Antibody Focusing to Conserved Sites of Vulnerability: The Immunological Pathways for ‘Universal’ Influenza Vaccines
Abstract
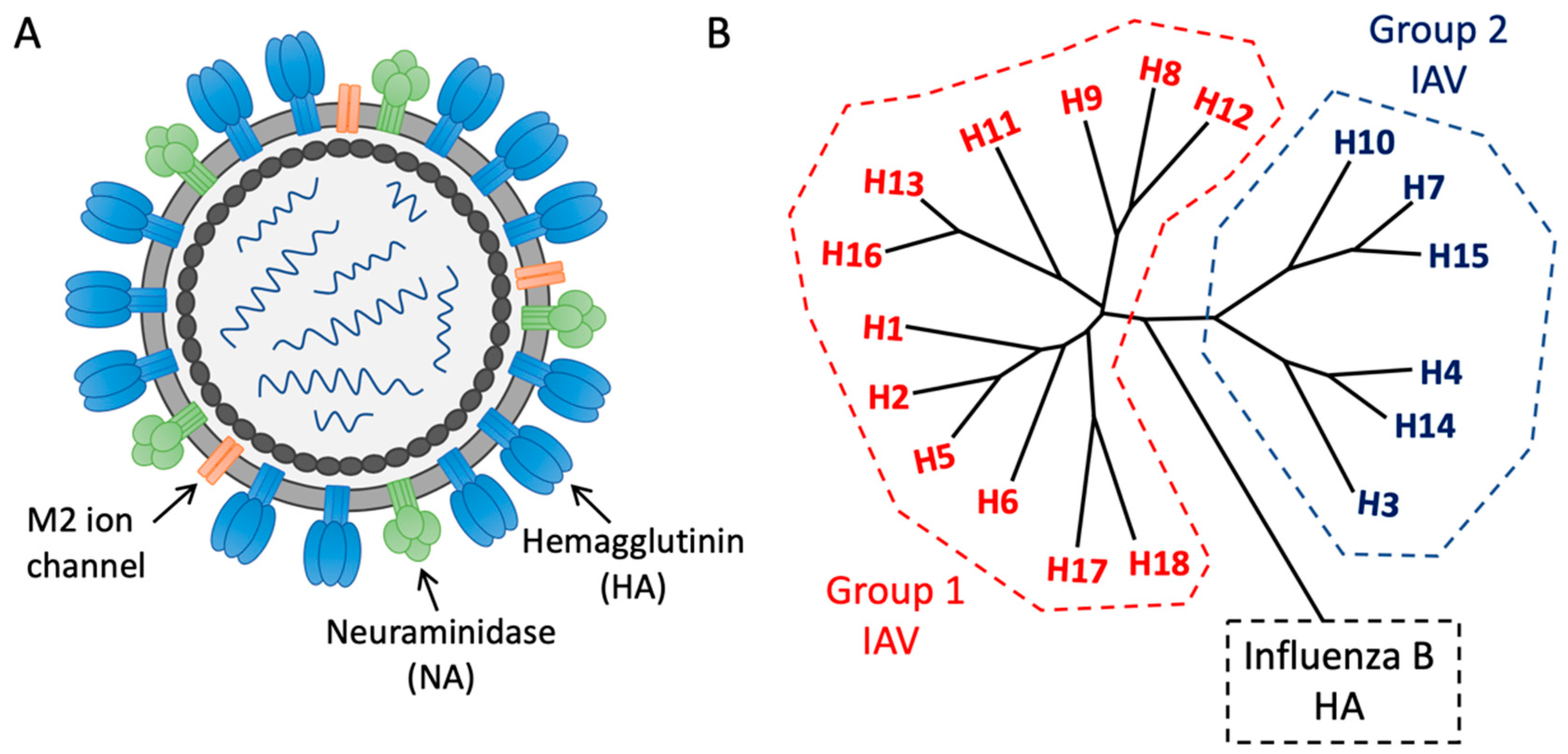
1. Introduction—The Need for Universal Influenza Vaccines
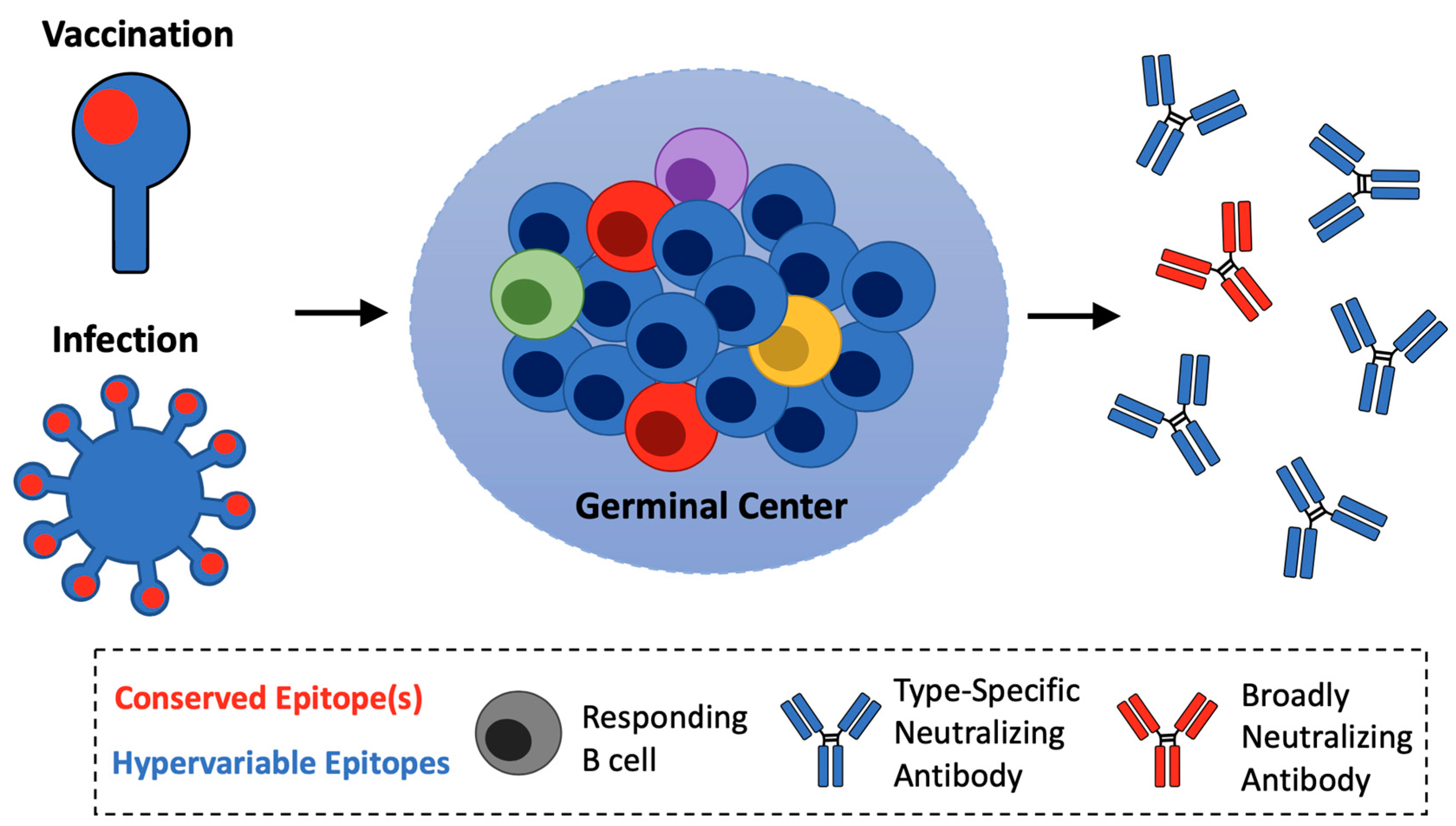
2. Subverting Natural Immunodominance Hierarchies: An Immunological Challenge for Universal Influenza Vaccines
3. Functionally Conserved Antibody Targets on Influenza Hemagglutinin
3.1. HA Head Epitopes
3.2. HA Stalk Epitopes
4. Alternative Universal Vaccine Targets
4.1. Neuraminidase
4.2. Matrix 2 Ectodomain
5. Refocusing Humoral Immunity Upon Conserved Sites of Vulnerability—Proof of Concept
5.1. HA Stalk-Based Immunogens
5.2. HA Head-Based Immunogens
6. Pathway Amplification as an Immunological Principle for Reproducible Vaccine Expansion of On-Target B cells
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nabel, G.J.; Fauci, A.S. Induction of unnatural immunity: Prospects for a broadly protective universal influenza vaccine. Nat. Med. 2010, 16, 1389–1391. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef]
- Tan, J.; Asthagiri Arunkumar, G.; Krammer, F. Universal influenza virus vaccines and therapeutics: Where do we stand with influenza B virus? Curr. Opin. Immunol. 2018, 53, 45–50. [Google Scholar] [CrossRef]
- Wei, C.J.; Crank, M.C.; Shiver, J.; Graham, B.S.; Mascola, J.R.; Nabel, G.J. Next-generation influenza vaccines: Opportunities and challenges. Nat. Rev. Drug Discov. 2020, 19, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Houser, K.; Subbarao, K. Influenza vaccines: Challenges and solutions. Cell Host Microbe 2015, 17, 295–300. [Google Scholar] [CrossRef]
- World Health Organization Influenza (Seasonal). Available online: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (accessed on 2 December 2020).
