Development of a SARS-CoV-2 Vaccine Candidate Using Plant-Based Manufacturing and a Tobacco Mosaic Virus-like Nano-Particle
Abstract
1. Introduction
2. Materials and Methods
2.1. Vaccine Construction
2.2. Transmission Electron Microscopy
2.3. Analytical Ultracentrifugation
2.4. In Vitro Binding Assays
2.5. Animal Immunization and Testing
2.6. IgG Detection by ELISA
2.7. IFNγ Detection by ELISpot
2.8. VaxArray Coronavirus SeroAssay
2.9. SARS-CoV-2 Neutralization Assays
2.10. Statistical Analysis
3. Results
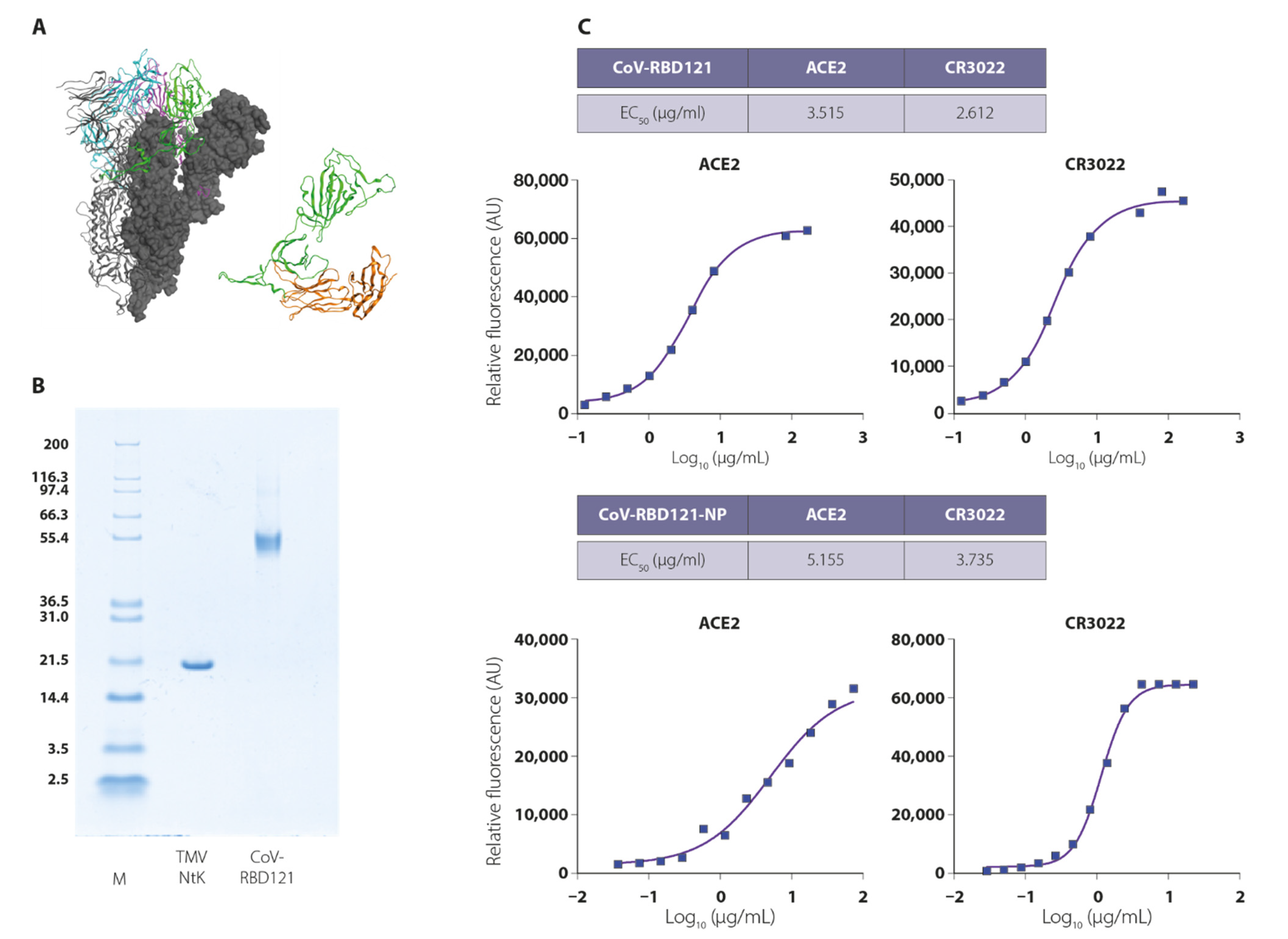
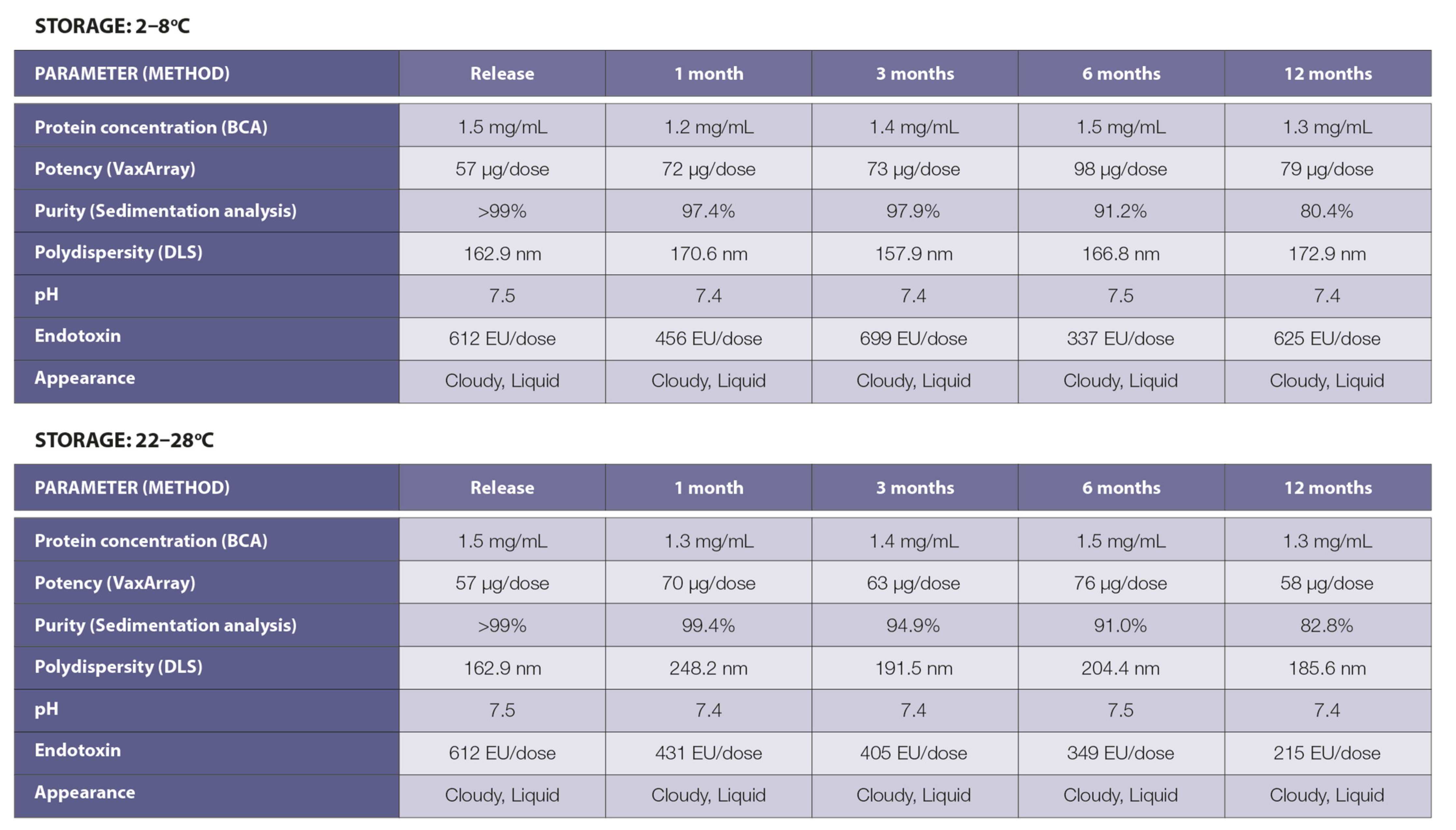
3.1. Vaccine Design, Construction, and Stability
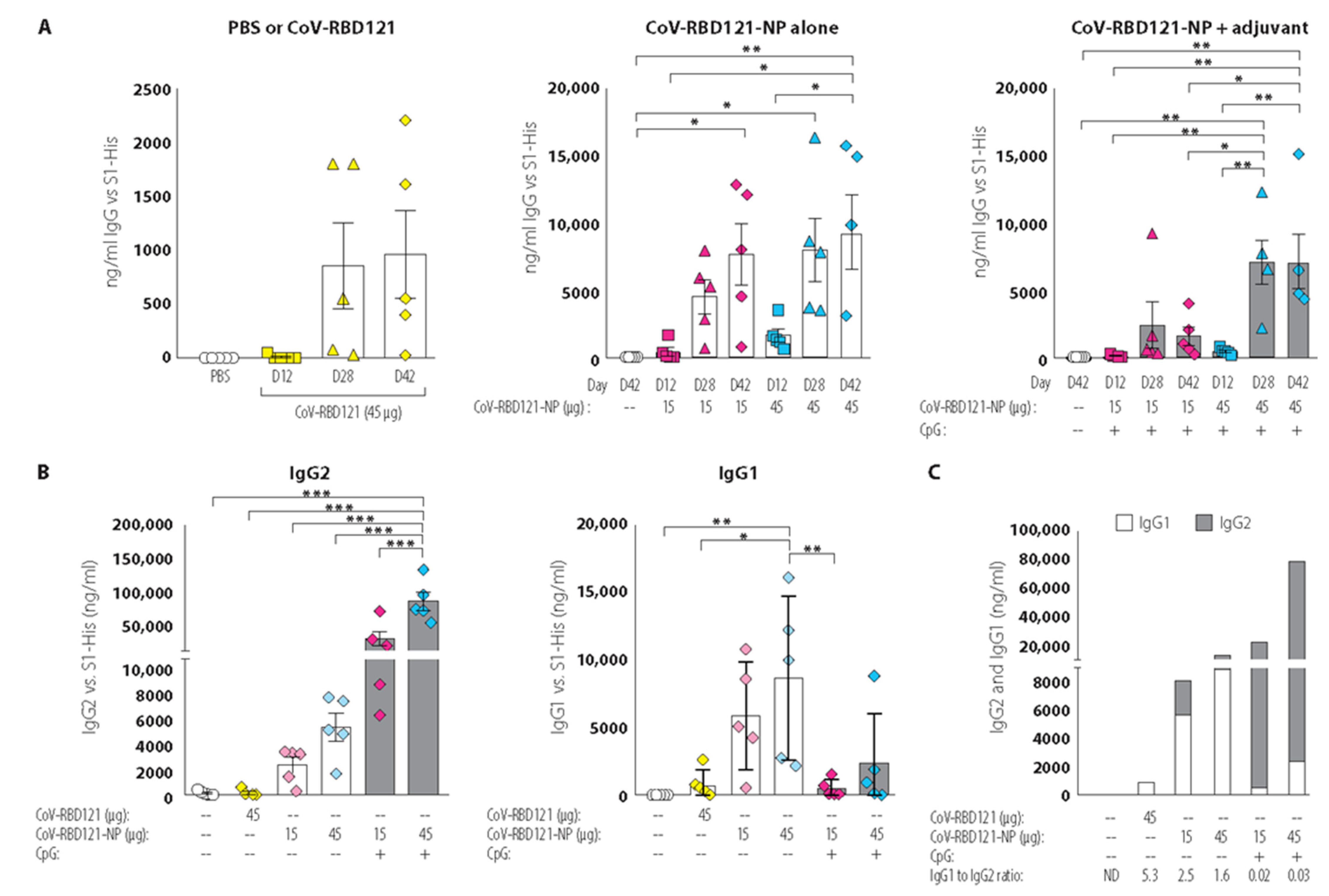
3.2. Vaccine Immunogenicity
3.3. Vaccine-Induced Neutralizing Antibody Titers
3.4. Reactivity with Diverse Coronavirus Spike Proteins
