A Five Collagen-Related Gene Signature to Estimate the Prognosis and Immune Microenvironment in Clear Cell Renal Cell Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Obtaining and Preprocessing
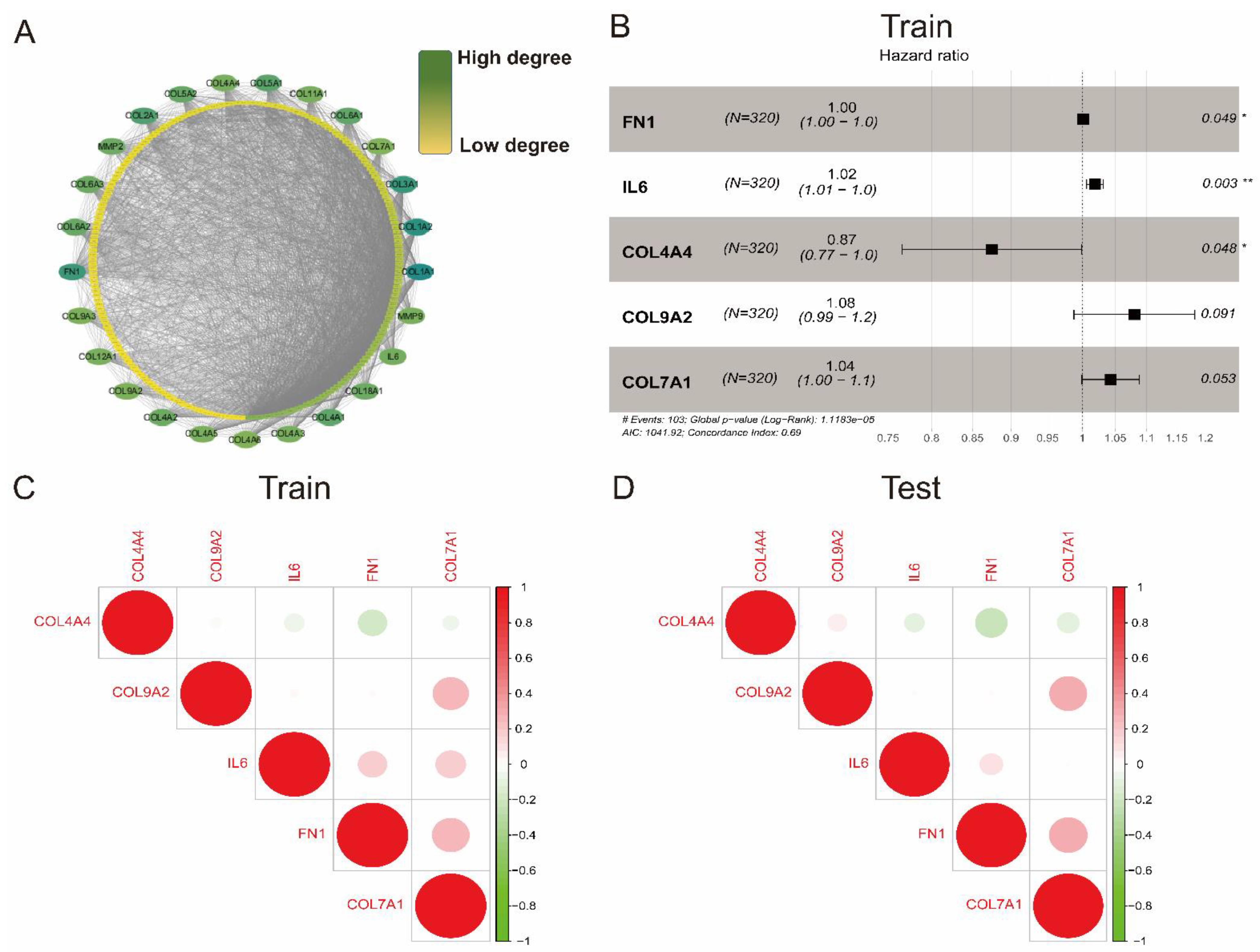
2.2. Establishment of a Protein–Protein Interaction Network
2.3. Collagen-Related Risk Model Establishment
2.4. GSEA Analysis and Immune Cell Type Fraction Estimation
2.5. Statistics
3. Results
3.1. Characteristics of Patients Separated into Training and Test Cohort
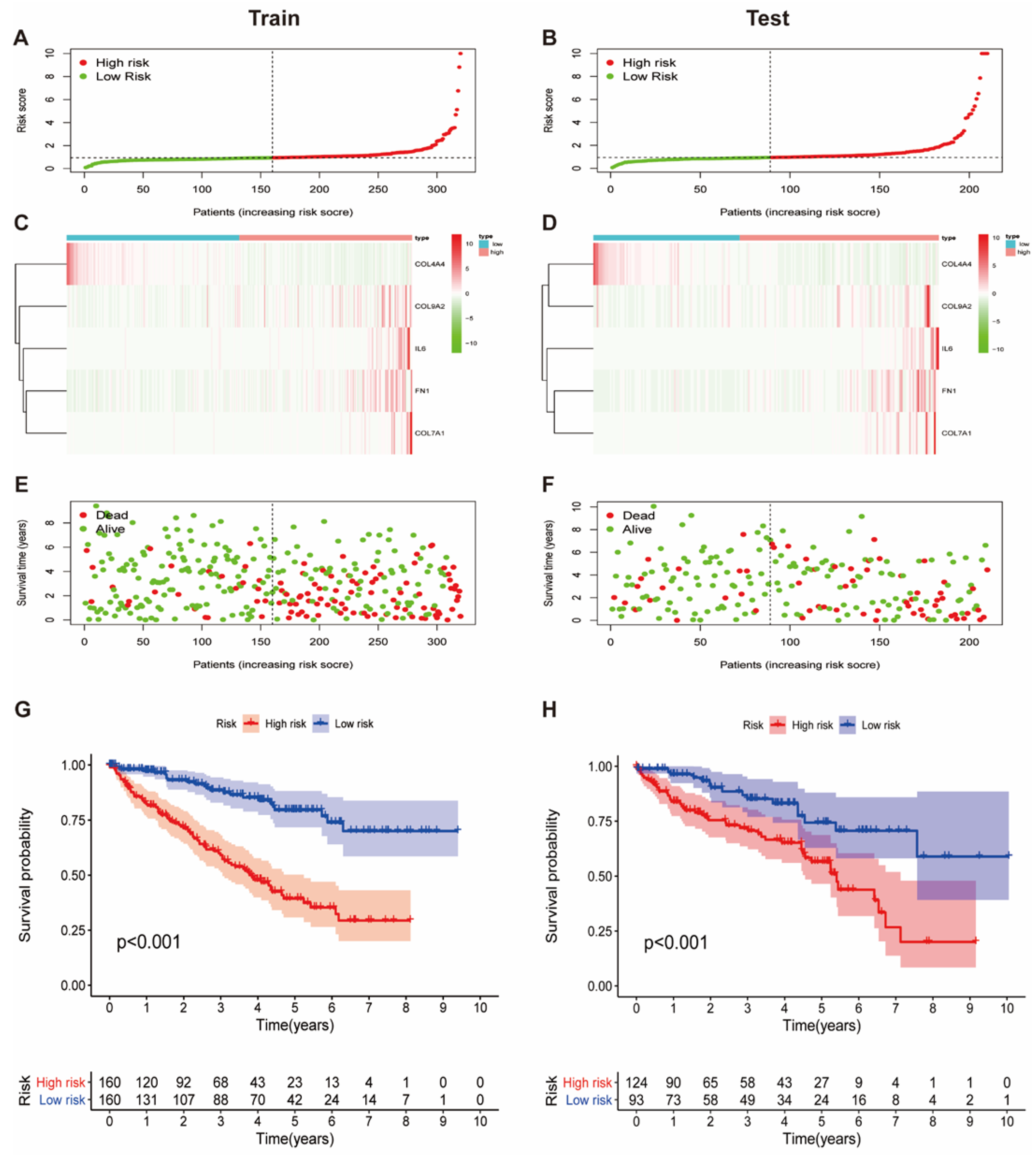
3.2. Establishment of a Collagen-Related Risk Model
3.3. Prognostic Value of the Collagen-Related Signature in ccRCC Patients
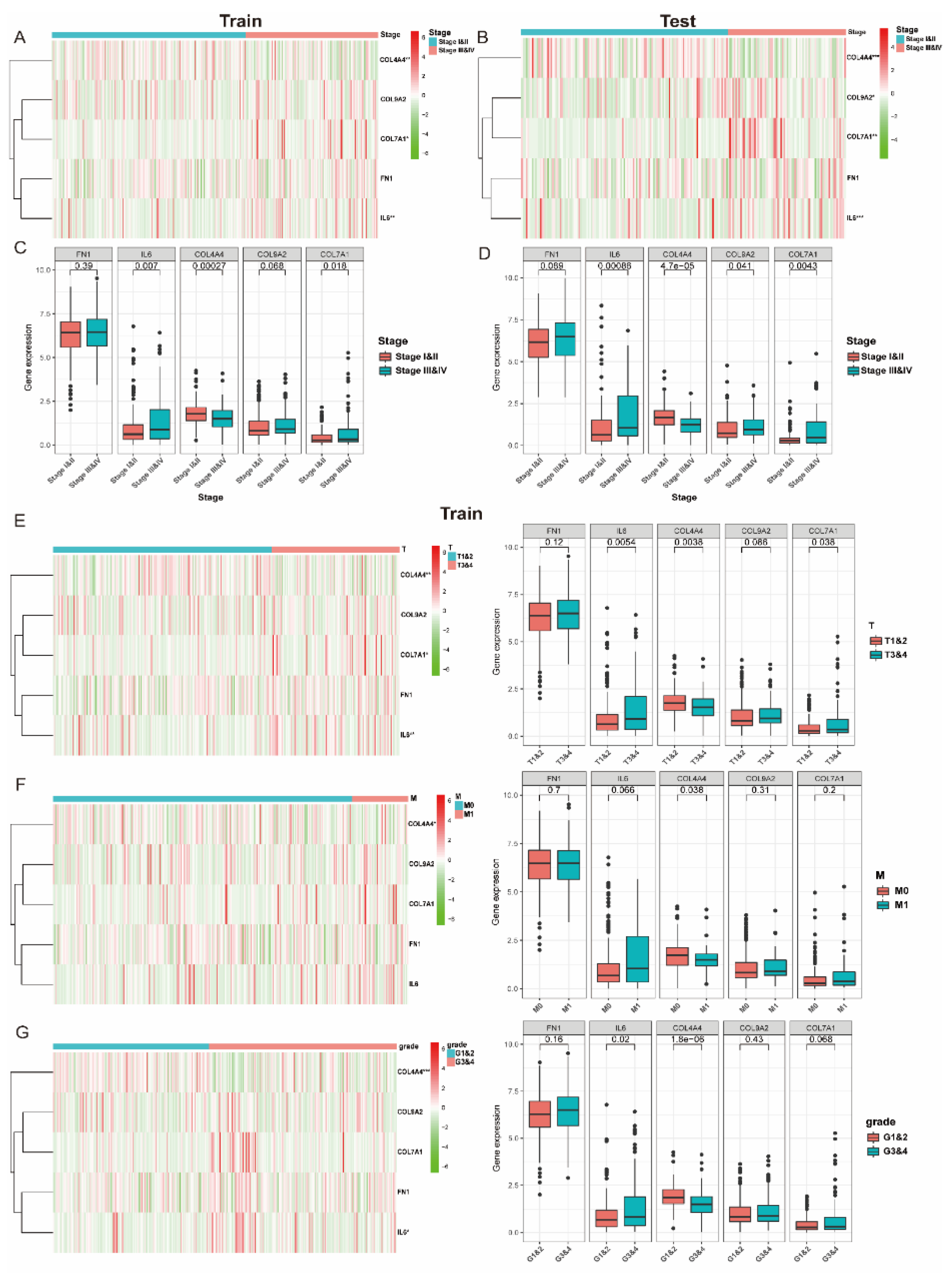
3.4. The Expression of Collagen-Related Genes Is Associated with Clinical and Pathological Characteristics
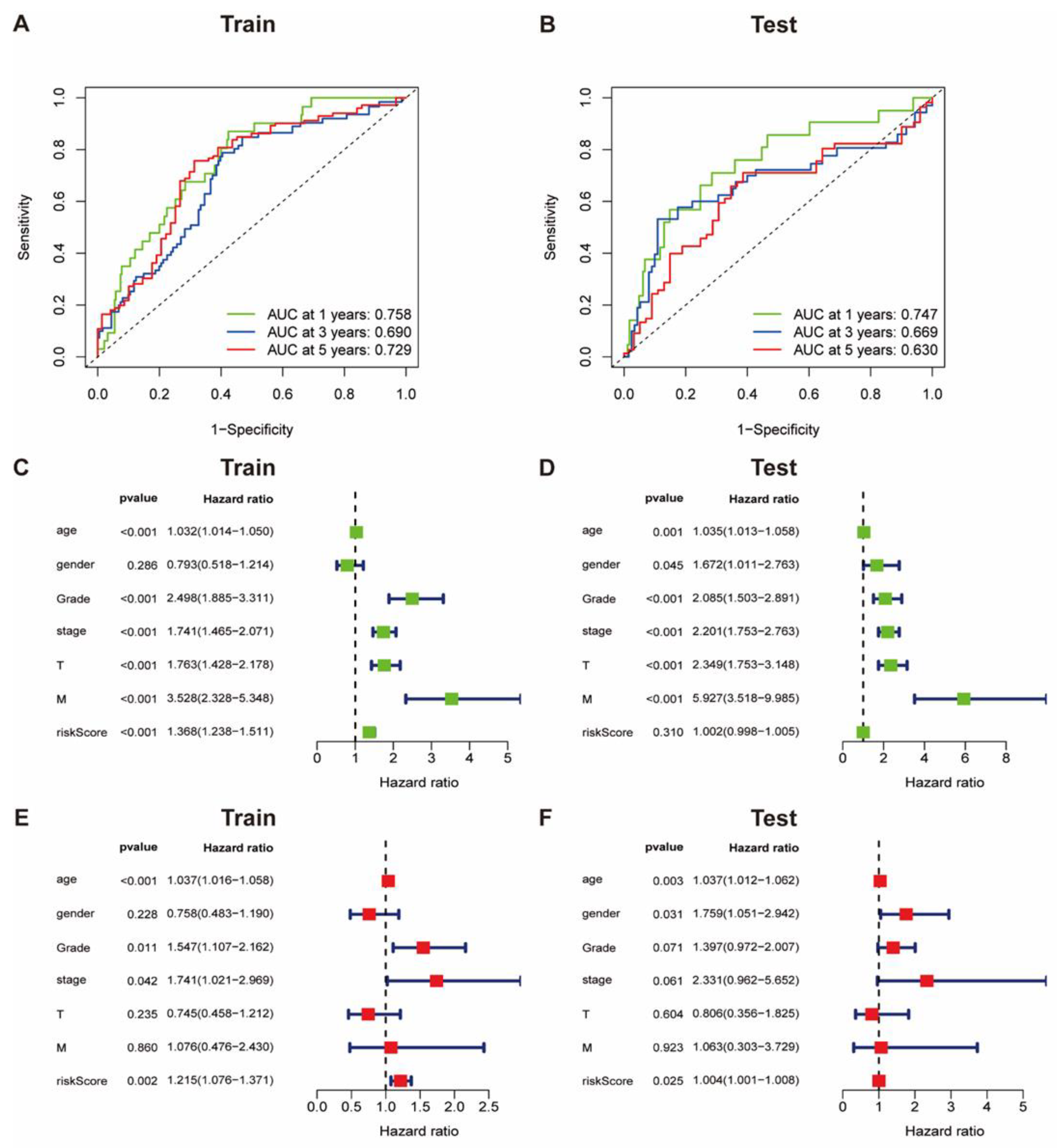
3.5. The Accuracy of Collagen-Related Signature for Prognosis Evaluation
3.6. GSEA Identifies Potential Signaling Pathways
3.7. Identification of Immune Cells Infiltrated in Patients with Different Risk Scores
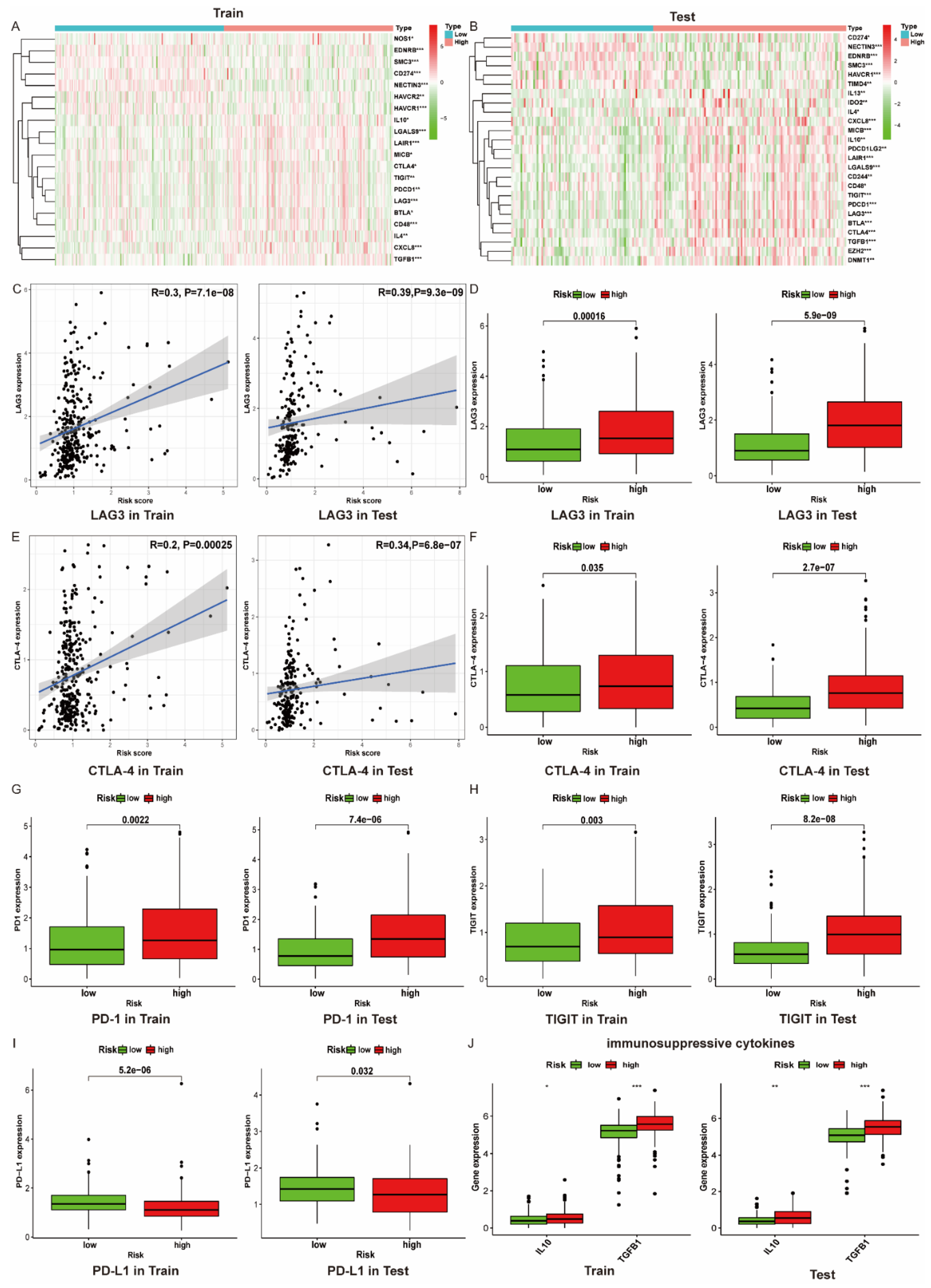
3.8. The Immune Microenvironment of Patients with High Risk Scores Tend to Be Suppressed
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Li, F.; Jin, Y.; Pei, X.; Guo, P.; Dong, K.; Wang, H.; Chen, Y.; Guo, P.; Meng, L.-B.; Wang, Z. Bioinformatics analysis and verification of gene targets for renal clear cell carcinoma. Comput. Biol. Chem. 2021, 92, 107453. [Google Scholar] [CrossRef]
- Win, A.Z.; Aparici, C.M. Clinical effectiveness of (18)f-fluorodeoxyglucose positron emission tomography/computed tomography in management of renal cell carcinoma: A single institution experience. World J. Nucl. Med. 2015, 14, 36–40. [Google Scholar] [CrossRef]
- Molina, A.M.; Lin, X.; Korytowsky, B.; Matczak, E.; Lechuga, M.J.; Wiltshire, R.; Motzer, R.J. Sunitinib objective response in metastatic renal cell carcinoma: Analysis of 1059 patients treated on clinical trials. Eur. J. Cancer 2014, 50, 351–358. [Google Scholar] [CrossRef]
