 217, 345–351. [Google Scholar] [CrossRef] [Green Version]
- Shirai, S.; Kawai, A.; Shibuya, M.; Munakata, L.; Omata, D.; Suzuki, R.; Yoshioka, Y. Lipid Nanoparticle Acts as a Potential Adjuvant for Influenza Split Vaccine without Inducing Inflammatory Responses. Vaccines 2020, 8, 433. [Google Scholar] [CrossRef]
- Swaminathan, G.; Thoryk, E.A.; Cox, K.S.; Meschino, S.; Dubey, S.A.; Vora, K.A.; Celano, R.; Gindy, M.; Casimiro, D.R.; Bett, A.J. A novel lipid nanoparticle adjuvant significantly enhances B cell and T cell responses to sub-unit vaccine antigens. Vaccine 2016, 34, 110–119. [Google Scholar] [CrossRef]
- Huysmans, H.; Zhong, Z.; De Temmerman, J.; Mui, B.L.; Tam, Y.K.; Mc Cafferty, S.; Gitsels, A.; Vanrompay, D.; Sanders, N.N. Expression Kinetics and Innate Immune Response after Electroporation and LNP-Mediated Delivery of a Self-Amplifying mRNA in the Skin. Mol. Ther. Nucleic Acids 2019, 17, 867–878. [Google Scholar] [CrossRef] [Green Version]
- Nikonov, A.; Molder, T.; Sikut, R.; Kiiver, K.; Mannik, A.; Toots, U.; Lulla, A.; Lulla, V.; Utt, A.; Merits, A.; et al. RIG-I and MDA-5 detection of viral RNA-dependent RNA polymerase activity restricts positive-strand RNA virus replication. PLoS Pathogens 2013, 9, e1003610. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Z.; Portela Catani, J.P.; Mc Cafferty, S.; Couck, L.; Van Den Broeck, W.; Gorle, N.; Vandenbroucke, R.E.; Devriendt, B.; Ulbert, S.; Cnops, L.; et al. Immunogenicity and Protection Efficacy of a Naked Self-Replicating mRNA-Based Zika Virus Vaccine. Vaccines 2019, 7, 96. [Google Scholar] [CrossRef] [Green Version]
- Bloom, K.; van den Berg, F.; Arbuthnot, P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 2020, 10. [Google Scholar] [CrossRef]
- Beissert, T.; Koste, L.; Perkovic, M.; Walzer, K.C.; Erbar, S.; Selmi, A.; Diken, M.; Kreiter, S.; Tureci, O.; Sahin, U. Improvement of In Vivo Expression of Genes Delivered by Self-Amplifying RNA Using Vaccinia Virus Immune Evasion Proteins. Hum. Gene Ther. 2017, 28, 1138–1146. [Google Scholar] [CrossRef] [Green Version]
- Yoshioka, N.; Gros, E.; Li, H.R.; Kumar, S.; Deacon, D.C.; Maron, C.; Muotri, A.R.; Chi, N.C.; Fu, X.D.; Yu, B.D.; et al. Efficient generation of human iPSCs by a synthetic self-replicative RNA. Cell Stem Cell 2013, 13, 246–254. [Google Scholar] [CrossRef] [Green Version]
- Diamond, M.S. Flaviviruses. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume One, pp. 747–792. [Google Scholar]
- Metz, S.W.; Thomas, A.; White, L.; Stoops, M.; Corten, M.; Hannemann, H.; de Silva, A.M. Dengue virus-like particles mimic the antigenic properties of the infectious dengue virus envelope. Virol. J. 2018, 15, 60. [Google Scholar] [CrossRef]
- Rey, F.A.; Stiasny, K.; Vaney, M.C.; Dellarole, M.; Heinz, F.X. The bright and the dark side of human antibody responses to flaviviruses: Lessons for vaccine design. EMBO Rep. 2018, 19, 206–224. [Google Scholar] [CrossRef]
- Cruz-Oliveira, C.; Freire, J.M.; Conceicao, T.M.; Higa, L.M.; Castanho, M.A.; Da Poian, A.T. Receptors and routes of dengue virus entry into the host cells. FEMS Microbiol Rev. 2015, 39, 155–170. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Lok, S.M.; Yu, I.M.; Zhang, Y.; Kuhn, R.J.; Chen, J.; Rossmann, M.G. The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science 2008, 319, 1830–1834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cherrier, M.V.; Kaufmann, B.; Nybakken, G.E.; Lok, S.M.; Warren, J.T.; Chen, B.R.; Nelson, C.A.; Kostyuchenko, V.A.; Holdaway, H.A.; Chipman, P.R.; et al. Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody. EMBO J. 2009, 28, 3269–3276. [Google Scholar] [CrossRef] [Green Version]