- Paules, C.; Subbarao, K. Influenza. Lancet 2017, 390, 697–708. [Google Scholar] [CrossRef]
- Bouvier, N.M.; Palese, P. The biology of influenza viruses. Vaccine 2008, 26 (Suppl. 4), D49–D53. [Google Scholar] [CrossRef] [PubMed]
- Nobusawa, E.; Sato, K. Comparison of the mutation rates of human influenza A and B viruses. J. Virol. 2006, 80, 3675–3678. [Google Scholar] [CrossRef]
- Wu, N.C.; Wilson, I.A. A Perspective on the Structural and Functional Constraints for Immune Evasion: Insights from Influenza Virus. J. Mol. Biol. 2017, 429, 2694–2709. [Google Scholar] [CrossRef] [PubMed]
- Center for Disease Control and Prevention Types of Influenza Viruses. Available online: https://www.cdc.gov/flu/about/viruses/types.htm (accessed on 3 December 2020).
- Grohskopf, L.A.; Alyanak, E.; Broder, K.R.; Walter, E.B.; Fry, A.M.; Jernigan, D.B. Prevention and Control of Seasonal Influenza with Vaccines: Recommendations of the Advisory Committee on Immunization Practices—United States, 2019–2020 Influenza Season. MMWR Recomm Rep. 2019, 68, 1–21. [Google Scholar] [CrossRef]
- Fiore, A.E.; Bridges, C.B.; Cox, N.J. Seasonal influenza vaccines. Curr. Top. Microbiol. Immunol. 2009, 333, 43–82. [Google Scholar]
- Center for Disease Control and Prevention Past Seasons Vaccine Effectiveness Estimates. Available online: https://www.cdc.gov/flu/vaccines-work/past-seasons-estimates.html (accessed on 3 December 2020).
- Carrat, F.; Flahault, A. Influenza vaccine: The challenge of antigenic drift. Vaccine 2007, 25, 6852–6862. [Google Scholar] [CrossRef]
- Yewdell, J.W. Viva la revolucion: Rethinking influenza a virus antigenic drift. Curr. Opin. Virol. 2011, 1, 177–183. [Google Scholar] [CrossRef]
- Archetti, I.; Horsfall, F.L., Jr. Persistent antigenic variation of influenza A viruses after incomplete neutralization in ovo with heterologous immune serum. J. Exp. Med. 1950, 92, 441–462. [Google Scholar] [CrossRef]
- Huang, K.Y.; Rijal, P.; Schimanski, L.; Powell, T.J.; Lin, T.Y.; McCauley, J.W.; Daniels, R.S.; Townsend, A.R. Focused antibody response to influenza linked to antigenic drift. J. Clin. Investig. 2015, 125, 2631–2645. [Google Scholar] [CrossRef]
- Chambers, B.S.; Parkhouse, K.; Ross, T.M.; Alby, K.; Hensley, S.E. Identification of Hemagglutinin Residues Responsible for H3N2 Antigenic Drift during the 2014–2015 Influenza Season. Cell Rep. 2015, 12, 1–6. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Recommendations on the Composition of Influenza Virus Vaccines. Available online: https://www.who.int/influenza/vaccines/virus/recommendations/en/ (accessed on 3 December 2020).
- De Jong, J.C.; Beyer, W.E.; Palache, A.M.; Rimmelzwaan, G.F.; Osterhaus, A.D. Mismatch between the 1997/1998 influenza vaccine and the major epidemic A(H3N2) virus strain as the cause of an inadequate vaccine-induced antibody response to this strain in the elderly. J. Med. Virol. 2000, 61, 94–99. [Google Scholar] [CrossRef]
- Xie, H.; Wan, X.F.; Ye, Z.; Plant, E.P.; Zhao, Y.; Xu, Y.; Li, X.; Finch, C.; Zhao, N.; Kawano, T.; et al. H3N2 Mismatch of 2014–15 Northern Hemisphere Influenza Vaccines and Head-to-head Comparison between Human and Ferret Antisera derived Antigenic Maps. Sci Rep. 2015, 5, 15279. [Google Scholar] [CrossRef] [PubMed]
- Francis, M.E.; King, M.L.; Kelvin, A.A. Back to the Future for Influenza Preimmunity-Looking Back at Influenza Virus History to Infer the Outcome of Future Infections. Viruses 2019, 11, 122. [Google Scholar] [CrossRef] [PubMed]
- Morens, D.M.; Taubenberger, J.K.; Harvey, H.A.; Memoli, M.J. The 1918 influenza pandemic: Lessons for 2009 and the future. Crit. Care Med. 2010, 38 (Suppl. 4), e10–e20. [Google Scholar] [CrossRef]
- Smith, G.J.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef]
- Trifonov, V.; Khiabanian, H.; Rabadan, R. Geographic dependence, surveillance, and origins of the 2009 influenza A (H1N1) virus. N. Engl. J. Med. 2009, 361, 115–119. [Google Scholar] [CrossRef]
- Reid, A.H.; Fanning, T.G.; Hultin, J.V.; Taubenberger, J.K. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl. Acad. Sci. USA 1999, 96, 1651–1656. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.C.; Wu, T.Z.; Liu, D.P.; Shao, P.L.; Chang, L.Y.; Lu, C.Y.; Lee, C.Y.; Huang, F.Y.; Huang, L.M. Influenza pandemics: Past, present and future. J. Formos. Med. Assoc. 2006, 105, 1–6. [Google Scholar] [CrossRef]
- Poovorawan, Y.; Pyungporn, S.; Prachayangprecha, S.; Makkoch, J. Global alert to avian influenza virus infection: From H5N1 to H7N9. Pathog Glob. Health 2013, 107, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Sutton, T.C. The Pandemic Threat of Emerging H5 and H7 Avian Influenza Viruses. Viruses 2018, 10, 461. [Google Scholar] [CrossRef] [PubMed]
- Erbelding, E.J.; Post, D.J.; Stemmy, E.J.; Roberts, P.C.; Augustine, A.D.; Ferguson, S.; Paules, C.I.; Graham, B.S.; Fauci, A.S. A Universal Influenza Vaccine: The Strategic Plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 2018, 218, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Paules, C.I.; Sullivan, S.G.; Subbarao, K.; Fauci, A.S. Chasing Seasonal Influenza—The Need for a Universal Influenza Vaccine. N. Engl. J. Med. 2018, 378, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Petrie, J.G.; Gordon, A. Epidemiological Studies to Support the Development of Next Generation Influenza Vaccines. Vaccines 2018, 6, 17. [Google Scholar] [CrossRef]
- Nachbagauer, R.; Palese, P. Development of next generation hemagglutinin-based broadly protective influenza virus vaccines. Curr. Opin. Immunol. 2018, 53, 51–57. [Google Scholar] [CrossRef]
- Krammer, F.; Garcia-Sastre, A.; Palese, P. Is It Possible to Develop a “Universal” Influenza Virus Vaccine? Potential Target Antigens and Critical Aspects for a Universal Influenza Vaccine. Cold Spring Harb. Perspect. Biol. 2018, 10, a028845. [Google Scholar] [CrossRef]
- Kanekiyo, M.; Graham, B.S. Next-Generation Influenza Vaccines. Cold Spring Harb. Perspect. Med. 2020, 19, 239–252. [Google Scholar] [CrossRef]
- Throsby, M.; van den Brink, E.; Jongeneelen, M.; Poon, L.L.; Alard, P.; Cornelissen, L.; Bakker, A.; Cox, F.; van Deventer, E.; Guan, Y.; et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE 2008, 3, e3942. [Google Scholar] [CrossRef]
- Sui, J.; Hwang, W.C.; Perez, S.; Wei, G.; Aird, D.; Chen, L.M.; Santelli, E.; Stec, B.; Cadwell, G.; Ali, M.; et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009, 16, 265–273. [Google Scholar] [CrossRef]
- Corti, D.; Suguitan, A.L., Jr.; Pinna, D.; Silacci, C.; Fernandez-Rodriguez, B.M.; Vanzetta, F.; Santos, C.; Luke, C.J.; Torres-Velez, F.J.; Temperton, N.J.; et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Investig. 2010, 120, 1663–1673. [Google Scholar] [CrossRef]
- Wrammert, J.; Koutsonanos, D.; Li, G.M.; Edupuganti, S.; Sui, J.; Morrissey, M.; McCausland, M.; Skountzou, I.; Hornig, M.; Lipkin, W.I.; et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. 2011, 208, 181–193. [Google Scholar] [CrossRef]
- Wyrzucki, A.; Dreyfus, C.; Kohler, I.; Steck, M.; Wilson, I.A.; Hangartner, L. Alternative recognition of the conserved stem epitope in influenza A virus hemagglutinin by a VH3-30-encoded heterosubtypic antibody. J. Virol. 2014, 88, 7083–7092. [Google Scholar] [CrossRef]