- García-Mendoza, M.G.; Inman, D.R.; Ponik, S.M.; Jeffery, J.J.; Sheerar, D.S.; van Doorn, R.R.; Keely, P.J. Neutrophils drive accelerated tumor progression in the collagen-dense mammary tumor microenvironment. Breast Cancer Res. 2016, 18, 49. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.; Polyak, K. Microenvironmental regulation of cancer development. Curr. Opin. Genet. Dev. 2008, 18, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Varol, C. Tumorigenic Interplay between Macrophages and Collagenous Matrix in the Tumor Microenvironment. Methods Mol. Biol. 2019, 1944, 203–220. [Google Scholar] [CrossRef]
- Liverani, C.; Mercatali, L.; Cristofolini, L.; Giordano, E.; Minardi, S.; Della Porta, G.; de Vita, A.; Miserocchi, G.; Spadazzi, C.; Tasciotti, E.; et al. Investigating the Mechanobiology of Cancer Cell-ECM Interaction Through Collagen-Based 3D Scaffolds. Cell. Mol. Bioeng. 2017, 10, 223–234. [Google Scholar] [CrossRef]
- Zhou, Z.-H.; Ji, C.-D.; Xiao, H.-L.; Zhao, H.-B.; Cui, Y.-H.; Bian, X.-W. Reorganized Collagen in the Tumor Microenvironment of Gastric Cancer and Its Association with Prognosis. J. Cancer 2017, 8, 1466–1476. [Google Scholar] [CrossRef] [Green Version]
- Martins Cavaco, A.C.; Dâmaso, S.; Casimiro, S.; Costa, L. Collagen biology making inroads into prognosis and treatment of cancer progression and metastasis. Cancer Metastasis Rev. 2020, 39, 603–623. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Fredericks, T.; Xiong, G.; Qi, Y.; Rychahou, P.G.; Li, J.-D.; Pihlajaniemi, T.; Xu, W.; Xu, R. Membrane associated collagen XIII promotes cancer metastasis and enhances anoikis resistance. Breast Cancer Res. 2018, 20, 116. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Dai, Y.; Mei, Y.; Yang, M.; Hu, L.; Yang, H.; Guan, X.; Li, J. Clinical significance and biological role of cancer-derived Type I collagen in lung and esophageal cancers. Thorac. Cancer 2019, 10, 277–288. [Google Scholar] [CrossRef] [Green Version]