- Plevka, P.; Battisti, A.J.; Junjhon, J.; Winkler, D.C.; Holdaway, H.A.; Keelapang, P.; Sittisombut, N.; Kuhn, R.J.; Steven, A.C.; Rossmann, M.G. Maturation of flaviviruses starts from one or more icosahedrally independent nucleation centres. EMBO Rep. 2011, 12, 602–606. [Google Scholar] [CrossRef] [Green Version]
- Allison, S.L.; Schalich, J.; Stiasny, K.; Mandl, C.W.; Kunz, C.; Heinz, F.X. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 1995, 69, 695–700. [Google Scholar] [CrossRef] [Green Version]
- Zaitseva, E.; Yang, S.T.; Melikov, K.; Pourmal, S.; Chernomordik, L.V. Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS Pathogens 2010, 6, e1001131. [Google Scholar] [CrossRef] [Green Version]
- Halstead, S.B.; Nimmannitya, S.; Cohen, S.N. Observations related to pathogenesis of dengue hemorrhagic fever. IV. Relation of disease severity to antibody response and virus recovered. Yale J. Biol. Med. 1970, 42, 311–328. [Google Scholar]
- Katzelnick, L.C.; Gresh, L.; Halloran, M.E.; Mercado, J.C.; Kuan, G.; Gordon, A.; Balmaseda, A.; Harris, E. Antibody-dependent enhancement of severe dengue disease in humans. Science 2017, 358, 929–932. [Google Scholar] [CrossRef] [Green Version]
- Balsitis, S.J.; Williams, K.L.; Lachica, R.; Flores, D.; Kyle, J.L.; Mehlhop, E.; Johnson, S.; Diamond, M.S.; Beatty, P.R.; Harris, E. Lethal antibody enhancement of dengue disease in mice is prevented by Fc modification. PLoS Pathogens 2010, 6, e1000790. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.J.; Yang, Y.C.; Lin, Y.S.; Huang, J.H.; Liu, H.S.; Yeh, T.M.; Chen, S.H.; Liu, C.C.; Lei, H.Y. The dual-specific binding of dengue virus and target cells for the antibody-dependent enhancement of dengue virus infection. J. Immun. 2006, 176, 2825–2832. [Google Scholar] [CrossRef] [Green Version]
- Chareonsirisuthigul, T.; Kalayanarooj, S.; Ubol, S. Dengue virus (DENV) antibody-dependent enhancement of infection upregulates the production of anti-inflammatory cytokines, but suppresses anti-DENV free radical and pro-inflammatory cytokine production, in THP-1 cells. J. Gen. Virol. 2007, 88, 365–375. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Jumnainsong, A.; Onsirisakul, N.; Fitton, P.; Vasanawathana, S.; Limpitikul, W.; Puttikhunt, C.; Edwards, C.; Duangchinda, T.; Supasa, S.; et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science 2010, 328, 745–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katzelnick, L.C.; Narvaez, C.; Arguello, S.; Lopez Mercado, B.; Collado, D.; Ampie, O.; Elizondo, D.; Miranda, T.; Bustos Carillo, F.; Mercado, J.C.; et al. Zika virus infection enhances future risk of severe dengue disease. Science 2020, 369, 1123–1128. [Google Scholar] [CrossRef]
- Henein, S.; Swanstrom, J.; Byers, A.M.; Moser, J.M.; Shaik, S.F.; Bonaparte, M.; Jackson, N.; Guy, B.; Baric, R.; de Silva, A.M. Dissecting Antibodies Induced by a Chimeric Yellow Fever-Dengue, Live-Attenuated, Tetravalent Dengue Vaccine (CYD-TDV) in Naive and Dengue-Exposed Individuals. J. Infect. Dis. 2017, 215, 351–358. [Google Scholar] [CrossRef] [Green Version]
- Halstead, S.B. Dengvaxia sensitizes seronegatives to vaccine enhanced disease regardless of age. Vaccine 2017, 35, 6355–6358. [Google Scholar] [CrossRef]
- Dengue Vaccine: WHO position paper—September 2018. Wkly. Epidemiol. Record 2018, 93, 457–476.
