Abstract
Human papillomavirus (HPV) 16 capsids have been chosen as a DNA delivery vehicle in many studies. Our preliminary studies suggest that HPV58 capsids could be better vehicles than HPV16 capsids to deliver encapsidated DNA in vitro and in vivo. In the current study, we compared HPV16, HPV58, and the cottontail rabbit papillomavirus (CRPV) capsids either as L1/L2 VLPs or pseudoviruses (PSVs) to deliver externally attached GFP-expressing DNA. Both rabbit and human cells were used to test whether there was a species-specific effect. DNA delivery efficiency was determined by quantifying either GFP-expressing cell populations or mean fluorescent intensities (MFI) by flow cytometry. Interestingly, CRPV and 58-VLPs and PSVs were significantly more efficient at delivering attached DNA when compared to 16-VLPs and PSVs. A capsid/DNA ratio of 2:1 showed the highest efficiency for delivering external DNA. The PSVs with papillomavirus DNA genomes also showed higher efficiency than those with irrelevant plasmid DNA. HPV16L1/58L2 hybrid VLPs displayed increased efficiency compared to HPV58L1/16L2 VLPs, suggesting that L2 may play a critical role in the delivery of attached DNA. Additionally, we demonstrated that VLPs increased in vivo infectivity of CRPV DNA in rabbits. We conclude that choosing CRPV or 58 capsids to deliver external DNA could improve DNA uptake in in vitro and in vivo models.
Keywords:
virus-like particles (VLPs); L1; L2; pseudovirus (PSV); papillomavirus; DNA delivery; GFP; flow cytometry; HPV16; HPV58; CRPV 1. Introduction
Papillomavirus capsids comprise a major capsid protein, L1, and a minor capsid protein, L2 [1,2]. Papillomavirus virus-like particles (VLPs) can be produced from L1 only or L1 and L2 [3]. L1 VLPs have been developed into successful prophylactic vaccines for the control of several human papillomavirus-induced diseases and cancers [4,5]. Papillomavirus L1 VLPs have been shown to bind to several different species of cells and cell types and be internalized by these same cells; therefore [6,7], they are potential vehicles to deliver encapsidated DNA. A previous study suggested that L1/L2 VLPs were more efficient for DNA delivery than L1 VLPs, suggesting that L2 positively impacted the delivery [8]. Pseudoviruses (PSVs, L1/L2 VLPs containing unrelated DNA) have been used to deliver genes, including GFP and luciferase, for in vitro and in vivo studies [9,10,11,12]. Apart from the capability of delivering encapsidated DNA, L1/L2 VLPs have also been shown to efficiently deliver externally attached DNA in vitro and in vivo [13]. This delivery method shows promise because it can deliver a larger amount of DNA when compared with limited copies of encapsidated DNA.
Papillomaviruses do not show a species-specificity in binding and uptake in in vitro cultures [7]. This suggests that different papillomavirus capsids can be used as vehicles for delivering DNA into a variety of cell types. HPV16 was chosen for most delivery studies; however, no studies have compared the delivery efficiency of different types of HPVs. Our unpublished and published studies showed that HPV58 with CRPV DNA genomes was more infectious than HPV16 in vitro and in vivo [14]. Therefore, we hypothesized that different HPV capsids may have diverse inherent efficiencies to also deliver attached DNA to the cells. To test this hypothesis, we designed a series of experiments to compare the delivery efficiency of VLPs (no genome) and PSVs (containing a plasmid or PV genome) made from HPV16 and HPV58 (16-VLP, 58-VLP, PSV-16, and PSV-58) as well as an animal papillomavirus (the cottontail rabbit papillomavirus, CRPV) (CRPV-VLP, PSV-CRPV) in both rabbit and human cell cultures. Our data clearly suggest different delivery efficiencies by several papillomavirus capsid types. Overall, HPV58 capsids were more efficient at delivering both externally attached DNA into rabbit and human cells than HPV16 capsids. CRPV capsids were more efficient for rabbit cells but comparable to HPV58 capsids in human cells. In agreement with previous studies [6,15,16,17,18], we also demonstrated that the L2 protein plays a critical role that contributed to the efficiency of the delivery of attached DNA. Furthermore, we showed that PSVs with a papillomavirus genome showed better delivery of externally attached DNA when compared with PSVs with a plasmid DNA. Our findings suggest that the conformational structure of these particles potentially plays a role in the delivery of attached DNA.
2. Material and Methods
2.1. L1 VLPs, L1/L2 VLPs, and PSV Production and Validation
L1, L1/L2 VLPs, and pseudoviruses (PSVs) were produced in 293TT cells as reported previously with some modifications (Table 1) [19]. In brief, 293TT cells maintained in DMEM complemented with hygromycin were used for transfection. No antibodies were added to the medium during DNA transfection using Lipofectamine 2000 (Invitrogen). The transfected cells were split 1:2 and cultured in a medium with 50 units/mL penicillin/streptomycin. Transfected cells were harvested at 48 h post-transfection and allowed to mature at 37 °C overnight. DNA contamination was eliminated by adding Benzonase (Sigma) and plasmid-safe exonuclease (NEB) during OptiPrep gradient ultracentrifugation. A panel of plasmid DNA with different sizes, including the PCX expression vector (5.5 kb), the secreted alkaline phosphatase (5.5 kb, SEAP), and the CRPV genomic DNA (7.8 kb), were used for PSV production (23–25). All plasmid DNA was prepared by the QIAGEN maxiprep kit. For PSVs, 20 µg of plasmid DNA was co-transfected with L1 and L2 DNA of the relevant HPV type (HPV16 and HPV58) or CRPV. Hybrid HPV16L1/58L2, HPV58L1/16L2 VLPs, and corresponding PSVs were also produced. VLPs were produced by transfecting L1 or L1/L2 plasmids without any genomic DNA plasmids. All VLPs and PSVs were purified by OptiPrep ultracentrifugation and different fractions were examined by ELISA for both L1 (in house monoclonal antibodies, H16.V5, H58.G5.1, and CRPV.A4 were used to detect HPV16, HPV58, and CRPV L1, respectively) and L2 (rabbit polyclonal antibody R972 responding to the conserved region of HPV L2) (Table 1). Figure 1A shows representative HPV58 VLP L1 and L2 detection by ELISA. The fractions from VLPs and PSVs positive for both L1 and L2 were pooled (fraction #5–7 in this example) as the stock for experiments in the current study. The protein concentration for each preparation was determined by Bio-Rad protein analysis using different concentrations of BSA as the standard curve (Figure 1A). We prepared several batches of VLPs and PSVs for repeated experiments. VLP particles were also examined by TEM using negative staining, as reported previously (Figure 1B) [20]. All preparations were stored in OptiPrep at 4 °C for short-term storage or −20 °C for long-term storage.