- Pankova, D.; Jiang, Y.; Chatzifrangkeskou, M.; Vendrell, I.; Buzzelli, J.; Ryan, A.; Brown, C.; O’Neill, E. RASSF1A controls tissue stiffness and cancer stem-like cells in lung adenocarcinoma. EMBO J. 2019, 38, e100532. [Google Scholar] [CrossRef]
- Afik, R.; Zigmond, E.; Vugman, M.; Klepfish, M.; Shimshoni, E.; Pasmanik-Chor, M.; Shenoy, A.; Bassat, E.; Halpern, Z.; Geiger, T.; et al. Tumor macrophages are pivotal constructors of tumor collagenous matrix. J. Exp. Med. 2016, 213, 2315–2331. [Google Scholar] [CrossRef]
- Ingman, W.V.; Wyckoff, J.; Gouon-Evans, V.; Condeelis, J.; Pollard, J.W. Macrophages promote collagen fibrillogenesis around terminal end buds of the developing mammary gland. Dev. Dyn. 2006, 235, 3222–3229. [Google Scholar] [CrossRef]
- Casimiro, S.; Ferreira, A.R.; Mansinho, A.; Alho, I.; Costa, L. Molecular Mechanisms of Bone Metastasis: Which Targets Came from the Bench to the Bedside? Int. J. Mol. Sci. 2016, 17, 1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Condeelis, J.; Segall, J.E. Intravital imaging of cell movement in tumours. Nat. Rev. Cancer 2003, 3, 921–930. [Google Scholar] [CrossRef]
- Shen, L.; Yang, M.; Lin, Q.; Zhang, Z.; Zhu, B.; Miao, C. COL11A1 is overexpressed in recurrent non-small cell lung cancer and promotes cell proliferation, migration, invasion and drug resistance. Oncol. Rep. 2016, 36, 877–885. [Google Scholar] [CrossRef] [Green Version]
- Owusu-Ansah, K.G.; Song, G.; Chen, R.; Edoo, M.I.A.; Li, J.; Chen, B.; Wu, J.; Zhou, L.; Xie, H.; Jiang, D.; et al. COL6A1 promotes metastasis and predicts poor prognosis in patients with pancreatic cancer. Int. J. Oncol. 2019, 55, 391–404. [Google Scholar] [CrossRef] [Green Version]