- Wheatley, A.K.; Whittle, J.R.; Lingwood, D.; Kanekiyo, M.; Yassine, H.M.; Ma, S.S.; Narpala, S.R.; Prabhakaran, M.S.; Matus-Nicodemos, R.A.; Bailer, R.T.; et al. H5N1 Vaccine-Elicited Memory B Cells Are Genetically Constrained by the IGHV Locus in the Recognition of a Neutralizing Epitope in the Hemagglutinin Stem. J. Immunol. 2015, 195, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Whittle, J.R.; Wheatley, A.K.; Wu, L.; Lingwood, D.; Kanekiyo, M.; Ma, S.S.; Narpala, S.R.; Yassine, H.M.; Frank, G.M.; Yewdell, J.W.; et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J. Virol. 2014, 88, 4047–4057. [Google Scholar] [CrossRef] [PubMed]
- Ekiert, D.C.; Friesen, R.H.; Bhabha, G.; Kwaks, T.; Jongeneelen, M.; Yu, W.; Ophorst, C.; Cox, F.; Korse, H.J.; Brandenburg, B.; et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 2011, 333, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.S.; Lee, P.S.; Hoffman, R.M.; Mazel-Sanchez, B.; Krammer, F.; Leon, P.E.; Ward, A.B.; Wilson, I.A.; Palese, P. Characterization of a broadly neutralizing monoclonal antibody that targets the fusion domain of group 2 influenza A virus hemagglutinin. J. Virol. 2014, 88, 13580–13592. [Google Scholar] [CrossRef] [PubMed]
- Friesen, R.H.; Lee, P.S.; Stoop, E.J.; Hoffman, R.M.; Ekiert, D.C.; Bhabha, G.; Yu, W.; Juraszek, J.; Koudstaal, W.; Jongeneelen, M.; et al. A common solution to group 2 influenza virus neutralization. Proc. Natl. Acad. Sci. USA 2014, 111, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Henry Dunand, C.J.; Leon, P.E.; Kaur, K.; Tan, G.S.; Zheng, N.Y.; Andrews, S.; Huang, M.; Qu, X.; Huang, Y.; Salgado-Ferrer, M.; et al. Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. J. Clin. Investig. 2015, 125, 1255–1268. [Google Scholar] [CrossRef]
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011, 333, 850–856. [Google Scholar] [CrossRef]
- Dreyfus, C.; Laursen, N.S.; Kwaks, T.; Zuijdgeest, D.; Khayat, R.; Ekiert, D.C.; Lee, J.H.; Metlagel, Z.; Bujny, M.V.; Jongeneelen, M.; et al. Highly conserved protective epitopes on influenza B viruses. Science 2012, 337, 1343–1348. [Google Scholar] [CrossRef]
- Wu, Y.; Cho, M.; Shore, D.; Song, M.; Choi, J.; Jiang, T.; Deng, Y.Q.; Bourgeois, M.; Almli, L.; Yang, H.; et al. A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus. Nat. Commun. 2015, 6, 7708. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, Z.; Sheehan, J.; Avnir, Y.; Ridenour, C.; Sachnik, T.; Sun, J.; Hossain, M.J.; Chen, L.M.; Zhu, Q.; et al. A broadly neutralizing anti-influenza antibody reveals ongoing capacity of haemagglutinin-specific memory B cells to evolve. Nat. Commun. 2016, 7, 12780. [Google Scholar] [CrossRef] [PubMed]
- Kallewaard, N.L.; Corti, D.; Collins, P.J.; Neu, U.; McAuliffe, J.M.; Benjamin, E.; Wachter-Rosati, L.; Palmer-Hill, F.J.; Yuan, A.Q.; Walker, P.A.; et al. Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes. Cell 2016, 166, 596–608. [Google Scholar] [CrossRef]
- Ekiert, D.C.; Bhabha, G.; Elsliger, M.A.; Friesen, R.H.; Jongeneelen, M.; Throsby, M.; Goudsmit, J.; Wilson, I.A. Antibody recognition of a highly conserved influenza virus epitope. Science 2009, 324, 246–251. [Google Scholar] [CrossRef]
- Lee, P.S.; Wilson, I.A. Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Curr. Top. Microbiol. Immunol. 2015, 386, 323–341. [Google Scholar]
- Yoshida, R.; Igarashi, M.; Ozaki, H.; Kishida, N.; Tomabechi, D.; Kida, H.; Ito, K.; Takada, A. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 2009, 5, e1000350. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.S.; Yoshida, R.; Ekiert, D.C.; Sakai, N.; Suzuki, Y.; Takada, A.; Wilson, I.A. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc. Natl. Acad. Sci. USA 2012, 109, 17040–17045. [Google Scholar] [CrossRef]
- Ekiert, D.C.; Kashyap, A.K.; Steel, J.; Rubrum, A.; Bhabha, G.; Khayat, R.; Lee, J.H.; Dillon, M.A.; O’Neil, R.E.; Faynboym, A.M.; et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 2012, 489, 526–532. [Google Scholar] [CrossRef]
- Lee, P.S.; Ohshima, N.; Stanfield, R.L.; Yu, W.; Iba, Y.; Okuno, Y.; Kurosawa, Y.; Wilson, I.A. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun. 2014, 5, 3614. [Google Scholar] [CrossRef]
- Hu, H.; Voss, J.; Zhang, G.; Buchy, P.; Zuo, T.; Wang, L.; Wang, F.; Zhou, F.; Wang, G.; Tsai, C.; et al. A human antibody recognizing a conserved epitope of H5 hemagglutinin broadly neutralizes highly pathogenic avian influenza H5N1 viruses. J. Virol. 2012, 86, 2978–2989. [Google Scholar] [CrossRef] [PubMed]
- Krause, J.C.; Tsibane, T.; Tumpey, T.M.; Huffman, C.J.; Albrecht, R.; Blum, D.L.; Ramos, I.; Fernandez-Sesma, A.; Edwards, K.M.; Garcia-Sastre, A.; et al. Human monoclonal antibodies to pandemic 1957 H2N2 and pandemic 1968 H3N2 influenza viruses. J. Virol. 2012, 86, 6334–6340. [Google Scholar] [CrossRef]
- Xu, R.; Krause, J.C.; McBride, R.; Paulson, J.C.; Crowe, J.E., Jr.; Wilson, I.A. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat. Struct. Mol. Biol. 2013, 20, 363–370. [Google Scholar] [CrossRef]
- Schmidt, A.G.; Xu, H.; Khan, A.R.; O’Donnell, T.; Khurana, S.; King, L.R.; Manischewitz, J.; Golding, H.; Suphaphiphat, P.; Carfi, A.; et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc. Natl. Acad. Sci. USA 2013, 110, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Whittle, J.R.; Zhang, R.; Khurana, S.; King, L.R.; Manischewitz, J.; Golding, H.; Dormitzer, P.R.; Haynes, B.F.; Walter, E.B.; Moody, M.A.; et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA 2011, 108, 14216–14221. [Google Scholar] [CrossRef]
- Hong, M.; Lee, P.S.; Hoffman, R.M.; Zhu, X.; Krause, J.C.; Laursen, N.S.; Yoon, S.I.; Song, L.; Tussey, L.; Crowe, J.E., Jr.; et al. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J. Virol. 2013, 87, 12471–12480. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, N.; Iba, Y.; Kubota-Koketsu, R.; Asano, Y.; Okuno, Y.; Kurosawa, Y. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J. Virol. 2011, 85, 11048–11057. [Google Scholar] [CrossRef]
- Kashyap, A.K.; Steel, J.; Oner, A.F.; Dillon, M.A.; Swale, R.E.; Wall, K.M.; Perry, K.J.; Faynboym, A.; Ilhan, M.; Horowitz, M.; et al. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc. Natl. Acad. Sci. USA 2008, 105, 5986–5991. [Google Scholar] [CrossRef]
- Nakamura, G.; Chai, N.; Park, S.; Chiang, N.; Lin, Z.; Chiu, H.; Fong, R.; Yan, D.; Kim, J.; Zhang, J.; et al. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza A antibodies. Cell Host Microbe 2013, 14, 93–103. [Google Scholar] [CrossRef]
- Joyce, M.G.; Wheatley, A.K.; Thomas, P.V.; Chuang, G.Y.; Soto, C.; Bailer, R.T.; Druz, A.; Georgiev, I.S.; Gillespie, R.A.; Kanekiyo, M.; et al. Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses. Cell 2016, 166, 609–623. [Google Scholar] [CrossRef]
- Ledgerwood, J.E.; Zephir, K.; Hu, Z.; Wei, C.J.; Chang, L.; Enama, M.E.; Hendel, C.S.; Sitar, S.; Bailer, R.T.; Koup, R.A.; et al. Prime-boost interval matters: A randomized phase 1 study to identify the minimum interval necessary to observe the H5 DNA influenza vaccine priming effect. J. Infect. Dis. 2013, 208, 418–422. [Google Scholar] [CrossRef]
- Wan, H.; Gao, J.; Xu, K.; Chen, H.; Couzens, L.K.; Rivers, K.H.; Easterbrook, J.D.; Yang, K.; Zhong, L.; Rajabi, M.; et al. Molecular basis for broad neuraminidase immunity: Conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J. Virol. 2013, 87, 9290–9300. [Google Scholar] [CrossRef]
- Wan, H.; Yang, H.; Shore, D.A.; Garten, R.J.; Couzens, L.; Gao, J.; Jiang, L.; Carney, P.J.; Villanueva, J.; Stevens, J.; et al. Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers. Nat. Commun. 2015, 6, 6114. [Google Scholar] [CrossRef]