Table 1.
Viral particles (VLPs) and pseudoviruses (PSVs) used in the current study.
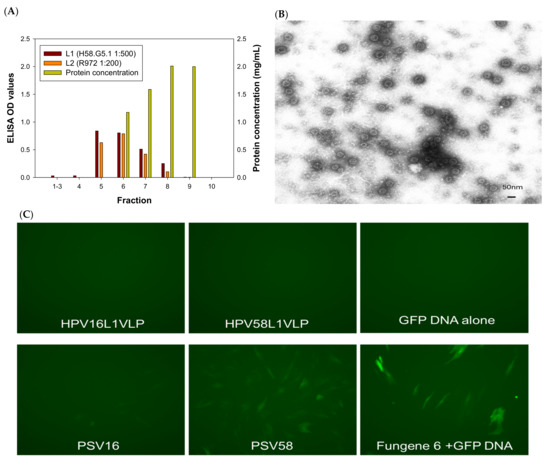
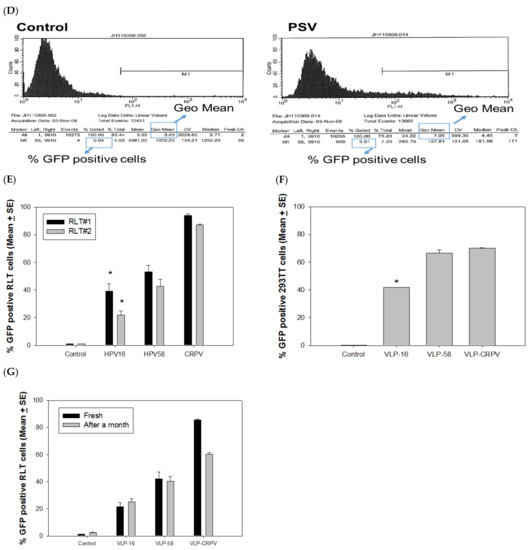
Figure 1.
(A) Using HPV58 VLPs as an example, L1 and L2 contents were determined by ELISA assay using type-specific antibodies against L1 (H58.G5) and a cross-reacting rabbit antibody against L2 (R972). The corresponding protein concentration in each fraction was also determined by Bio-Rad protein assay (right axis mg/mL). (B) VLPs visualized by TEM should be empty particles, but many particles were filled with cellular DNAs and appear full. (C) GFP expression in rabbit cells (RLT) by HPV 16 and 58 L1-only VLPs and PSVs. As shown in the top panel, HPV 16 and 58 L1-only VLPs failed to deliver attached GFP into cultured cells. By contrast, GFP-positive cells were found when 16-PSV and 58-PSV delivered attached GFP DNA. 58-PSV showed comparable GFP expression to the commercial transfection kit. (D) Flow cytometry analysis was used to quantify GFP signals. Two parameters, the percentage of GFP positive population and mean fluorescent intensity (MFI) by geometric mean were analyzed, as shown here in a typical dataset. These two parameters correlated with each other and thus were used interchangeably for comparison among groups. (E) Significantly fewer rabbit cells (cell lines from two different rabbits) were GFP-positive by VLP-16 delivery when compared to HPV58- and CRPV-VLPs (p < 0.05, unpaired Mann Whitney test, indicated by *). (F) We tested delivery of GFP DNA by HPV58-, 16-, and CRPV-VLPs in 293TT cells. CRPV-VLPs showed comparative efficiency when compared to 58-VLP (p > 0.05, unpaired Mann Whitney test). The 16-VLPs showed significantly lower efficacy to deliver DNA when compared with both HPV 58- and CRPV-VLPs (p < 0.05, unpaired Mann Whitney test, indicated by *). (G) To determine the stability of VLPs and PSVs, we tested fresh VLPs with those stored at −20 °C for a month. We observed that CRPV-VLP delivery efficiency decreased, while HPV16- and 58-VLPs maintained the same level of potency for delivery.
2.2. Cells for Externally Attached DNA Delivery
Rabbit cells (primary cultures from two individual rabbits or SV40LT immortalized cell lines based on the primary cultures designated as RLT, and an inbred rabbit cancer cell line, I3) and human cell lines (293TT cells and SiHa cells) were maintained in DMEM. Mouse dendritic cells (mDCs, cultured from previously harvested mouse bone marrow cells as described previously [21,22]) were maintained in RPMI1640. Both media were supplemented with 10% FBS, 50 units/mL penicillin/streptomycin, 2 mM glutamine, and 1 mM Na pyruvate. Cells were cultured at 37 °C with 5% CO2. All cell lines were seeded at a density of 3 × 105/well into 24-well plates and tested after reaching 70–80% confluence.
2.3. External DNA Delivery Using VLPs and PSVs in In Vitro Cell Cultures
Green fluorescent protein (GFP) DNA was cloned into a mammalian expression vector PCR3 (Invitrogen) and confirmed by DNA sequencing at the Core Facility of Penn State College of Medicine. Different amounts of VLPs or PSVs were incubated with different amounts of GFP DNA at room temperature for 5 min for the delivery of attached DNA. Triplicate wells in a 24-well plate were used for each condition. The mixtures were then added to the cells and cultured for 48–72 h. The cells were subsequently harvested and fixed in 1 × PBS with 2% paraformaldehyde for flow cytometry using one-color (FITC) analysis.
2.4. Fluorescent Detection and Quantification of Delivery Efficacy Using Flow Cytometry Analysis
The GFP-positive cells were visualized under a fluorescent microscope, and a representative image is shown in Figure 1C at 48–72 h post-transfection. The cells were also collected and fixed with 2% paraformaldehyde for further analysis by flow cytometry at the Core Facility of Pennsylvania State University College of Medicine. To quantify the efficacy of DNA delivery, we measured two parameters: the percentage of GFP positive population and mean fluorescent intensity (MFI) using the geometric mean. An example dataset is shown (Figure 1D). These two parameters correlated with each other and thus were used interchangeably for our comparison among groups in the following experiments. FlowJo software was used for single-color data analysis.