- Wan, F.; Wang, H.; Shen, Y.; Zhang, H.; Shi, G.; Zhu, Y.; Dai, B.; Ye, D. Upregulation of COL6A1 is predictive of poor prognosis in clear cell renal cell carcinoma patients. Oncotarget 2015, 6, 27378–27387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, S.L.; Tan, M.L.; Hollows, R.J.; Robinson, M.; Ibrahim, M.; Margielewska, S.; Parkinson, E.K.; Ramanathan, A.; Zain, R.B.; Mehanna, H.; et al. Collagen Induces a More Proliferative, Migratory and Chemoresistant Phenotype in Head and Neck Cancer via DDR1. Cancers 2019, 11, 1766. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Wu, S.; Chen, X.; Ye, Y.; Weng, Y.; Pan, Y.; Chen, Z.; Chen, L.; Qiu, X.; Qiu, S. Characterization of Hypoxia Signature to Evaluate the Tumor Immune Microenvironment and Predict Prognosis in Glioma Groups. Front. Oncol. 2020, 10, 796. [Google Scholar] [CrossRef]
- Li, B.; Chan, H.L.; Chen, P. Immune Checkpoint Inhibitors: Basics and Challenges. Curr. Med. Chem. 2019, 26, 3009–3025. [Google Scholar] [CrossRef]
- Darvin, P.; Toor, S.M.; Sasidharan Nair, V.; Elkord, E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Dennis, K.L.; Blatner, N.R.; Gounari, F.; Khazaie, K. Current status of interleukin-10 and regulatory T-cells in cancer. Curr. Opin. Oncol. 2013, 25, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.-T.; Liu, X.-P.; Liu, T.-Z.; Wang, X.-H. The clinical significance of COL5A2 in patients with bladder cancer: A retrospective analysis of bladder cancer gene expression data. Medicine 2018, 97, e0091. [Google Scholar] [CrossRef] [PubMed]
- Miyake, M.; Hori, S.; Morizawa, Y.; Tatsumi, Y.; Toritsuka, M.; Ohnishi, S.; Shimada, K.; Furuya, H.; Khadka, V.S.; Deng, Y.; et al. Collagen type IV alpha 1 (COL4A1) and collagen type XIII alpha 1 (COL13A1) produced in cancer cells promote tumor budding at the invasion front in human urothelial carcinoma of the bladder. Oncotarget 2017, 8, 36099–36114. [Google Scholar] [CrossRef] [Green Version]