- Mandl, C.W.; Aberle, J.H.; Aberle, S.W.; Holzmann, H.; Allison, S.L.; Heinz, F.X. In vitro-synthesized infectious RNA as an attenuated live vaccine in a flavivirus model. Nat. Med. 1998, 4, 1438–1440. [Google Scholar] [CrossRef] [PubMed]
- Kofler, R.M.; Aberle, J.H.; Aberle, S.W.; Allison, S.L.; Heinz, F.X.; Mandl, C.W. Mimicking live flavivirus immunization with a noninfectious RNA vaccine. Proc. Natl. Acad. Sci. USA 2004, 101, 1951–1956. [Google Scholar] [CrossRef] [Green Version]
- Aberle, J.H.; Aberle, S.W.; Kofler, R.M.; Mandl, C.W. Humoral and cellular immune response to RNA immunization with flavivirus replicons derived from tick-borne encephalitis virus. J. Virol. 2005, 79, 15107–15113. [Google Scholar] [CrossRef] [Green Version]
- Fleeton, M.N.; Chen, M.; Berglund, P.; Rhodes, G.; Parker, S.E.; Murphy, M.; Atkins, G.J.; Liljestrom, P. Self-replicative RNA vaccines elicit protection against influenza A virus, respiratory syncytial virus, and a tickborne encephalitis virus. J. Infect. Dis. 2001, 183, 1395–1398. [Google Scholar] [CrossRef] [Green Version]
- Musso, D.; Ko, A.I.; Baud, D. Zika Virus Infection—After the Pandemic. N. Engl. J. Med. 2019, 381, 1444–1457. [Google Scholar] [CrossRef]
- Do Rosario, M.S.; de Jesus, P.A.; Vasilakis, N.; Farias, D.S.; Novaes, M.A.; Rodrigues, S.G.; Martins, L.C.; Vasconcelos, P.F.; Ko, A.I.; Alcantara, L.C.; et al. Guillain-Barre Syndrome After Zika Virus Infection in Brazil. Am. J. Trop Med. Hyg. 2016, 95, 1157–1160. [Google Scholar] [CrossRef] [Green Version]
- Barbi, L.; Coelho, A.V.C.; Alencar, L.C.A.; Crovella, S. Prevalence of Guillain-Barre syndrome among Zika virus infected cases: A systematic review and meta-analysis. Braz J. Infect. Dis. 2018, 22, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Song, H.; Ming, G.L. How does Zika virus cause microcephaly? Genes Dev. 2017, 31, 849–861. [Google Scholar] [CrossRef] [Green Version]
- Congenital Zika Syndrome & Other Birth Defects. Available online: https://www.cdc.gov/pregnancy/zika/testing-follow-up/zika-syndrome-birth-defects.html (accessed on 7 January 2021).
- Richner, J.M.; Diamond, M.S. Zika virus vaccines: Immune response, current status, and future challenges. Curr. Opin. Immunol. 2018, 53, 130–136. [Google Scholar] [CrossRef]
- Richner, J.M.; Himansu, S.; Dowd, K.A.; Butler, S.L.; Salazar, V.; Fox, J.M.; Julander, J.G.; Tang, W.W.; Shresta, S.; Pierson, T.C.; et al. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell 2017, 168, 1114–1125 e1110. [Google Scholar] [CrossRef] [Green Version]
- Richner, J.M.; Jagger, B.W.; Shan, C.; Fontes, C.R.; Dowd, K.A.; Cao, B.; Himansu, S.; Caine, E.A.; Nunes, B.T.D.; Medeiros, D.B.A.; et al. Vaccine Mediated Protection Against Zika Virus-Induced Congenital Disease. Cell 2017, 170, 273–283 e212. [Google Scholar] [CrossRef] [Green Version]
- Interim Phase 1 Data for Zika Virus Vaccine (mRNA-1893). Available online: https://investors.modernatx.com/news-releases/news-release-details/moderna-highlights-opportunity-mrna-vaccines-its-first-vaccines (accessed on 14 April 2020).