2.5. VLP Delivery of Viral DNA for In Vivo Infection of Rabbits
All work on rabbits was approved by the Institutional Animal Care and Use Committee of the Pennsylvania State University College of Medicine (PSUCOM), and all procedures were performed in strict accordance with guidelines and regulations. Two New Zealand White Rabbits were used to test the delivery of DNA with CRPV-VLPs in vivo. We used several low doses (10, 20, and 40 ng) of CRPV DNA alone or DNA mixed with 20, 40, and 80 ng of CRPV-VLPs and delivered the DNA onto the pre-wounded back skin sites of rabbits as described previously [23]. Tumor outgrowth was monitored and recorded in millimeters as length × width × height. A geometric mean was calculated for each tumor. The tumor appearance (tumor sites/infected sites) of each combination was recorded.
2.6. Statistics
Mean fluorescent intensity (MFI) and the geo mean and percentage (%) of GFP positive cells were compared among different groups for each experiment according to types of VLPs and PSVs. Data were entered into SigmaPlot, and means and standard errors (SEM) for each test group were calculated. Plots were generated using SigmaPlot. Unpaired Mann Whitney tests were used to identify the significance (p < 0.05 was considered a significant difference between samples).
3. Results
3.1. L2 Is Required for Delivering Externally Attached DNA to the Cells In Vitro
Previous studies demonstrated that HPV16 L1 VLPs failed to carry attached DNA into human cells [13]. To investigate whether this was due to a papillomavirus type-specific characteristic, we tested 16- and 58-L1 VLPs by mixing the corresponding L1 VLPs with a GFP-expressing DNA and incubating them at room temperature for 5 min before adding the mixtures to RLT cells [12,24,25]. GFP expression in RLT cells was examined by fluorescence microscopy 48 h later. Consistent with previous findings for 16 L1-VLPs, no GFP expression was found from any of the L1 VLPs tested (Figure 1C). By contrast, PSV16 and PSV58 showed GFP-positive populations. PSV58 showed similar GFP expression when compared to GFP DNA transfection by the transfection reagent FuGENE 6. More GFP-positive cells were found in the PSV58 and FuGENE 6 control group when compared with the PSV16 group (Figure 1C, bottom panel). To quantify the GFP-positive cells, we harvested cells for flow cytometry, as shown in Figure 1D. We used both the percentage of GFP-positive (an arbitrary measure set for all samples in one test) and the Geo Mean for the mean fluorescent intensity (MFI) to compare groups. These two parameters correlated with each other, and both were used in the analysis interchangeably. All capsids used in subsequent studies contained L2, and thus 16-VLP indicates L1/L2 VLPs and all delivery tests were externally attached GFP DNA.
3.2. PSVs and VLPs Deliver Attached DNA to Both Rabbit and Human Cells
The N-terminus of HPV16 L2 interacts with HPV16 DNA [7], and the N-terminus of BPV-1 L2 also displays on the surface of the BPV-1 capsid [16]. These observations suggest that the N-terminus of L2 on the capsid surface could interact with attached DNA, leading to DNA delivery to cells [13]. Our preliminary study demonstrated that HPV58 capsids containing the cottontail rabbit papillomavirus (CRPV) DNA genome showed higher infectivity when compared with corresponding HPV16 capsids (unpublished observations). We hypothesize that 58-VLPs may be more efficient than 16-VLPs to deliver attached DNA in vitro and in vivo.
To compare the delivery efficiency of different capsids, we incubated 1 µg of 16-, 58- and CRPV-VLPs with 0.5 µg of GFP-expressing DNA. The mixtures were then added to two SV40 LT immortalized rabbit cell lines (RLT#1 and RLT#2). All three VLP types successfully delivered GFP DNA into the rabbit cells as determined by a GF- positive population (Figure 1E). Interestingly, CRPV-VLPs showed the highest delivery efficiency, and a significant difference was also found between 58- and 16-VLPs, with 16-VLPs delivering the least amount of DNA (Figure 1E, p < 0.05, unpaired Mann Whitney test). Therefore, 58-VLPs are more efficient than 16-VLPs for the delivery of attached DNA. The results suggest that VLPs could be used as vectors to deliver attached DNA to cells efficiently and that the delivery efficiency is papillomavirus type-dependent.
Our studies described above were performed using rabbit cells, and therefore, we questioned whether these different VLPs displayed a similar pattern in human cells. We used 293TT cells to test delivery by adding GFP DNA attached to 58-, 16-, and CRPV-VLPs (the same batch of VLPs we used for the above experiment, as shown in Figure 1E). Three days after, the cells were tested for GFP expression. CRPV-VLPs showed comparable efficiency with 58-VLPs in 293TT cells (Figure 1F). In both rabbit and 293TT cells, 16-VLPs were the least efficient for DNA delivery (Figure 1E,F, p < 0.05, unpaired Mann Whitney test). Therefore, 58-VLPs are superior to 16-VLPs for the delivery of attached DNA in both human and rabbit cells.
All particles were stored in OptiPrep at 4 °C for short-term storage or −20 °C for long-term storage. We further tested the stability of these VLPs after storage for a month at −20 °C. We conducted the delivery experiment in the same rabbit cell line in which we tested the fresh VLP stocks above. As shown in Figure 1G, CRPV-VLPs showed slightly decreased delivery efficiency, while both 16- and 58-VLPs maintained the same potency.
3.3. PSVs Were More Efficient in Delivering DNA When Encapisidating a Papillomavirus Genome DNA
PSV58 encapsidating an unrelated PCX plasmid (5.5 kb) (designated as PSV58/5.5 kb) showed improved delivery of attached DNA when compared to 58-VLPs (Figure 2A, p < 0.05, unpaired Mann Whitney test) in the rabbit cells. The signal was comparable with CRPV-VLPs. This suggests that PSV58 was more efficient than corresponding VLPs for delivering external DNA. The experiment was repeated to compare PSV58 with the commercial transfection reagent FuGENE 6. We detected significantly higher MFI in the group delivered by PSV58 when compared to those transfected by FuGENE 6 (Figure 2B, p < 0.05, unpaired Mann Whitney test). This finding suggests that PSV58 could be used as a vehicle for DNA transfection in vitro.
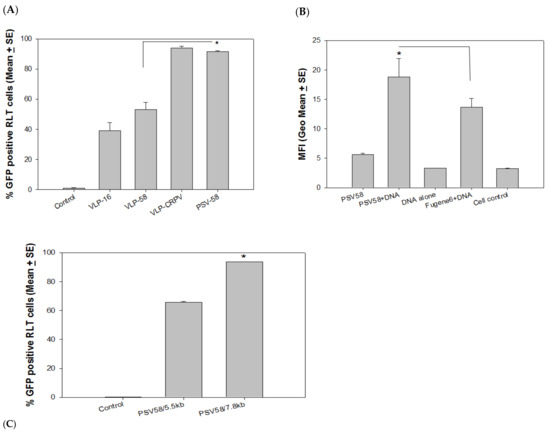
Figure 2.