- Banyard, J.; Bao, L.; Hofer, M.D.; Zurakowski, D.; Spivey, K.A.; Feldman, A.S.; Hutchinson, L.M.; Kuefer, R.; Rubin, M.A.; Zetter, B.R. Collagen XXIII expression is associated with prostate cancer recurrence and distant metastases. Clin. Cancer Res. 2007, 13, 2634–2642. [Google Scholar] [CrossRef] [Green Version]
- Ritzenthaler, J.D.; Han, S.; Roman, J. Stimulation of lung carcinoma cell growth by fibronectin-integrin signalling. Mol. Biosyst. 2008, 4, 1160–1169. [Google Scholar] [CrossRef]
- Cai, X.; Liu, C.; Zhang, T.-N.; Zhu, Y.-W.; Dong, X.; Xue, P. Down-regulation of FN1 inhibits colorectal carcinogenesis by suppressing proliferation, migration, and invasion. J. Cell. Biochem. 2018, 119, 4717–4728. [Google Scholar] [CrossRef]
- Kumari, N.; Dwarakanath, B.S.; Das, A.; Bhatt, A.N. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumour Biol. 2016, 37, 11553–11572. [Google Scholar] [CrossRef]
- Legendre, F.; Dudhia, J.; Pujol, J.-P.; Bogdanowicz, P. JAK/STAT but not ERK1/ERK2 pathway mediates interleukin (IL)-6/soluble IL-6R down-regulation of Type II collagen, aggrecan core, and link protein transcription in articular chondrocytes. Association with a down-regulation of SOX9 expression. J. Biol. Chem. 2003, 278, 2903–2912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heichler, C.; Scheibe, K.; Schmied, A.; Geppert, C.I.; Schmid, B.; Wirtz, S.; Thoma, O.-M.; Kramer, V.; Waldner, M.J.; Büttner, C.; et al. STAT3 activation through IL-6/IL-11 in cancer-associated fibroblasts promotes colorectal tumour development and correlates with poor prognosis. Gut 2020, 69, 1269–1282. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Noma, K.; Ohara, T.; Kashima, H.; Katsura, Y.; Sato, H.; Komoto, S.; Katsube, R.; Ninomiya, T.; Tazawa, H.; et al. Cancer-Associated Fibroblasts Affect Intratumoral CD8+ and FoxP3+ T Cells Via IL6 in the Tumor Microenvironment. Clin. Cancer Res. 2018, 24, 4820–4833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Hu, P.; Xu, H.; Yuan, J.; Yuan, L.; Xiong, W.; Deng, X.; Deng, H. A novel heterozygous COL4A4 missense mutation in a Chinese family with focal segmental glomerulosclerosis. J. Cell. Mol. Med. 2016, 20, 2328–2332. [Google Scholar] [CrossRef] [Green Version]