- Jiang, L.; Fantoni, G.; Couzens, L.; Gao, J.; Plant, E.; Ye, Z.; Eichelberger, M.C.; Wan, H. Comparative Efficacy of Monoclonal Antibodies That Bind to Different Epitopes of the 2009 Pandemic H1N1 Influenza Virus Neuraminidase. J. Virol. 2016, 90, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Wohlbold, T.J.; Nachbagauer, R.; Xu, H.; Tan, G.S.; Hirsh, A.; Brokstad, K.A.; Cox, R.J.; Palese, P.; Krammer, F. Vaccination with adjuvanted recombinant neuraminidase induces broad heterologous, but not heterosubtypic, cross-protection against influenza virus infection in mice. mBio 2015, 6, e02556. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Q.; Wohlbold, T.J.; Zheng, N.Y.; Huang, M.; Huang, Y.; Neu, K.E.; Lee, J.; Wan, H.; Rojas, K.T.; Kirkpatrick, E.; et al. Influenza Infection in Humans Induces Broadly Cross-Reactive and Protective Neuraminidase-Reactive Antibodies. Cell 2018, 173, 417–429.e10. [Google Scholar] [CrossRef] [PubMed]
- Stadlbauer, D.; Zhu, X.; McMahon, M.; Turner, J.S.; Wohlbold, T.J.; Schmitz, A.J.; Strohmeier, S.; Yu, W.; Nachbagauer, R.; Mudd, P.A.; et al. Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science 2019, 366, 499–504. [Google Scholar] [CrossRef]
- Madsen, A.; Dai, Y.N.; McMahon, M.; Schmitz, A.J.; Turner, J.S.; Tan, J.; Lei, T.; Alsoussi, W.B.; Strohmeier, S.; Amor, M.; et al. Human Antibodies Targeting Influenza B Virus Neuraminidase Active Site Are Broadly Protective. Immunity 2020, 53, 852–863.e7. [Google Scholar] [CrossRef] [PubMed]
- Doyle, T.M.; Hashem, A.M.; Li, C.; Van Domselaar, G.; Larocque, L.; Wang, J.; Smith, D.; Cyr, T.; Farnsworth, A.; He, R.; et al. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antiviral Res. 2013, 100, 567–574. [Google Scholar] [CrossRef]
- Altman, M.O.; Angeletti, D.; Yewdell, J.W. Antibody Immunodominance: The Key to Understanding Influenza Virus Antigenic Drift. Viral Immunol. 2018, 31, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Altman, M.O.; Bennink, J.R.; Yewdell, J.W.; Herrin, B.R. Lamprey VLRB response to influenza virus supports universal rules of immunogenicity and antigenicity. Elife 2015, 4, e07467. [Google Scholar] [CrossRef]
- Zost, S.J.; Wu, N.C.; Hensley, S.E.; Wilson, I.A. Immunodominance and Antigenic Variation of Influenza Virus Hemagglutinin: Implications for Design of Universal Vaccine Immunogens. J. Infect. Dis. 2019, 219 (Suppl. 1), S38–S45. [Google Scholar] [CrossRef]
- Angeletti, D.; Gibbs, J.S.; Angel, M.; Kosik, I.; Hickman, H.D.; Frank, G.M.; Das, S.R.; Wheatley, A.K.; Prabhakaran, M.; Leggat, D.J.; et al. Defining B cell immunodominance to viruses. Nat. Immunol. 2017, 18, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Dugan, H.L.; Guthmiller, J.J.; Arevalo, P.; Huang, M.; Chen, Y.Q.; Neu, K.E.; Henry, C.; Zheng, N.Y.; Lan, L.Y.; Tepora, M.E.; et al. Preexisting immunity shapes distinct antibody landscapes after influenza virus infection and vaccination in humans. Sci. Transl. Med. 2020, 12, eabd3601. [Google Scholar] [CrossRef]
- Tan, H.X.; Jegaskanda, S.; Juno, J.A.; Esterbauer, R.; Wong, J.; Kelly, H.G.; Liu, Y.; Tilmanis, D.; Hurt, A.C.; Yewdell, J.W.; et al. Subdominance and poor intrinsic immunogenicity limit humoral immunity targeting influenza HA stem. J. Clin. Invest. 2019, 129, 850–862. [Google Scholar] [CrossRef]
- Abbott, R.K.; Lee, J.H.; Menis, S.; Skog, P.; Rossi, M.; Ota, T.; Kulp, D.W.; Bhullar, D.; Kalyuzhniy, O.; Havenar-Daughton, C.; et al. Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity 2018, 48, 133–146e6. [Google Scholar] [CrossRef]
- Amitai, A.; Sangesland, M.; Barnes, R.M.; Rohrer, D.; Lonberg, N.; Lingwood, D.; Chakraborty, A.K. Defining and Manipulating B Cell Immunodominance Hierarchies to Elicit Broadly Neutralizing Antibody Responses against Influenza Virus. Cell Syst. 2020, 11, 573–588e9. [Google Scholar] [CrossRef]
- Sangesland, M.; Ronsard, L.; Kazer, S.W.; Bals, J.; Boyoglu-Barnum, S.; Yousif, A.S.; Barnes, R.; Feldman, J.; Quirindongo-Crespo, M.; McTamney, P.M.; et al. Germline-Encoded Affinity for Cognate Antigen Enables Vaccine Amplification of a Human Broadly Neutralizing Response against Influenza Virus. Immunity 2019, 51, 735–749e8. [Google Scholar] [CrossRef]
- Dosenovic, P.; Kara, E.E.; Pettersson, A.K.; McGuire, A.T.; Gray, M.; Hartweger, H.; Thientosapol, E.S.; Stamatatos, L.; Nussenzweig, M.C. Anti-HIV-1 B cell responses are dependent on B cell precursor frequency and antigen-binding affinity. Proc. Natl. Acad. Sci. USA 2018, 115, 4743–4748. [Google Scholar] [CrossRef] [PubMed]
- Peterhoff, D.; Wagner, R. Guiding the long way to broad HIV neutralization. Curr. Opin. HIV AIDS 2017, 12, 257–264. [Google Scholar] [CrossRef]
- Huang, D.; Abbott, R.K.; Havenar-Daughton, C.; Skog, P.D.; Al-Kolla, R.; Groschel, B.; Blane, T.R.; Menis, S.; Tran, J.T.; Thinnes, T.C.; et al. B cells expressing authentic naive human VRC01-class BCRs can be recruited to germinal centers and affinity mature in multiple independent mouse models. Proc. Natl. Acad. Sci. USA 2020, 117, 22920–22931. [Google Scholar] [CrossRef] [PubMed]
- Steichen, J.M.; Lin, Y.-C.; Havenar-Daughton, C.; Pecetta, S.; Ozorowski, G.; Willis, J.R.; Toy, L.; Sok, D.; Liguori, A.; Kratochvil, S.; et al. A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses. Science 2019, 366, eaax4380. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ray, R.; Kratochvil, S.; Melzi, E.; Lin, Y.C.; Giguere, S.; Xu, L.; Warner, J.; Cheon, D.; Liguori, A.; et al. Multiplexed CRISPR/CAS9-mediated engineering of pre-clinical mouse models bearing native human B cell receptors. EMBO J. 2020, 40, e105926. [Google Scholar]
- Abbott, R.K.; Crotty, S. Factors in B cell competition and immunodominance. Immunol. Rev. 2020, 296, 120–131. [Google Scholar] [CrossRef]
- Koutsakos, M.; Nguyen, T.H.O.; Kedzierska, K. With a Little Help from T Follicular Helper Friends: Humoral Immunity to Influenza Vaccination. J. Immunol. 2019, 202, 360–367. [Google Scholar] [CrossRef]
- Wu, N.C.; Wilson, I.A. Structural Biology of Influenza Hemagglutinin: An Amaranthine Adventure. Viruses 2020, 12, 1053. [Google Scholar] [CrossRef]
- Wu, N.C.; Wilson, I.A. Influenza Hemagglutinin Structures and Antibody Recognition. Cold Spring Harb. Perspect. Med. 2020, 10, a038778. [Google Scholar] [CrossRef]
- Wilson, I.A.; Skehel, J.J.; Wiley, D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 1981, 289, 366–373. [Google Scholar] [CrossRef]
- Skehel, J.J.; Wiley, D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef]
- Ren, H.; Zhou, P. Epitope-focused vaccine design against influenza A and B viruses. Curr. Opin. Immunol. 2016, 42, 83–90. [Google Scholar] [CrossRef]
- Weis, W.; Brown, J.H.; Cusack, S.; Paulson, J.C.; Skehel, J.J.; Wiley, D.C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 1988, 333, 426–431. [Google Scholar] [CrossRef]
- Hobson, D.; Curry, R.L.; Beare, A.S.; Ward-Gardner, A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J. Hyg. 1972, 70, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Klein, S.L. Host Factors Impact Vaccine Efficacy: Implications for Seasonal and Universal Influenza Vaccine Programs. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Knossow, M.; Gaudier, M.; Douglas, A.; Barrere, B.; Bizebard, T.; Barbey, C.; Gigant, B.; Skehel, J.J. Mechanism of neutralization of influenza virus infectivity by antibodies. Virology 2002, 302, 294–298. [Google Scholar] [CrossRef] [PubMed]