(A) PSV58 encapsidating an unrelated 5.5 kb DNA plasmid (PSV58/5.5 kb) showed improved delivery over VLP-58 (p < 0.05, unpaired Mann Whitney test, indicated by *) that was also comparable to VLP-CRPV. (B) External GFP DNA delivered by PSV58 was more efficient than a commercial transfection reagent (p < 0.05, unpaired Mann Whitney test, indicated by *). No difference was found between PSV58 alone, GFP DNA alone, and medium control (p > 0.05, unpaired Mann Whitney test). (C) Efficiency of external DNA delivery is significantly higher by PSV58 containing the CRPV genome when compared to PSV58/5.5 kb (PCX expression vector, 5.5 kb) genome (p < 0.05, unpaired Mann Whitney test, indicated by *).
Papillomavirus genomes are preferred to be packed inside papillomavirus capsids in vitro [26,27]. To determine whether including a papillomavirus genome would further improve the delivery efficiency of attached DNA, we produced PSVs encapsidating the CRPV genome (identified as PSV58/7.8 kb) to compare with PSV58/5.5 kb in rabbit cell cultures. Significantly more GFP-expressing cells were found to be delivered by PSV58/7.8 kb when compared with those delivered by PSV58/5.5 kb (Figure 2C, p < 0.05, unpaired Mann Whitney test). Therefore, PSVs with a papillomavirus genome DNA appear to deliver attached DNA more efficiently than particles encapsidating a plasmid DNA (5.5 kb). We also tested a panel of PSV58 encapsidating different sizes of plasmid DNA but did not observe increased delivery of external DNA with an increase in DNA size (data not shown). This may indicate that PSVs containing the papillomavirus genome improve external DNA binding and delivery, as the size of encapsidated DNA alone is not necessarily correlated with improved delivery.
4. Optimal VLP/External DNA Ratio for Delivery
Since we observed that VLPs and PSVs are promising delivery vehicles for external DNA, we sought to determine the optimal ratio between capsid and GFP DNA. For this experiment, we chose PSV58 to optimize the conditions, as this PSV58 was more efficient than PSV16 for external DNA delivery. We first determined the highest amount of PSV needed for the delivery of 0.5 µg GFP DNA and identified that 1 µg PSV58 achieved close to saturation levels of expression in a 24-well plate (Figure 3A, p < 0.05, unpaired Mann Whitney test). Next, we mixed 1 µg of PSV58 with 0, 0.5, 1, or 2 µg of GFP DNA (a ratio of 2:1, 1:1, or 1:2) and tested the different combinations in RLT cultures. An increased amount of GFP DNA did not show increased delivery efficiency (Figure 3B, p > 0.05, unpaired Mann Whitney test). The capsid/DNA ratio of 2:1 induced the strongest GFP expression when compared with the other ratios. The following experiments were conducted with the optimal 2:1 VLPs/DNA ratio.
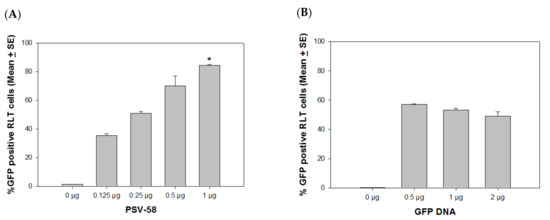
Figure 3.
Dose curve to determine the optimal capsid/DNA ratio. (A) Different amounts of PSV58 (0.125, 0.25, 0.5, and 1 µg) were mixed with 0.5 µg of GFP DNA and added to SV40 LT immortalized rabbit cells (RLT). The 1 µg PSV-58 group showed increased numbers of positive cells when compared with the 0.125 and 0.25 µg groups (p < 0.05, unpaired Mann Whitney test, indicated by *) (B) A range of GFP DNA was delivered by 1 µg of PSV58 to test for optimum delivery. An increased amount of GFP DNA did not show increased delivery efficiency; 1 µg PSV58 delivering 0.5 µg GFP DNA was the most efficient ratio for external DNA delivery.
4.1. PSVs Deliver DNA More Efficiently in Immortalized Cells vs. Normal Cells
VLPs delivering external DNA could be a promising future gene therapy for the treatment of different diseases. To examine whether the capsids have advantages in delivering GFP DNA to immortalized cells compared to primary cells, we compared the efficiency of PSV16, PSV58, and PSVCRPV to deliver external GFP DNA to both primary and immortalized rabbit cells with the same method described above. Again, PSV58 was more efficient in delivering GFP DNA in both normal and transformed cells when compared with PSV16. Most interestingly, we observed that significantly higher levels of GFP were found in immortalized cells (RLT) when compared with primary cells for both PSV58 and PSV16 (Figure 4A, p < 0.05, unpaired Mann Whitney test), and again, PSV58 delivered more GFP when compared with PSV16. We also examined whether these PSVs could deliver external DNA to antigen-presenting cells and used cultured mouse dendritic cells for this experiment. Although the delivery efficiency of all VLPs was much lower in DCs when compared with RLT or 293TT cells, we did find a similar pattern as seen in the primary rabbit cells in that PSV58 was more efficient than PSV16 (Figure 4B, p < 0.05, unpaired Mann Whitney test).
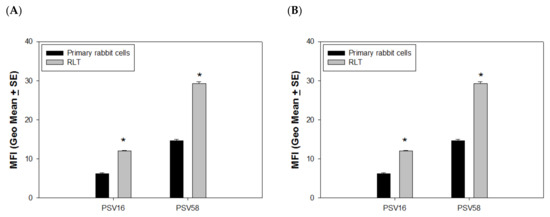
Figure 4.
Delivery efficacy was determined by cell status. (A) Higher delivery efficacy was found in immortalized cells when compared to primary cells (p < 0.05, unpaired Mann Whitney test, indicated by *). (B) Significantly higher delivery efficacy was found in PSV58 when compared with PSV16 (p < 0.05, unpaired Mann Whitney test, indicated by *) in mouse dendritic cells.