- Chattopadhyay, I.; Phukan, R.; Singh, A.; Vasudevan, M.; Purkayastha, J.; Hewitt, S.; Kataki, A.; Mahanta, J.; Kapur, S.; Saxena, S. Molecular profiling to identify molecular mechanism in esophageal cancer with familial clustering. Oncol. Rep. 2009, 21, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, X.; Zheng, K.; Liu, Y.; Li, J.; Wang, S.; Liu, K.; Song, X.; Li, N.; Xie, S.; et al. The clinical significance of collagen family gene expression in esophageal squamous cell carcinoma. PeerJ 2019, 7, e7705. [Google Scholar] [CrossRef]
- Kita, Y.; Mimori, K.; Tanaka, F.; Matsumoto, T.; Haraguchi, N.; Ishikawa, K.; Matsuzaki, S.; Fukuyoshi, Y.; Inoue, H.; Natsugoe, S.; et al. Clinical significance of LAMB3 and COL7A1 mRNA in esophageal squamous cell carcinoma. Eur. J. Surg. Oncol. 2009, 35, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Pourreyron, C.; Chen, M.; McGrath, J.A.; Salas-Alanis, J.C.; South, A.P.; Leigh, I.M. High levels of type VII collagen expression in recessive dystrophic epidermolysis bullosa cutaneous squamous cell carcinoma keratinocytes increases PI3K and MAPK signalling, cell migration and invasion. Br. J. Dermatol. 2014, 170, 1256–1265. [Google Scholar] [CrossRef]
- Gloushankova, N.A.; Zhitnyak, I.Y.; Rubtsova, S.N. Role of Epithelial-Mesenchymal Transition in Tumor Progression. Biochemistry 2018, 83, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Jing, Y.; Han, Z.; Zhang, S.; Liu, Y.; Wei, L. Epithelial-Mesenchymal Transition in tumor microenvironment. Cell Biosci. 2011, 1, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, D.H.; Rodriguez, B.L.; Diao, L.; Chen, L.; Wang, J.; Byers, L.A.; Wei, Y.; Chapman, H.A.; Yamauchi, M.; Behrens, C.; et al. Collagen promotes anti-PD-1/PD-L1 resistance in cancer through LAIR1-dependent CD8+ T cell exhaustion. Nat. Commun. 