- Raymond, D.D.; Stewart, S.M.; Lee, J.; Ferdman, J.; Bajic, G.; Do, K.T.; Ernandes, M.J.; Suphaphiphat, P.; Settembre, E.C.; Dormitzer, P.R.; et al. Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain. Nat. Med. 2016, 22, 1465–1469. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.G.; Therkelsen, M.D.; Stewart, S.; Kepler, T.B.; Liao, H.X.; Moody, M.A.; Haynes, B.F.; Harrison, S.C. Viral receptor-binding site antibodies with diverse germline origins. Cell 2015, 161, 1026–1034. [Google Scholar] [CrossRef]
- Cheung, C.S.; Fruehwirth, A.; Paparoditis, P.C.G.; Shen, C.H.; Foglierini, M.; Joyce, M.G.; Leung, K.; Piccoli, L.; Rawi, R.; Silacci-Fregni, C.; et al. Identification and Structure of a Multidonor Class of Head-Directed Influenza-Neutralizing Antibodies Reveal the Mechanism for Its Recurrent Elicitation. Cell Rep. 2020, 32, 108088. [Google Scholar] [CrossRef]
- Bangaru, S.; Zhang, H.; Gilchuk, I.M.; Voss, T.G.; Irving, R.P.; Gilchuk, P.; Matta, P.; Zhu, X.; Lang, S.; Nieusma, T.; et al. A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA. Nat. Commun. 2018, 9, 2669. [Google Scholar] [CrossRef]
- Chai, N.; Swem, L.R.; Park, S.; Nakamura, G.; Chiang, N.; Estevez, A.; Fong, R.; Kamen, L.; Kho, E.; Reichelt, M.; et al. A broadly protective therapeutic antibody against influenza B virus with two mechanisms of action. Nat. Commun. 2017, 8, 14234. [Google Scholar] [CrossRef]
- Raymond, D.D.; Bajic, G.; Ferdman, J.; Suphaphiphat, P.; Settembre, E.C.; Moody, M.A.; Schmidt, A.G.; Harrison, S.C. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc. Natl. Acad. Sci. USA 2018, 115, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Bajic, G.; Maron, M.J.; Adachi, Y.; Onodera, T.; McCarthy, K.R.; McGee, C.E.; Sempowski, G.D.; Takahashi, Y.; Kelsoe, G.; Kuraoka, M.; et al. Influenza Antigen Engineering Focuses Immune Responses to a Subdominant but Broadly Protective Viral Epitope. Cell Host Microbe 2019, 25, 827–835e6. [Google Scholar] [CrossRef]
- Watanabe, A.; McCarthy, K.R.; Kuraoka, M.; Schmidt, A.G.; Adachi, Y.; Onodera, T.; Tonouchi, K.; Caradonna, T.M.; Bajic, G.; Song, S.; et al. Antibodies to a Conserved Influenza Head Interface Epitope Protect by an IgG Subtype-Dependent Mechanism. Cell 2019, 177, 1124–1135e16. [Google Scholar] [CrossRef]
- Bangaru, S.; Lang, S.; Schotsaert, M.; Vanderven, H.A.; Zhu, X.; Kose, N.; Bombardi, R.; Finn, J.A.; Kent, S.J.; Gilchuk, P.; et al. A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface. Cell 2019, 177, 1136–1152.e18. [Google Scholar] [CrossRef] [PubMed]
- Das, D.K.; Govindan, R.; Nikic-Spiegel, I.; Krammer, F.; Lemke, E.A.; Munro, J.B. Direct Visualization of the Conformational Dynamics of Single Influenza Hemagglutinin Trimers. Cell 2018, 174, 926–937.e12. [Google Scholar] [CrossRef]
- DiLillo, D.J.; Palese, P.; Wilson, P.C.; Ravetch, J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Investig. 2016, 126, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Bournazos, S.; Corti, D.; Virgin, H.W.; Ravetch, J.V. Fc-optimized antibodies elicit CD8 immunity to viral respiratory infection. Nature 2020, 588, 485–490. [Google Scholar] [CrossRef]
- Cox, F.; Kwaks, T.; Brandenburg, B.; Koldijk, M.H.; Klaren, V.; Smal, B.; Korse, H.J.; Geelen, E.; Tettero, L.; Zuijdgeest, D.; et al. HA Antibody-Mediated FcgammaRIIIa Activity Is Both Dependent on FcR Engagement and Interactions between HA and Sialic Acids. Front. Immunol. 2016, 7, 399. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Tan, G.S.; Mullarkey, C.E.; Lee, A.J.; Lam, M.M.; Krammer, F.; Henry, C.; Wilson, P.C.; Ashkar, A.A.; Palese, P.; et al. Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. Proc. Natl. Acad. Sci. USA 2016, 113, 11931–11936. [Google Scholar] [CrossRef] [PubMed]
- DiLillo, D.J.; Tan, G.S.; Palese, P.; Ravetch, J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat. Med. 2014, 20, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Leon, P.E.; He, W.; Mullarkey, C.E.; Bailey, M.J.; Miller, M.S.; Krammer, F.; Palese, P.; Tan, G.S. Optimal activation of Fc-mediated effector functions by influenza virus hemagglutinin antibodies requires two points of contact. Proc. Natl. Acad. Sci. USA 2016, 113, E5944–E5951. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Liu, F.; Wilson, J.R.; Holiday, C.; Li, Z.N.; Bai, Y.; Tzeng, W.P.; Stevens, J.; York, I.A.; Levine, M.Z. Antibody-Dependent Cell-Mediated Cytotoxicity to Hemagglutinin of Influenza A Viruses After Influenza Vaccination in Humans. Open Forum Infect. Dis. 2016, 3, ofw102. [Google Scholar] [CrossRef]
- Gao, R.; Sheng, Z.; Sreenivasan, C.C.; Wang, D.; Li, F. Influenza A Virus Antibodies with Antibody-Dependent Cellular Cytotoxicity Function. Viruses 2020, 12, 276. [Google Scholar] [CrossRef]
- Koopman, G.; Mortier, D.; Michels, S.; Hofman, S.; Fagrouch, Z.; Remarque, E.J.; Verschoor, E.J.; Mooij, P.; Bogers, W. Influenza virus infection as well as immunization with DNA encoding haemagglutinin protein induces potent antibody-dependent phagocytosis (ADP) and monocyte infection-enhancing responses in macaques. J. Gen. Virol. 2019, 100, 738–751. [Google Scholar] [CrossRef]
- Krammer, F.; Palese, P. Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov. 2015, 14, 167–182. [Google Scholar] [CrossRef]
- Hamilton, B.S.; Whittaker, G.R.; Daniel, S. Influenza virus-mediated membrane fusion: Determinants of hemagglutinin fusogenic activity and experimental approaches for assessing virus fusion. Viruses 2012, 4, 1144–1168. [Google Scholar] [CrossRef]
- Okuno, Y.; Isegawa, Y.; Sasao, F.; Ueda, S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 1993, 67, 2552–2558. [Google Scholar] [CrossRef]
- Yusuf, M.; Konc, J.; Sy Bing, C.; Trykowska Konc, J.; Ahmad Khairudin, N.B.; Janezic, D.; Wahab, H.A. Structurally conserved binding sites of hemagglutinin as targets for influenza drug and vaccine development. J. Chem. Inf. Model. 2013, 53, 2423–2436. [Google Scholar] [CrossRef]
- Balazs, A.B.; Bloom, J.D.; Hong, C.M.; Rao, D.S.; Baltimore, D. Broad protection against influenza infection by vectored immunoprophylaxis in mice. Nat. Biotechnol. 2013, 31, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Vanderven, H.A.; Jegaskanda, S.; Wheatley, A.K.; Kent, S.J. Antibody-dependent cellular cytotoxicity and influenza virus. Curr. Opin. Virol. 2017, 22, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Kosik, I.; Angeletti, D.; Gibbs, J.S.; Angel, M.; Takeda, K.; Kosikova, M.; Nair, V.; Hickman, H.D.; Xie, H.; Brooke, C.B.; et al. Neuraminidase inhibition contributes to influenza A virus neutralization by anti-hemagglutinin stem antibodies. J. Exp. Med. 2019, 216, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Q.; Lan, L.Y.; Huang, M.; Henry, C.; Wilson, P.C. Hemagglutinin Stalk-Reactive Antibodies Interfere with Influenza Virus Neuraminidase Activity by Steric Hindrance. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Palese, P.; Compans, R.W. Inhibition of influenza virus replication in tissue culture by 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA): Mechanism of action. J. Gen. Virol. 1976, 33, 159–163. [Google Scholar] [CrossRef]
- Palese, P.; Schulman, J.L.; Bodo, G.; Meindl, P. Inhibition of influenza and parainfluenza virus replication in tissue culture by 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA). Virology 1974, 59, 490–498. [Google Scholar] [CrossRef]
- Matrosovich, M.N.; Matrosovich, T.Y.; Gray, T.; Roberts, N.A.; Klenk, H.D. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J. Virol. 2004, 78, 12665–12667. [Google Scholar] [CrossRef]