4.2. L2 Played a Critical Role in Delivery Efficiency
The previous experiments demonstrated that L1-only VLPs failed to deliver external DNA, but VLP-58 was advantageous when compared to VLP-16 in delivering external DNA in both rabbit and human cells. Therefore, we hypothesized that L2 played a critical role in this delivery method. To test this hypothesis, we produced two hybrid VLPs (HPV58L1/16L2 and HPV16L1/58L2). These two hybrid VLPs were tested for the delivery of external GFP DNA to RLT cells. Hybrid VLP-16L1/58L2 showed significantly higher delivery efficiency when compared with VLP-58L1/16L2 (Figure 5A, p < 0.05, unpaired Mann Whitney test), although the delivery of both was significantly lower than that of VLP-58. We also tested these hybrid VLPs in two immortalized cell lines, including rabbit (I3) and human (SiHa) cell lines. Consistent with what is shown in Figure 5A, hybrid VLP-16L1/58L2 showed significantly higher delivery efficiency when compared with VLP-58L1/16L2 (Figure 5B, p < 0.05, unpaired Mann Whitney test) in both human and rabbit cells. Our results suggest that the L1/L2 VLP delivery efficiency is potentially an L2-dependent phenomenon.
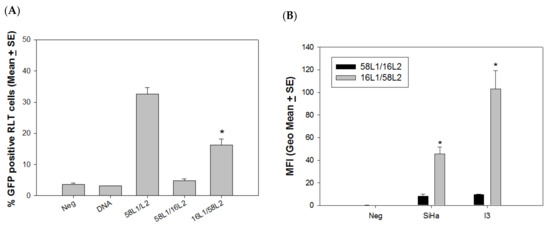
Figure 5.
Delivery efficacy of external DNA is L2-dependent. Two hybrid VLPs, 16L1/58L2 and 58L1/16L2, were tested for attached DNA delivery with GFP DNA in rabbit and human cells. (A) The delivery efficiency of hybrid VLP-16L1/58L2 was partially restored in RLT cells when compared with VLP-58L1/16L2. Significantly more cells expressed GFP when delivered by VLP-16L1/58L2 (similar to VLP-58) when compared to VLP-58L1/16L2 (similar to VLP-16) in rabbit cells (p < 0.05, unpaired Mann Whitney test, indicated by *). (B) Additionally, two more cell cultures (a rabbit cell line (I3) and a human cell line (SiHa)) were tested for hybrid VLP delivery. A similar pattern was found, as shown in RLT cells in the above experiments (p < 0.05, unpaired Mann Whitney test, indicated by *).
4.3. VLP-CRPV Increased Viral Infectivity In Vivo
To further determine whether VLP-CRPV could deliver DNA in vivo, we tested different low doses of CRPV DNA with and without VLPs for delivery and infectivity in a rabbit model. VLP-CRPV was mixed in a 2:1 ratio with 10, 20, and 40 ng of CRPV DNA and infected at two pre-wounded back skin sites of New Zealand White Rabbits as described previously [23]. The rabbits were monitored for tumor growth and recorded by photography. As shown in Table 2, more tumors were observed in groups where the DNA was delivered by VLPs compared to the DNA alone group. This effect was greater in the groups with a lower dose of DNA. However, the difference in the groups was not significant because of the small sample size (p > 0.05, unpaired Mann Whitney test).

Table 2.
CRPVL1/L2 VLPs increase the infectivity of DNA infection in vivo.
5. Discussion
Previous studies have demonstrated that HPV capsids assembled as pseudoviruses can effectively deliver encapsidated DNA into cells and are considered promising vehicles for gene therapy and DNA vaccination because HPV VLPs and PSVs can be taken up by different cell types [7,28,29,30]. More recently, L1/L2 VLPs but not L1 VLPs have been found to effectively deliver externally attached DNA in vitro and in vivo [13]. HPV16 capsids were used for most DNA delivery studies. Our previous unpublished and published studies suggested that HPV58 L1/L2 encapsidating CRPV DNA was more infectious than HPV16 L1/L2 encapsidating CRPV DNA [14]. In the current study, we tested our hypothesis that different HPVs may have diverse inherent efficiencies to deliver attached DNA to the cells using VLPs or PSVs (containing a plasmid DNA). We tested HPV16, HPV58, and CRPV and a variety of cell lines, including rabbit, human, and mouse dendritic cells. Consistent with previous findings, L1-only VLPs from all PV types failed to deliver attached DNA into cells, indicating that L1 alone is not sufficient. By contrast, all L1/L2 VLPs and PSVs tested successfully delivered externally attached DNA into both rabbit and human cells despite the difference in delivery efficiencies. VLP-58 showed significantly higher delivery efficacy when compared to VLP-16. Interestingly, VLP-CRPV showed higher delivery to rabbit cells, but the delivery was comparable to 58-VLPs in human cells. In addition, the delivery efficacy of attached DNA was L2-dependent because hybrid VLP-16L1/58L2 showed increased efficacy when compared to hybrid VLP-58 L1/16L2 in both rabbit and human cells.
Both L1 and L2 contain DNA binding sites [6,7,15,31]. At the C-terminus, nine amino acids have been demonstrated essential for DNA binding and gene delivery by L1 VLPs [32]. Unrelated DNA can be encapsidated into L1-only particles by reassembly in vitro and can thus be delivered into cells [33,34]. The efficacy of this delivery has been characterized in vitro and in vivo [5,28,35,36]. Although L1 can successfully encapsidate DNA and form infectious PSVs, it failed to bind external DNA in previous studies and in our studies, indicating that the DNA binding sites of L1 might be embedded inside the particles [13]. The L2 protein has been found to be critical for the infectivity of PSVs [37]. Several DNA binding sites have been identified in HPV L2. Three conserved regions that are important for neutralizing and cross-neutralizing antibody targets have been identified in L2: (1) an N-terminal external loop of L2 (L2/17-36aa) has been identified as the target of neutralizing and cross-neutralizing antibodies [18,38]; (2) an N-terminal region (L2/108-120aa) has been identified as a surface epitope that shares a common neutralizing epitope for HPVs [37,39,40]; and (3) a C-terminal region of L2 has a lysine–arginine-rich sequence (RKRRKR in HPV16 L2). This latter region is also identified as an important L1-L2 interaction region [6,41]. Higher efficacy in delivering externally attached DNA by HPV58 L2 compared to HPV16 L2 implicates that one of these conserved regions of L2 might play a critical role [42]. A 23 amino acid membrane-destabilizing L2 peptide located at the C-terminus of L2 has been found to be critical for external DNA binding [8,13]. It would be interesting to identify the critical DNA binding sites in this region for HPV58. We found that the L1 and L2 interaction between hybrid HPV16 and HPV58 L1/L2 VLPs was not interrupted in terms of capsid formation and encapsidating DNA. The compatibility of L1 and L2 between HPV16 and HPV18 in encapsidating DNA genomes has been reported previously [43], as these capsids contain highly conserved regions among the HPV L2 proteins. We postulate that L2, rather than the interaction between L1 and L2, may play a predominant role in this externally attached DNA delivery because the two hybrid VLPs HPV16/HPV58 were both capable of delivering DNA to cells. The identification of the nucleus-guided sequences from L2 would also be valuable for the design of future DNA carriers for gene therapy.