- Pardi, N.; Hogan, M.J.; Pelc, R.S.; Muramatsu, H.; Andersen, H.; DeMaso, C.R.; Dowd, K.A.; Sutherland, L.L.; Scearce, R.M.; Parks, R.; et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 2017, 543, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.J.; Naradikian, M.S.; Parkhouse, K.; Cain, D.W.; Jones, L.; Moody, M.A.; Verkerke, H.P.; Myles, A.; Willis, E.; et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018, 215, 1571–1588. [Google Scholar] [CrossRef]
- Chahal, J.S.; Fang, T.; Woodham, A.W.; Khan, O.F.; Ling, J.; Anderson, D.G.; Ploegh, H.L. An RNA nanoparticle vaccine against Zika virus elicits antibody and CD8+ T cell responses in a mouse model. Sci. Rep. 2017, 7, 252. [Google Scholar] [CrossRef] [PubMed]
- Luisi, K.; Morabito, K.M.; Burgomaster, K.E.; Sharma, M.; Kong, W.P.; Foreman, B.M.; Patel, S.; Fisher, B.; Aleshnick, M.A.; Laliberte, J.; et al. Development of a potent Zika virus vaccine using self-amplifying messenger RNA. Sci. Adv. 2020, 6, eaba5068. [Google Scholar] [CrossRef] [PubMed]
- Erasmus, J.H.; Khandhar, A.P.; Guderian, J.; Granger, B.; Archer, J.; Archer, M.; Gage, E.; Fuerte-Stone, J.; Larson, E.; Lin, S.; et al. A Nanostructured Lipid Carrier for Delivery of a Replicating Viral RNA Provides Single, Low-Dose Protection against Zika. Mol. Ther. 2018, 26, 2507–2522. [Google Scholar] [CrossRef] [Green Version]
- Stettler, K.; Beltramello, M.; Espinosa, D.A.; Graham, V.; Cassotta, A.; Bianchi, S.; Vanzetta, F.; Minola, A.; Jaconi, S.; Mele, F.; et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 2016, 353, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Bardina, S.V.; Bunduc, P.; Tripathi, S.; Duehr, J.; Frere, J.J.; Brown, J.A.; Nachbagauer, R.; Foster, G.A.; Krysztof, D.; Tortorella, D.; et al. Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science 2017, 356, 175–180. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- Roth, C.; Cantaert, T.; Colas, C.; Prot, M.; Casademont, I.; Levillayer, L.; Thalmensi, J.; Langlade-Demoyen, P.; Gerke, C.; Bahl, K.; et al. A Modified mRNA Vaccine Targeting Immunodominant NS Epitopes Protects Against Dengue Virus Infection in HLA Class I Transgenic Mice. Front. Immunol. 2019, 10, 1424. [Google Scholar] [CrossRef]
- Zhang, M.; Sun, J.; Li, M.; Jin, X. Modified mRNA-LNP Vaccines Confer Protection against Experimental DENV-2 Infection in Mice. Mol. Ther. Methods Clin. Dev. 2020, 18, 702–712. [Google Scholar] [CrossRef]
- Wollner, C.J.; Richner, M.; Hassert, M.A.; Pinto, A.K.; Brien, J.D.; Richner, J.M. A mRNA-LNP vaccine against Dengue Virus elicits robust, serotype-specific immunity. bioRxiv 2021. [CrossRef]
- Powassan Statistics & Maps. Available online: https://www.cdc.gov/powassan/statistics.html (accessed on 7 January 2021).
- Ebel, G.D. Update on Powassan virus: Emergence of a North American tick-borne flavivirus. Annu. Rev. Entomol. 2010, 55, 95–110. [Google Scholar] [CrossRef]
- Hermance, M.E.; Thangamani, S. Powassan Virus: An Emerging Arbovirus of Public Health Concern in North America. Vector Borne Zoonotic Dis. 2017, 17, 453–462. [Google Scholar] [CrossRef] [PubMed]
- VanBlargan, L.A.; Himansu, S.; Foreman, B.M.; Ebel, G.D.; Pierson, T.C.; Diamond, M.S. An mRNA Vaccine Protects Mice against Multiple Tick-Transmitted Flavivirus Infections. Cell Rep. 2018, 25, 3382–3392.e3383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierson, T.C.; Diamond, M.S. The continued threat of emerging flaviviruses. Nat. Microbiol. 2020, 5, 796–812. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Dejnirattisai, W.; Wongwiwat, W.; Supasa, S.; Zhang, X.; Dai, X.; Rouvinski, A.; Jumnainsong, A.; Edwards, C.; Quyen, N.T.H.; Duangchinda, T.; et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 2015, 16, 170–177. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wollner, C.J.; Richner, J.M. mRNA Vaccines against Flaviviruses. Vaccines 2021, 9, 148. https://doi.org/10.3390/vaccines9020148
Wollner CJ, Richner JM. mRNA Vaccines against Flaviviruses. Vaccines. 2021; 9(2):148. https://doi.org/10.3390/vaccines9020148
Chicago/Turabian StyleWollner, Clayton J., and Justin M. Richner. 2021. "mRNA Vaccines against Flaviviruses" Vaccines 9, no. 2: 148. https://doi.org/10.3390/vaccines9020148
APA StyleWollner, C. J., & Richner, J. M. (2021). mRNA Vaccines against Flaviviruses. Vaccines, 9(2), 148. https://doi.org/10.3390/vaccines9020148