- Kilbourne, E.D.; Johansson, B.E.; Grajower, B. Independent and disparate evolution in nature of influenza A virus hemagglutinin and neuraminidase glycoproteins. Proc. Natl. Acad. Sci. USA 1990, 87, 786–790. [Google Scholar] [CrossRef]
- Westgeest, K.B.; de Graaf, M.; Fourment, M.; Bestebroer, T.M.; van Beek, R.; Spronken, M.I.J.; de Jong, J.C.; Rimmelzwaan, G.F.; Russell, C.A.; Osterhaus, A.; et al. Genetic evolution of the neuraminidase of influenza A (H3N2) viruses from 1968 to 2009 and its correspondence to haemagglutinin evolution. J. Gen. Virol. 2012, 93, 1996–2007. [Google Scholar] [CrossRef]
- Abed, Y.; Hardy, I.; Li, Y.; Boivin, G. Divergent evolution of hemagglutinin and neuraminidase genes in recent influenza A:H3N2 viruses isolated in Canada. J. Med. Virol. 2002, 67, 589–595. [Google Scholar] [CrossRef]
- Sandbulte, M.R.; Westgeest, K.B.; Gao, J.; Xu, X.; Klimov, A.I.; Russell, C.A.; Burke, D.F.; Smith, D.J.; Fouchier, R.A.; Eichelberger, M.C. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc. Natl. Acad. Sci. USA 2011, 108, 20748–20753. [Google Scholar] [CrossRef]
- Couch, R.B.; Kasel, J.A.; Gerin, J.L.; Schulman, J.L.; Kilbourne, E.D. Induction of partial immunity to influenza by a neuraminidase-specific vaccine. J. Infect. Dis. 1974, 129, 411–420. [Google Scholar] [CrossRef]
- Memoli, M.J.; Shaw, P.A.; Han, A.; Czajkowski, L.; Reed, S.; Athota, R.; Bristol, T.; Fargis, S.; Risos, K.; Powers, J.H.; et al. Evaluation of Antihemagglutinin and Antineuraminidase Antibodies as Correlates of Protection in an Influenza A/H1N1 Virus Healthy Human Challenge Model. mBio 2016, 7, e00417-16. [Google Scholar] [CrossRef] [PubMed]
- Monto, A.S.; Kendal, A.P. Effect of neuraminidase antibody on Hong Kong influenza. Lancet 1973, 1, 623–625. [Google Scholar] [CrossRef]
- Eichelberger, M.C.; Monto, A.S. Neuraminidase, the Forgotten Surface Antigen, Emerges as an Influenza Vaccine Target for Broadened Protection. J. Infect. Dis. 2019, 219 (Suppl. 1), S75–S80. [Google Scholar] [CrossRef]
- Liu, W.C.; Lin, C.Y.; Tsou, Y.T.; Jan, J.T.; Wu, S.C. Cross-Reactive Neuraminidase-Inhibiting Antibodies Elicited by Immunization with Recombinant Neuraminidase Proteins of H5N1 and Pandemic H1N1 Influenza A Viruses. J. Virol. 2015, 89, 7224–7234. [Google Scholar] [CrossRef]
- Marcelin, G.; DuBois, R.; Rubrum, A.; Russell, C.J.; McElhaney, J.E.; Webby, R.J. A contributing role for anti-neuraminidase antibodies on immunity to pandemic H1N1 2009 influenza A virus. PLoS ONE 2011, 6, e26335. [Google Scholar] [CrossRef] [PubMed]
- Wohlbold, T.J.; Podolsky, K.A.; Chromikova, V.; Kirkpatrick, E.; Falconieri, V.; Meade, P.; Amanat, F.; Tan, J.; tenOever, B.R.; Tan, G.S.; et al. Broadly protective murine monoclonal antibodies against influenza B virus target highly conserved neuraminidase epitopes. Nat. Microbiol. 2017, 2, 1415–1424. [Google Scholar] [CrossRef]
- Lamb, R.A.; Choppin, P.W. Identification of a second protein (M2) encoded by RNA segment 7 of influenza virus. Virology 1981, 112, 729–737. [Google Scholar] [CrossRef]
- Lamb, R.A.; Zebedee, S.L.; Richardson, C.D. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 1985, 40, 627–633. [Google Scholar] [CrossRef]
- Schnell, J.R.; Chou, J.J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature 2008, 451, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Schotsaert, M.; De Filette, M.; Fiers, W.; Saelens, X. Universal M2 ectodomain-based influenza A vaccines: Preclinical and clinical developments. Expert Rev. Vaccines 2009, 8, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Kolpe, A.; Schepens, B.; Fiers, W.; Saelens, X. M2-based influenza vaccines: Recent advances and clinical potential. Expert Rev. Vaccines 2017, 16, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Treanor, J.J.; Tierney, E.L.; Zebedee, S.L.; Lamb, R.A.; Murphy, B.R. Passively transferred monoclonal antibody to the M2 protein inhibits influenza A virus replication in mice. J. Virol. 1990, 64, 1375–1377. [Google Scholar] [CrossRef]
- Zebedee, S.L.; Lamb, R.A. Influenza A virus M2 protein: Monoclonal antibody restriction of virus growth and detection of M2 in virions. J. Virol. 1988, 62, 2762–2772. [Google Scholar] [CrossRef]
- Black, R.A.; Rota, P.A.; Gorodkova, N.; Klenk, H.D.; Kendal, A.P. Antibody response to the M2 protein of influenza A virus expressed in insect cells. J. Gen. Virol. 1993, 74, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhang, M.; Mozdzanowska, K.; Zharikova, D.; Hoff, H.; Wunner, W.; Couch, R.B.; Gerhard, W. Influenza A virus infection engenders a poor antibody response against the ectodomain of matrix protein 2. Virol. J. 2006, 3, 102. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Reed, C.; Blair, P.J.; Katz, J.M.; Hancock, K.; Influenza Serology Working, G. Serum antibody response to matrix protein 2 following natural infection with 2009 pandemic influenza A(H1N1) virus in humans. J. Infect. Dis. 2014, 209, 986–994. [Google Scholar] [CrossRef]
- Deng, L.; Cho, K.J.; Fiers, W.; Saelens, X. M2e-Based Universal Influenza A Vaccines. Vaccines 2015, 3, 105–136. [Google Scholar] [CrossRef]
- Andrews, S.F.; Huang, Y.; Kaur, K.; Popova, L.I.; Ho, I.Y.; Pauli, N.T.; Henry Dunand, C.J.; Taylor, W.M.; Lim, S.; Huang, M.; et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 2015, 7, 316ra192. [Google Scholar] [CrossRef]
- Abreu, R.B.; Kirchenbaum, G.A.; Clutter, E.F.; Sautto, G.A.; Ross, T.M. Preexisting subtype immunodominance shapes memory B cell recall response to influenza vaccination. JCI Insight 2020, 5, e132155. [Google Scholar] [CrossRef]
- Krammer, F.; Pica, N.; Hai, R.; Margine, I.; Palese, P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol. 2013, 87, 6542–6550. [Google Scholar] [CrossRef] [PubMed]
- Margine, I.; Krammer, F.; Hai, R.; Heaton, N.S.; Tan, G.S.; Andrews, S.A.; Runstadler, J.A.; Wilson, P.C.; Albrecht, R.A.; Garcia-Sastre, A.; et al. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J. Virol. 2013, 87, 10435–10446. [Google Scholar] [CrossRef]
- Ermler, M.E.; Kirkpatrick, E.; Sun, W.; Hai, R.; Amanat, F.; Chromikova, V.; Palese, P.; Krammer, F. Chimeric Hemagglutinin Constructs Induce Broad Protection against Influenza B Virus Challenge in the Mouse Model. J. Virol. 2017, 91, e00286-17. [Google Scholar] [CrossRef]
- Nachbagauer, R.; Kinzler, D.; Choi, A.; Hirsh, A.; Beaulieu, E.; Lecrenier, N.; Innis, B.L.; Palese, P.; Mallett, C.P.; Krammer, F. A chimeric haemagglutinin-based influenza split virion vaccine adjuvanted with AS03 induces protective stalk-reactive antibodies in mice. NPJ Vaccines 2016, 1, 16015. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.; Palm, A.E.; Krammer, F.; Wilson, P.C. From Original Antigenic Sin to the Universal Influenza Virus Vaccine. Trends Immunol. 2018, 39, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F. Novel universal influenza virus vaccine approaches. Curr. Opin. Virol. 2016, 17, 95–103. [Google Scholar] [CrossRef]
- Guthmiller, J.J.; Wilson, P.C. Harnessing immune history to combat influenza viruses. Curr. Opin. Immunol. 2018, 53, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Nachbagauer, R.; Feser, J.; Naficy, A.; Bernstein, D.I.; Guptill, J.; Walter, E.B.; Berlanda-Scorza, F.; Stadlbauer, D.; Wilson, P.C.; Aydillo, T.; et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 2021, 27, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Ellebedy, A.H.; Krammer, F.; Li, G.M.; Miller, M.S.; Chiu, C.; Wrammert, J.; Chang, C.Y.; Davis, C.W.; McCausland, M.; Elbein, R.; et al. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc. Natl. Acad. Sci. USA 2014, 111, 13133–13138. [Google Scholar] [CrossRef]