The difference in DNA binding between VLPs and PSVs has not been studied extensively. In this study, we demonstrated PSVs displayed higher delivery of attached DNA when compared with corresponding VLPs, while PSVs containing papillomavirus DNA were superior to PSVs containing 5.5 kb DNA in delivering attached DNA. This suggests a possible conformation structure difference among particle types, which may play a role in the external binding of DNA. In the current study, we know both VLPs and PSVs could potentially contain cellular DNA from production in the 293TT culture system. However, we argue that PSVs have more particles containing the target DNA, leading to a more rigid structure when compared with the less “filled” capsids (VLPs), and this may contribute to the higher binding/delivery [20]. How different sizes of encapsidated DNA influences the delivery efficacy of attached DNA needs further investigation.
Another interesting finding is that much higher DNA delivery was detected by externally attached DNA vs. the delivery of genome DNA encapsidating as PSVs (data not shown). Our unpublished study showed that only one copy of full-length DNA can be encapsidated because of the restriction of the space inside the capsid. For externally attached DNA delivery, excess DNA was incubated with the VLPs or PSVs, and therefore, significantly higher amounts of DNA could be attached to the particles. The mechanism by which attached DNA is carried into the cells without being damaged by nucleases is unclear. Our recently published study demonstrated that the papillomavirus genome could be delivered intravenously into the circulatory system and generate local lesions in the rabbit model [44]. We hypothesize that VLPs and PSVs may have provided some protection for externally attached DNA from nuclease digestion.
A previous study showed that a ratio of 1:5 between VLP-16: GFP DNA attained the highest level of GFP expression in animals 7 days after intradermal injection [13]. In our in vitro culture studies, we tested different doses of GFP DNA and identified a saturating dose that led to high expression in RLT cells. We further tested VLP-58 and GFP DNA for optimal delivery and demonstrated that a ratio of 2:1 was optimal for in vitro delivery. We used this ratio for the VLP-CRPV/CRPV genome for in vivo delivery. Whether the difference in our findings vs. those of the previous study resulted from the differences between in vitro (cell types, the length of incubation time, etc.) and in vivo systems (gene delivery vs. infection-induced tumor growth) needs further investigation.
6. Conclusions
In conclusion, our study demonstrated that HPV58 and CRPV capsids are advantageous to HPV16 for delivering attached DNA, and the delivery efficacy is L2-dependent. This finding agrees with our previous observation that the HPV58 capsid delivered attached DNA more efficiently than HPV16. The HPV L1/L2 VLPs/externally attached DNA method can be applied to both in vitro transfection studies (because comparative transfection efficiencies were found between VLPs and the commercial transfection reagents) and in vivo viral DNA infection, as higher and more consistent infectivity could be achieved. Our findings further demonstrated that the delivery of attached DNA by HPV VLPs was not species-specific, and delivery was preferential in immortalized cells compared to normal cells. Chimeric HPV VLPs have been tested for their function in activating antigen-presenting cells, such as Langerhans and dendritic cells, in vitro and in vivo [13,45,46,47,48,49]. These characteristics can be potentially used for therapeutic DNA vaccination and tumor treatment using different HPV capsids.
Author Contributions
Conceptualization, S.B., N.C. (Nancy Cladel), N.C. (Neil Christensen), S.A., C.M. and J.H.; methodology, S.B., N.C. (Nancy Cladel), K.B., J.H.; validation, S.B., N.C. (Nancy Cladel), K.B. and J.H.; formal analysis, S.B. and J.H.; investigation, S.B., N.C. (Nancy Cladel), K.B. and J.H.; resources, N.C. (Nancy Cladel) and J.H.; data curation, S.B., N.C. (Nancy Cladel), K.B. and J.H.; writing—original draft preparation, S.B., N.C. (Nancy Cladel), N.C. (Neil Christensen) and J.H.; writing—review and editing, S.B., S.A., C.M. and J.H.; visualization, S.B., N.C. (Nancy Cladel), K.B, and J.H.; supervision, J.H., N.C. (Neil Christensen), and C.M.; project administration, J.H.; funding acquisition, J.H. and N.C. (Neil Christensen). All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Cancer Institute grant R01 CA47622 from the National Institutes of Health to NDC and the Jake Gittlen Memorial Golf Tournament.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, J.W.; Roden, R.B. L2, the minor capsid protein of papillomavirus. Virology 2013, 445, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Buck, C.B.; Day, P.M.; Trus, B.L. The papillomavirus major capsid protein L1. Virology 2013, 445, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Schiller, J.T.; Lowy, D.R. Papillomavirus-like particle vaccines. J. Natl. Cancer Inst. Monogr. 2001, 2000, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Dochez, C.; Bogers, J.J.; Verhelst, R.; Rees, H. HPV vaccines to prevent cervical cancer and genital warts: An update. Vaccine 2014, 32, 1595–1601. [Google Scholar] [CrossRef]
- Peng, S.; Frazer, I.H.; Fernando, G.J.; Zhou, J. Papillomavirus virus-like particles can deliver defined CTL epitopes to the MHC class I pathway. Virology 1998, 240, 147–157. [Google Scholar] [CrossRef]