2020, 11, 4520. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.M.H.; Kuczek, D.E.; Kalvisa, A.; Siersbæk, M.S.; Thorseth, M.-L.; Johansen, A.Z.; Carretta, M.; Grøntved, L.; Vang, O.; Madsen, D.H. Collagen Density Modulates the Immunosuppressive Functions of Macrophages. J. Immunol. 2020, 205, 1461–1472. [Google Scholar] [CrossRef]
- Qiu, S.; Deng, L.; Liao, X.; Nie, L.; Qi, F.; Jin, K.; Tu, X.; Zheng, X.; Li, J.; Liu, L.; et al. Tumor-associated macrophages promote bladder tumor growth through PI3K/AKT signal induced by collagen. Cancer Sci. 2019, 110, 2110–2118. [Google Scholar] [CrossRef]
- Barbi, J.; Pardoll, D.; Pan, F. Treg functional stability and its responsiveness to the microenvironment. Immunol. Rev. 2014, 259, 115–139. [Google Scholar] [CrossRef]
- Colak, S.; Ten Dijke, P. Targeting TGF-β Signaling in Cancer. Trends Cancer 2017, 3, 56–71. [Google Scholar] [CrossRef]
- Haque, S.; Morris, J.C. Transforming growth factor-β: A therapeutic target for cancer. Hum. Vaccines Immunother. 2017, 13, 1741–1750. [Google Scholar] [CrossRef]



| Variables | Training Cohort (n = 320) | Test Cohort (n = 217) | p Value |
|---|---|---|---|
| Age | |||
| Mean (SD) | 60.03 (11.81) | 61.42 (12.63) | 0.194 |
| Median (min,max) | 60.00 (29,90) | 61 (26,90) | |
| Gender | |||
| MALE | 209 (65.31%) | 137 (63.13%) | 0.605 |
| FEMALE | 111 (34.69%) | 80 (36.87%) | |
| Overall Survival time | |||
| Mean (SD) | 1130.68 (803.79) | 1146.57 (831.64) | 0.824 |
| Median (min,max) | 1026.50 (3,3431) | 1106.00 (2,3668) | |
| Unknown | 3 | 2 | |
| Survival State | |||
| Alive | 217 (67.81%) | 150 (69.12%) | 0.748 |
| Dead | 103 (32.19%) | 67 (30.88%) | |
| Histologic grade | |||
| G1 | 10 (3.13%) | 4 (1.84%) | 0.822 |
| G2 | 133 (41.56%) | 97 (44.70%) | |