- Andrews, S.F.; Joyce, M.G.; Chambers, M.J.; Gillespie, R.A.; Kanekiyo, M.; Leung, K.; Yang, E.S.; Tsybovsky, Y.; Wheatley, A.K.; Crank, M.C.; et al. Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans. Sci. Immunol. 2017, 2, eaan2676. [Google Scholar] [CrossRef]
- Impagliazzo, A.; Milder, F.; Kuipers, H.; Wagner, M.V.; Zhu, X.; Hoffman, R.M.; van Meersbergen, R.; Huizingh, J.; Wanningen, P.; Verspuij, J.; et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 2015, 349, 1301–1306. [Google Scholar] [CrossRef]
- Yassine, H.M.; Boyington, J.C.; McTamney, P.M.; Wei, C.J.; Kanekiyo, M.; Kong, W.P.; Gallagher, J.R.; Wang, L.; Zhang, Y.; Joyce, M.G.; et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 2015, 21, 1065–1070. [Google Scholar] [CrossRef]
- Corbett, K.S.; Moin, S.M.; Yassine, H.M.; Cagigi, A.; Kanekiyo, M.; Boyoglu-Barnum, S.; Myers, S.I.; Tsybovsky, Y.; Wheatley, A.K.; Schramm, C.A.; et al. Design of Nanoparticulate Group 2 Influenza Virus Hemagglutinin Stem Antigens That Activate Unmutated Ancestor B Cell Receptors of Broadly Neutralizing Antibody Lineages. mBio 2019, 10, e02810-18. [Google Scholar] [CrossRef]
- Boyoglu-Barnum, S.; Hutchinson, G.B.; Boyington, J.C.; Moin, S.M.; Gillespie, R.A.; Tsybovsky, Y.; Stephens, T.; Vaile, J.R.; Lederhofer, J.; Corbett, K.S.; et al. Glycan repositioning of influenza hemagglutinin stem facilitates the elicitation of protective cross-group antibody responses. Nat. Commun. 2020, 11, 791. [Google Scholar] [CrossRef]
- Schneemann, A.; Speir, J.A.; Tan, G.S.; Khayat, R.; Ekiert, D.C.; Matsuoka, Y.; Wilson, I.A. A virus-like particle that elicits cross-reactive antibodies to the conserved stem of influenza virus hemagglutinin. J. Virol. 2012, 86, 11686–11697. [Google Scholar] [CrossRef]
- Mallajosyula, V.V.; Citron, M.; Ferrara, F.; Lu, X.; Callahan, C.; Heidecker, G.J.; Sarma, S.P.; Flynn, J.A.; Temperton, N.J.; Liang, X.; et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc. Natl. Acad. Sci. USA 2014, 111, E2514–E2523. [Google Scholar] [CrossRef] [PubMed]
- Angeletti, D.; Kosik, I.; Santos, J.J.S.; Yewdell, W.T.; Boudreau, C.M.; Mallajosyula, V.V.A.; Mankowski, M.C.; Chambers, M.; Prabhakaran, M.; Hickman, H.D.; et al. Outflanking immunodominance to target subdominant broadly neutralizing epitopes. Proc. Natl. Acad. Sci. USA 2019, 116, 13474–13479. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Abbott, R.K.; Freeman, B.L.; Haupt, S.; Groschel, B.; Silva, M.; Menis, S.; Irvine, D.J.; Schief, W.R.; Crotty, S. Multifaceted Effects of Antigen Valency on B Cell Response Composition and Differentiation In Vivo. Immunity 2020, 53, 548–563e8. [Google Scholar] [CrossRef] [PubMed]
- Kelly, H.G.; Tan, H.X.; Juno, J.A.; Esterbauer, R.; Ju, Y.; Jiang, W.; Wimmer, V.C.; Duckworth, B.C.; Groom, J.R.; Caruso, F.; et al. Self-assembling influenza nanoparticle vaccines drive extended germinal center activity and memory B cell maturation. JCI Insight 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Lingwood, D.; McTamney, P.M.; Yassine, H.M.; Whittle, J.R.; Guo, X.; Boyington, J.C.; Wei, C.J.; Nabel, G.J. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature 2012, 489, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Weaver, G.C.; Villar, R.F.; Kanekiyo, M.; Nabel, G.J.; Mascola, J.R.; Lingwood, D. In vitro reconstitution of B cell receptor-antigen interactions to evaluate potential vaccine candidates. Nat. Protoc. 2016, 11, 193–213. [Google Scholar] [CrossRef] [PubMed]
- Cirelli, K.M.; Carnathan, D.G.; Nogal, B.; Martin, J.T.; Rodriguez, O.L.; Upadhyay, A.A.; Enemuo, C.A.; Gebru, E.H.; Choe, Y.; Viviano, F.; et al. Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance. Cell 2019, 177, 1153–1171e28. [Google Scholar] [CrossRef]
- Pauthner, M.G.; Nkolola, J.P.; Havenar-Daughton, C.; Murrell, B.; Reiss, S.M.; Bastidas, R.; Prevost, J.; Nedellec, R.; von Bredow, B.; Abbink, P.; et al. Vaccine-Induced Protection from Homologous Tier 2 SHIV Challenge in Nonhuman Primates Depends on Serum-Neutralizing Antibody Titers. Immunity 2019, 50, 241–252e6. [Google Scholar] [CrossRef]
- Tam, H.H.; Melo, M.B.; Kang, M.; Pelet, J.M.; Ruda, V.M.; Foley, M.H.; Hu, J.K.; Kumari, S.; Crampton, J.; Baldeon, A.D.; et al. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proc. Natl. Acad. Sci. USA 2016, 113, E6639–E6648. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.J.; Boyington, J.C.; McTamney, P.M.; Kong, W.P.; Pearce, M.B.; Xu, L.; Andersen, H.; Rao, S.; Tumpey, T.M.; Yang, Z.Y.; et al. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science 2010, 329, 1060–1064. [Google Scholar] [CrossRef]
- Wei, C.J.; Yassine, H.M.; McTamney, P.M.; Gall, J.G.; Whittle, J.R.; Boyington, J.C.; Nabel, G.J. Elicitation of broadly neutralizing influenza antibodies in animals with previous influenza exposure. Sci. Transl. Med. 2012, 4, 147ra114. [Google Scholar] [CrossRef]
- Weidenbacher, P.A.; Kim, P.S. Protect, modify, deprotect (PMD): A strategy for creating vaccines to elicit antibodies targeting a specific epitope. Proc. Natl. Acad. Sci. USA 2019, 116, 9947–9952. [Google Scholar] [CrossRef]
- Bajic, G.; Maron, M.J.; Caradonna, T.M.; Tian, M.; Mermelstein, A.; Fera, D.; Kelsoe, G.; Kuraoka, M.; Schmidt, A.G. Structure-Guided Molecular Grafting of a Complex Broadly Neutralizing Viral Epitope. ACS Infect. Dis. 2020, 6, 1182–1191. [Google Scholar] [CrossRef]
- Krammer, F.; Palese, P. Universal Influenza Virus Vaccines That Target the Conserved Hemagglutinin Stalk and Conserved Sites in the Head Domain. J. Infect. Dis 2019, 219 (Suppl. 1), S62–S67. [Google Scholar] [CrossRef] [PubMed]
- Broecker, F.; Liu, S.T.H.; Suntronwong, N.; Sun, W.; Bailey, M.J.; Nachbagauer, R.; Krammer, F.; Palese, P. A mosaic hemagglutinin-based influenza virus vaccine candidate protects mice from challenge with divergent H3N2 strains. NPJ Vaccines 2019, 4, 31. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Kirkpatrick, E.; Ermler, M.; Nachbagauer, R.; Broecker, F.; Krammer, F.; Palese, P. Development of Influenza B Universal Vaccine Candidates Using the “Mosaic” Hemagglutinin Approach. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Kanekiyo, M.; Joyce, M.G.; Gillespie, R.A.; Gallagher, J.R.; Andrews, S.F.; Yassine, H.M.; Wheatley, A.K.; Fisher, B.E.; Ambrozak, D.R.; Creanga, A.; et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 2019, 20, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Skehel, J.J.; Stevens, D.J.; Daniels, R.S.; Douglas, A.R.; Knossow, M.; Wilson, I.A.; Wiley, D.C. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc. Natl. Acad. Sci. USA 1984, 81, 1779–1783. [Google Scholar] [CrossRef]
- Wei, C.J.; Boyington, J.C.; Dai, K.; Houser, K.V.; Pearce, M.B.; Kong, W.P.; Yang, Z.Y.; Tumpey, T.M.; Nabel, G.J. Cross-neutralization of 1918 and 2009 influenza viruses: Role of glycans in viral evolution and vaccine design. Sci. Transl. Med. 2010, 2, 24ra21. [Google Scholar] [CrossRef] [PubMed]
- Medina, R.A.; Stertz, S.; Manicassamy, B.; Zimmermann, P.; Sun, X.; Albrecht, R.A.; Uusi-Kerttula, H.; Zagordi, O.; Belshe, R.B.; Frey, S.E.; et al. Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses. Sci. Transl. Med. 2013, 5, 187ra70. [Google Scholar] [CrossRef]
- Eggink, D.; Goff, P.H.; Palese, P. Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J. Virol. 2014, 88, 699–704. [Google Scholar] [CrossRef]