- Zhou, J.; Sun, X.-Y.; Louis, K.; Frazer, I.H. Interaction of human papillomavirus (HPV) type 16 capsid proteins with HPV DNA requires an intact L2 N-terminal sequence. J. Virol. 1994, 68, 619–625. [Google Scholar] [CrossRef]
- Müller, M.; Gissmann, L.; Cristiano, R.J.; Sun, X.-Y.; Frazer, I.H.; Jenson, A.B.; Alonso, A.; Zentgraf, H.; Zhou, J. Papillomavirus capsid binding and uptake by cells from different tissues and species. J. Virol. 1995, 69, 948–954. [Google Scholar] [CrossRef]
- Kamper, N.; Day, P.M.; Nowak, T.; Selinka, H.C.; Florin, L.; Bolscher, J.; Hilbig, L.; Schiller, J.T.; Sapp, M. A membrane-destabilizing peptide in capsid protein L2 is required for egress of papillomavirus genomes from endosomes. J. Virol. 2006, 80, 759–768. [Google Scholar] [CrossRef]
- Handisurya, A.; Day, P.M.; Thompson, C.D.; Buck, C.B.; Kwak, K.; Roden, R.B.; Lowy, D.R.; Schiller, J.T. Murine skin and vaginal mucosa are similarly susceptible to infection by pseudovirions of different papillomavirus classifications and species. Virology 2012, 433, 385–394. [Google Scholar] [CrossRef]
- Kines, R.C.; Zarnitsyn, V.; Johnson, T.R.; Pang, Y.Y.; Corbett, K.S.; Nicewonger, J.D.; Gangopadhyay, A.; Chen, M.; Liu, J.; Prausnitz, M.R.; et al. Vaccination with human papillomavirus pseudovirus-encapsidated plasmids targeted to skin using microneedles. PLoS ONE 2015, 10, e0120797. [Google Scholar] [CrossRef]
- Cuburu, N.; Graham, B.S.; Buck, C.B.; Kines, R.C.; Pang, Y.Y.; Day, P.M.; Lowy, D.R.; Schiller, J.T. Intravaginal immunization with HPV vectors induces tissue-resident CD8+ T cell responses. J. Clin. Invest. 2012, 122, 4606–4620. [Google Scholar] [CrossRef]
- Mitsunaga, M.; Kosaka, N.; Kines, R.C.; Roberts, J.N.; Lowy, D.R.; Schiller, J.T.; Ishihara, Y.; Hasegawa, A.; Choyke, P.L.; Kobayashi, H. In vivo longitudinal imaging of experimental human papillomavirus infection in mice with a multicolor fluorescence mini-endoscopy system. Cancer Prev. Res. 2011, 4, 767–773. [Google Scholar] [CrossRef]
- Malboeuf, C.M.; Simon, D.A.; Lee, Y.E.; Lankes, H.A.; Dewhurst, S.; Frelinger, J.G.; Rose, R.C. Human papillomavirus-like particles mediate functional delivery of plasmid DNA to antigen presenting cells in vivo. Vaccine 2007, 25, 3270–3276. [Google Scholar] [CrossRef]
- Broutian, T.R.; Brendle, S.A.; Christensen, N.D. Differential binding patterns to host cells associated with particles of several human alphapapillomavirus types. J. Gen Virol. 2010, 91 Pt 2, 531–540. [Google Scholar] [CrossRef]
- Touze, A.; Coursaget, P. In vitro gene transfer using human papillomavirus-like particles. Nucleic Acids Res. 1998, 26, 1317–1323. [Google Scholar] [CrossRef]
- Liu, W.J.; Gissmann, L.; Sun, X.Y.; Kanjanahaluethai, A.; Muller, M.; Doorbar, J.; Zhou, J. Sequence close to the N-terminus of L2 protein is displayed on the surface of bovine papillomavirus type 1 virions. Virology 1997, 227, 474. [Google Scholar] [CrossRef]
- Holmgren, S.C.; Patterson, N.A.; Ozbun, M.A.; Lambert, P.F. The minor capsid protein L2 contributes to two steps in the human papillomavirus type 31 life cycle. J. Virol. 2005, 79, 3938–3948. [Google Scholar] [CrossRef]
- Campos, S.K.; Ozbun, M.A. Two highly conserved cysteine residues in HPV16 L2 form an intramolecular disulfide bond and are critical for infectivity in human keratinocytes. PLoS ONE 2009, 4, e4463. [Google Scholar] [CrossRef]
- Culp, T.D.; Cladel, N.M.; Balogh, K.K.; Budgeon, L.R.; Mejia, A.F.; Christensen, N.D. Papillomavirus particles assembled in 293TT cells are infectious in vivo. J. Virol. 2006, 80, 11381–11384. [Google Scholar] [CrossRef]
- Goetschius, D.J.; Hartmann, S.R.; Subramanian, S.; Bator, C.M.; Christensen, N.D.; Hafenstein, S.L. High resolution cryo EM analysis of HPV16 identifies minor structural protein L2 and describes capsid flexibility. Sci. Rep. 2021, 11, 3498. [Google Scholar] [CrossRef]
- Schell, T.D.; Lippolis, J.D.; Tevethia, S.S. Cytotoxic T lymphocytes from HLA-A2.1 transgenic mice define a potential human epitope from simian virus 40 large T antigen. Cancer Res. 2001, 61, 873–879. [Google Scholar]
- Hu, J.; Peng, X.; Schell, T.D.; Budgeon, L.R.; Cladel, N.M.; Christensen, N.D. An HLA-A2.1-Transgenic Rabbit Model to Study Immunity to Papillomavirus Infection. J. Immunol. 2006, 177, 8037–8045. [Google Scholar] [CrossRef]
- Cladel, N.M.; Hu, J.; Balogh, K.; Mejia, A.; Christensen, N.D. Wounding prior to challenge substantially improves infectivity of cottontail rabbit papillomavirus and allows for standardization of infection. J. Virol. Methods 2008, 148, 34–39. [Google Scholar] [CrossRef][Green Version]
- Roden, R.B.; Kirnbauer, R.; Jenson, A.B.; Lowy, D.R.; Schiller, J.T. Interaction of papillomaviruses with the cell surface. J. Virol. 1994, 68, 7260–7266. [Google Scholar] [CrossRef]
- Roberts, J.N.; Buck, C.B.; Thompson, C.D.; Kines, R.; Bernardo, M.; Choyke, P.L.; Lowy, D.R.; Schiller, J.T. Genital transmission of HPV in a mouse model is potentiated by nonoxynol-9 and inhibited by carrageenan. Nat. Med. 2007, 13, 857–861. [Google Scholar] [CrossRef]
- Zhao, K.N.; Sun, X.Y.; Frazer, I.H.; Zhou, J. DNA packaging by L1 and L2 capsid proteins of bovine papillomavirus type 1. Virology 1998, 243, 482–491. [Google Scholar] [CrossRef]
- Roden, R.B.; Greenstone, H.L.; Kirnbauer, R.; Booy, F.P.; Jessie, J.; Lowy, D.R.; Schiller, J.T. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J. Virol. 1996, 70, 5875–5883. [Google Scholar] [CrossRef]