| G3 | 127 (39.69%) | 80 (36.87%) | |
| G4 | 45 (14.06%) | 33 (15.21%) | |
| Unknown | 5 (1.56%) | 3 (1.38%) | |
| T stage | |||
| T1 | 159 (49.69%) | 116 (53.46%) | 0.851 |
| T2 | 43 (13.44%) | 26 (11.98%) | |
| T3 | 111 (34.69%) | 71 (32.72%) | |
| T4 | 7 (2.19%) | 4 (1.84%) | |
| Unknown | 0 (0%) | 0 (0%) | |
| N stage | |||
| N1 | 12 (3.75%) | 5 (2.30%) | 0.221 |
| N0 | 134 (41.88%) | 106 (48.85%) | |
| Unknown | 174 (54.38%) | 106 (48.85%) | |
| M stage | |||
| M1 | 48 (15.00%) | 31 (14.29%) | 0.519 |
| M0 | 256 (80.00%) | 170 (78.34%) | |
| Unknown | 16 (5.00%) | 16 (7.37%) | |
| Clinical stage | |||
| Stage I | 155 (48.44%) | 114 (52.53%) | 0.8803 |
| Stage II | 34 (10.63%) | 23 (10.60%) | |
| Stage III | 79 (24.69%) | 46 (21.20%) | |
| Stage IV | 50 (15.63%) | 33 (15.21%) | |
| Unknown | 2 (0.63%) | 1 (0.46%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, X.; Zhou, X.; Yue, C.; Gao, S.; Sun, Z.; Lu, C.; Zuo, L. A Five Collagen-Related Gene Signature to Estimate the Prognosis and Immune Microenvironment in Clear Cell Renal Cell Cancer. Vaccines 2021, 9, 1510. https://doi.org/10.3390/vaccines9121510
Shi X, Zhou X, Yue C, Gao S, Sun Z, Lu C, Zuo L. A Five Collagen-Related Gene Signature to Estimate the Prognosis and Immune Microenvironment in Clear Cell Renal Cell Cancer. Vaccines. 2021; 9(12):1510. https://doi.org/10.3390/vaccines9121510
Chicago/Turabian StyleShi, Xiaokai, Xiao Zhou, Chuang Yue, Shenglin Gao, Zhiqin Sun, Chao Lu, and Li Zuo. 2021. "A Five Collagen-Related Gene Signature to Estimate the Prognosis and Immune Microenvironment in Clear Cell Renal Cell Cancer" Vaccines 9, no. 12: 1510. https://doi.org/10.3390/vaccines9121510
APA StyleShi, X., Zhou, X., Yue, C., Gao, S., Sun, Z., Lu, C., & Zuo, L. (2021). A Five Collagen-Related Gene Signature to Estimate the Prognosis and Immune Microenvironment in Clear Cell Renal Cell Cancer. Vaccines, 9(12), 1510. https://doi.org/10.3390/vaccines9121510