- Xu, J.L.; Davis, M.M. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 2000, 13, 37–45. [Google Scholar] [CrossRef]
- Schroeder, H.W., Jr.; Cavacini, L. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 2010, 125 (Suppl. 2), S41–S52. [Google Scholar] [CrossRef]
- Wedemayer, G.J.; Patten, P.A.; Wang, L.H.; Schultz, P.G.; Stevens, R.C. Structural insights into the evolution of an antibody combining site. Science 1997, 276, 1665–1669. [Google Scholar] [CrossRef]
- Saada, R.; Weinberger, M.; Shahaf, G.; Mehr, R. Models for antigen receptor gene rearrangement: CDR3 length. Immunol. Cell Biol. 2007, 85, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Morea, V.; Tramontano, A.; Rustici, M.; Chothia, C.; Lesk, A.M. Conformations of the third hypervariable region in the VH domain of immunoglobulins. J. Mol. Biol. 1998, 275, 269–294. [Google Scholar] [CrossRef]
- Oliva, B.; Bates, P.A.; Querol, E.; Aviles, F.X.; Sternberg, M.J. Automated classification of antibody complementarity determining region 3 of the heavy chain (H3) loops into canonical forms and its application to protein structure prediction. J. Mol. Biol. 1998, 279, 1193–1210. [Google Scholar] [CrossRef]
- Shirai, H.; Kidera, A.; Nakamura, H. H3-rules: Identification of CDR-H3 structures in antibodies. FEBS Lett. 1999, 455, 188–197. [Google Scholar] [CrossRef]
- Kuroda, D.; Shirai, H.; Kobori, M.; Nakamura, H. Structural classification of CDR-H3 revisited: A lesson in antibody modeling. Proteins 2008, 73, 608–620. [Google Scholar] [CrossRef]
- North, B.; Lehmann, A.; Dunbrack, R.L., Jr. A new clustering of antibody CDR loop conformations. J. Mol. Biol. 2011, 406, 228–256. [Google Scholar] [CrossRef]
- Lerner, R.A. Rare antibodies from combinatorial libraries suggests an S.O.S. component of the human immunological repertoire. Mol. Biosyst. 2011, 7, 1004–1012. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Lynch, R.M.; Chen, L.; Acharya, P.; Wu, X.; Doria-Rose, N.A.; Joyce, M.G.; Lingwood, D.; Soto, C.; Bailer, R.T.; et al. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell 2015, 161, 1280–1292. [Google Scholar] [CrossRef] [PubMed]
- Flyak, A.I.; Ruiz, S.; Colbert, M.D.; Luong, T.; Crowe, J.E., Jr.; Bailey, J.R.; Bjorkman, P.J. HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design. Cell Host Microbe 2018, 24, 703–716e3. [Google Scholar] [CrossRef]
- Pappas, L.; Foglierini, M.; Piccoli, L.; Kallewaard, N.L.; Turrini, F.; Silacci, C.; Fernandez-Rodriguez, B.; Agatic, G.; Giacchetto-Sasselli, I.; Pellicciotta, G.; et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature 2014, 516, 418–422. [Google Scholar] [CrossRef]
- Avnir, Y.; Tallarico, A.S.; Zhu, Q.; Bennett, A.S.; Connelly, G.; Sheehan, J.; Sui, J.; Fahmy, A.; Huang, C.Y.; Cadwell, G.; et al. Molecular signatures of hemagglutinin stem-directed heterosubtypic human neutralizing antibodies against influenza A viruses. PLoS Pathog. 2014, 10, e1004103. [Google Scholar] [CrossRef]
- Tzarum, N.; Giang, E.; Kong, L.; He, L.; Prentoe, J.; Augestad, E.; Hua, Y.; Castillo, S.; Lauer, G.M.; Bukh, J.; et al. Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies. Sci. Adv. 2019, 5, eaav1882. [Google Scholar] [CrossRef]
- Scheid, J.F.; Mouquet, H.; Ueberheide, B.; Diskin, R.; Klein, F.; Oliveira, T.Y.; Pietzsch, J.; Fenyo, D.; Abadir, A.; Velinzon, K.; et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 2011, 333, 1633–1637. [Google Scholar] [CrossRef]
- Wec, A.Z.; Haslwanter, D.; Abdiche, Y.N.; Shehata, L.; Pedreno-Lopez, N.; Moyer, C.L.; Bornholdt, Z.A.; Lilov, A.; Nett, J.H.; Jangra, R.K.; et al. Longitudinal dynamics of the human B cell response to the yellow fever 17D vaccine. Proc. Natl. Acad. Sci. USA 2020, 117, 6675–6685. [Google Scholar] [CrossRef] [PubMed]
- Golsaz-Shirazi, F.; Amiri, M.M.; Bahadori, M.; Bayat, A.A.; Mohammadi, H.; Farid, S.; Maddah, M.; Khoshnoodi, J.; Zarnani, A.H.; Jeddi-Tehrani, M.; et al. Molecular Characterization of Murine Monoclonal Antibody Variable Regions Specific for Hepatitis B Surface Antigen. Viral Immunol. 2015, 28, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Lucas, A.H.; Langley, R.J.; Granoff, D.M.; Nahm, M.H.; Kitamura, M.Y.; Scott, M.G. An idiotypic marker associated with a germ-line encoded kappa light chain variable region that predominates the vaccine-induced human antibody response to the Haemophilus influenzae b polysaccharide. J. Clin. Investig. 1991, 88, 1811–1818. [Google Scholar] [CrossRef]
- Zhou, T.; Zhu, J.; Wu, X.; Moquin, S.; Zhang, B.; Acharya, P.; Georgiev, I.S.; Altae-Tran, H.R.; Chuang, G.Y.; Joyce, M.G.; et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 2013, 39, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhou, T.; Zhu, J.; Zhang, B.; Georgiev, I.; Wang, C.; Chen, X.; Longo, N.S.; Louder, M.; McKee, K.; et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 2011, 333, 1593–1602. [Google Scholar] [CrossRef] [PubMed]
- Hehle, V.; Beretta, M.; Bourgine, M.; Ait-Goughoulte, M.; Planchais, C.; Morisse, S.; Vesin, B.; Lorin, V.; Hieu, T.; Stauffer, A.; et al. Potent human broadly neutralizing antibodies to hepatitis B virus from natural controllers. J. Exp. Med. 2020, 217, e20200840. [Google Scholar] [CrossRef]
- Henry Dunand, C.J.; Wilson, P.C. Restricted, canonical, stereotyped and convergent immunoglobulin responses. Philos Trans. R Soc. Lond. B Biol. Sci. 2015, 370, 20140238. [Google Scholar] [CrossRef] [PubMed]
- Sangesland, M.; Yousif, A.S.; Ronsard, L.; Kazer, S.W.; Zhu, A.L.; Gatter, G.J.; Hayward, M.R.; Barnes, R.M.; Quirindongo-Crespo, M.; Rohrer, D.; et al. A Single Human VH-gene Allows for a Broad-Spectrum Antibody Response Targeting Bacterial Lipopolysaccharides in the Blood. Cell Rep. 2020, 32, 108065. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Liu, H.; Wu, N.C.; Lee, C.D.; Zhu, X.; Zhao, F.; Huang, D.; Yu, W.; Hua, Y.; Tien, H.; et al. Structural basis of a shared antibody response to SARS-CoV-2. Science 2020, 369, 1119–1123. [Google Scholar] [CrossRef] [PubMed]
- Wec, A.Z.; Wrapp, D.; Herbert, A.S.; Maurer, D.P.; Haslwanter, D.; Sakharkar, M.; Jangra, R.K.; Dieterle, M.E.; Lilov, A.; Huang, D.; et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 2020, 369, 731–736. [Google Scholar] [CrossRef] [PubMed]
- Villar, R.F.; Patel, J.; Weaver, G.C.; Kanekiyo, M.; Wheatley, A.K.; Yassine, H.M.; Costello, C.E.; Chandler, K.B.; McTamney, P.M.; Nabel, G.J.; et al. Reconstituted B cell receptor signaling reveals carbohydrate-dependent mode of activation. Sci. Rep. 2016, 6, 36298. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sangesland, M.; Lingwood, D. Antibody Focusing to Conserved Sites of Vulnerability: The Immunological Pathways for ‘Universal’ Influenza Vaccines. Vaccines 2021, 9, 125. https://doi.org/10.3390/vaccines9020125
Sangesland M, Lingwood D. Antibody Focusing to Conserved Sites of Vulnerability: The Immunological Pathways for ‘Universal’ Influenza Vaccines. Vaccines. 2021; 9(2):125. https://doi.org/10.3390/vaccines9020125
Chicago/Turabian StyleSangesland, Maya, and Daniel Lingwood. 2021. "Antibody Focusing to Conserved Sites of Vulnerability: The Immunological Pathways for ‘Universal’ Influenza Vaccines" Vaccines 9, no. 2: 125. https://doi.org/10.3390/vaccines9020125
APA StyleSangesland, M., & Lingwood, D. (2021). Antibody Focusing to Conserved Sites of Vulnerability: The Immunological Pathways for ‘Universal’ Influenza Vaccines. Vaccines, 9(2), 125. https://doi.org/10.3390/vaccines9020125