- Shi, W.; Liu, J.; Huang, Y.; Qiao, L. Papillomavirus pseudovirus: A novel vaccine to induce mucosal and systemic cytotoxic T-lymphocyte responses. J. Virol. 2001, 75, 10139–10148. [Google Scholar] [CrossRef]
- Liu, W.J.; Zhao, K.N.; Gao, F.G.; Leggatt, G.R.; Fernando, G.J.; Frazer, I.H. Polynucleotide viral vaccines: Codon optimisation and ubiquitin conjugation enhances prophylactic and therapeutic efficacy. Vaccine 2001, 20, 862–869. [Google Scholar] [CrossRef]
- Ohlschlager, P.; Osen, W.; Dell, K.; Faath, S.; Garcea, R.L.; Jochmus, I.; Muller, M.; Pawlita, M.; Schafer, K.; Sehr, P.; et al. Human papillomavirus type 16 L1 capsomeres induce L1-specific cytotoxic T lymphocytes and tumor regression in C57BL/6 mice. J. Virol. 2003, 77, 4635–4645. [Google Scholar] [CrossRef]
- Sun, X.Y.; Frazer, I.; Müller, M.; Gissmann, L.; Zhou, J. Sequences required for the nuclear targeting and accumulation of human papillomavirus type 6B L2 protein. Virology 1995, 213, 321–327. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Touze, A.; Mahe, D.; El Mehdaoui, S.; Dupuy, C.; Combita-Rojas, A.L.; Bousarghin, L.; Sizaret, P.Y.; Coursaget, P. The nine C-terminal amino acids of the major capsid protein of the human papillomavirus type 16 are essential for DNA binding and gene transfer capacity. FEMS Microbiol. Lett. 2000, 189, 121–127. [Google Scholar] [CrossRef]
- Mach, H.; Volkin, D.B.; Troutman, R.D.; Wang, B.; Luo, Z.; Jansen, K.U.; Shi, L. Disassembly and reassembly of yeast-derived recombinant human papillomavirus virus-like particles (HPV VLPs). J. Pharm. Sci. 2006, 95, 2195–2206. [Google Scholar] [CrossRef] [PubMed]
- Garcea, R.L.; Gissmann, L. Virus-like particles as vaccines and vessels for the delivery of small molecules. Curr. Opin. Biotechnol. 2004, 15, 513–517. [Google Scholar] [CrossRef]
- Petry, H.; Goldmann, C.; Ast, O.; Luke, W. The use of virus-like particles for gene transfer. Curr. Opin. Mol. Ther. 2003, 5, 524–528. [Google Scholar]
- Schwarz, K.; Meijerink, E.; Speiser, D.E.; Tissot, A.C.; Cielens, I.; Renhof, R.; Dishlers, A.; Pumpens, P.; Bachmann, M.F. Efficient homologous prime-boost strategies for T cell vaccination based on virus-like particles. Eur. J. Immunol. 2005, 35, 816–821. [Google Scholar] [CrossRef]
- Yang, R.; Day, P.M.; Yutzy, W.H.; Lin, K.Y.; Hung, C.F.; Roden, R.B. Cell surface-binding motifs of L2 that facilitate papillomavirus infection. J. Virol. 2003, 77, 3531–3541. [Google Scholar] [CrossRef]
- Conway, M.J.; Alam, S.; Christensen, N.D.; Meyers, C. Overlapping and independent structural roles for human papillomavirus type 16 L2 conserved cysteines. Virology 2009, 393, 295–303. [Google Scholar] [CrossRef]
- Kawana, K.; Yoshikawa, H.; Taketani, Y.; Yoshiike, K.; Kanda, T. Common neutralization epitope in minor capsid protein L2 of human papillomavirus types 16 and 6. J. Virol. 1999, 73, 6188–6190. [Google Scholar] [CrossRef]
- Rubio, I.; Seitz, H.; Canali, E.; Sehr, P.; Bolchi, A.; Tommasino, M.; Ottonello, S.; Muller, M. The N-terminal region of the human papillomavirus L2 protein contains overlapping binding sites for neutralizing, cross-neutralizing and non-neutralizing antibodies. Virology 2011, 409, 348–359. [Google Scholar] [CrossRef]
- Lowe, J.; Panda, D.; Rose, S.; Jensen, T.; Hughes, W.A.; Tso, F.Y.; Angeletti, P.C. Evolutionary and structural analyses of alpha-papillomavirus capsid proteins yields novel insights into L2 structure and interaction with L1. Virol. J. 2008, 5, 150. [Google Scholar] [CrossRef]
- Buck, C.B.; Cheng, N.; Thompson, C.D.; Lowy, D.R.; Steven, A.C.; Schiller, J.T.; Trus, B.L. Arrangement of L2 within the papillomavirus capsid. J. Virol. 2008, 82, 5190–5197. [Google Scholar] [CrossRef]
- Meyers, C.; Bromberg-White, J.L.; Zhang, J.; Kaupas, M.E.; Bryan, J.T.; Lowe, R.S.; Jansen, K.U. Infectious virions produced from a human papillomavirus type 18/16 genomic DNA chimera. J. Virol. 2002, 76, 4723–4733. [Google Scholar] [CrossRef]
- Cladel, N.M.; Jiang, P.; Li, J.J.; Peng, X.; Cooper, T.K.; Majerciak, V.; Balogh, K.K.; Meyer, T.J.; Brendle, S.A.; Budgeon, L.R.; et al. Papillomavirus can be transmitted through the blood and produce infections in blood recipients: Evidence from two animal models. Emerg. Microbes Infect. 2019, 8, 1108–1121. [Google Scholar] [CrossRef]
- Greenstone, H.L.; Nieland, J.D.; de Visser, K.E.; De Bruijn, M.L.; Kirnbauer, R.; Roden, R.B.; Lowy, D.R.; Kast, W.M.; Schiller, J.T. Chimeric papillomavirus virus-like particles elicit antitumor immunity against the E7 oncoprotein in an HPV16 tumor model. Proc. Natl. Acad. Sci. USA 1998, 95, 1800–1805. [Google Scholar] [CrossRef]
- Fausch, S.C.; Da Silva, D.M.; Kast, W.M. Differential uptake and cross-presentation of human papillomavirus virus-like particles by dendritic cells and Langerhans cells. Cancer Res. 2003, 63, 3478–3482. [Google Scholar]
- Lenz, P.; Thompson, C.D.; Day, P.M.; Bacot, S.M.; Lowy, D.R.; Schiller, J.T. Interaction of papillomavirus virus-like particles with human myeloid antigen-presenting cells. Clin. Immunol. 2003, 106, 231–237. [Google Scholar] [CrossRef]
- Lenz, P.; Day, P.M.; Pang, Y.Y.; Frye, S.A.; Jensen, P.N.; Lowy, D.R.; Schiller, J.T. Papillomavirus-like particles induce acute activation of dendritic cells. J. Immunol. 2001, 166, 5346–5355. [Google Scholar] [CrossRef]
- Fausch, S.C.; Da Silva, D.M.; Kast, W.M. Heterologous papillomavirus virus-like particles and human papillomavirus virus-like particle immune complexes activate human Langerhans cells. Vaccine 2005, 23, 1720–1729. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).