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus Biology and Replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Petrosillo, N.; Viceconte, G.; Ergonul, O.; Ippolito, G.; Petersen, E. COVID-19, SARS and MERS: Are They Closely Related? Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2020, 26, 729–734. [Google Scholar] [CrossRef]
- COVID-19 Map. Available online: https://coronavirus.jhu.edu/map.html (accessed on 13 June 2021).
- Andreano, E.; D’Oro, U.; Rappuoli, R.; Finco, O. Vaccine Evolution and Its Application to Fight Modern Threats. Front. Immunol. 2019, 10, 1722. [Google Scholar] [CrossRef]
- Krammer, F. SARS-CoV-2 Vaccines in Development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef]
- Gregory, A.E.; Titball, R.; Williamson, D. Vaccine Delivery Using Nanoparticles. Front. Cell. Infect. Microbiol. 2013, 3, 13. [Google Scholar] [CrossRef]
- Gheibi Hayat, S.M.; Darroudi, M. Nanovaccine: A Novel Approach in Immunization. J. Cell. Physiol. 2019, 234, 12530–12536. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Zha, L.; Cabral-Miranda, G.; Bachmann, M.F. Major Findings and Recent Advances in Virus-like Particle (VLP)-Based Vaccines. Semin. Immunol. 2017, 34, 123–132. [Google Scholar] [CrossRef]
- Marcandalli, J.; Fiala, B.; Ols, S.; Perotti, M.; de van der Schueren, W.; Snijder, J.; Hodge, E.; Benhaim, M.; Ravichandran, R.; Carter, L.; et al. Induction of Potent Neutralizing Antibody Responses by a Designed Protein Nanoparticle Vaccine for Respiratory Syncytial Virus. Cell 2019, 176, 1420–1431.e17. [Google Scholar] [CrossRef]
- Boyoglu-Barnum, S.; Ellis, D.; Gillespie, R.A.; Hutchinson, G.B.; Park, Y.-J.; Moin, S.M.; Acton, O.J.; Ravichandran, R.; Murphy, M.; Pettie, D.; et al. Quadrivalent Influenza Nanoparticle Vaccines Induce Broad Protection. Nature 2021, 592, 623–628. [Google Scholar] [CrossRef]
- Bruun, T.U.J.; Andersson, A.-M.C.; Draper, S.J.; Howarth, M. Engineering a Rugged Nanoscaffold To Enhance Plug-and-Display Vaccination. ACS Nano 2018, 12, 8855–8866. [Google Scholar] [CrossRef]
- Steele, J.F.C.; Peyret, H.; Saunders, K.; Castells-Graells, R.; Marsian, J.; Meshcheriakova, Y.; Lomonossoff, G.P. Synthetic Plant Virology for Nanobiotechnology and Nanomedicine. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017, 9, e1447. [Google Scholar] [CrossRef]
- McCormick, A.A.; Corbo, T.A.; Wykoff-Clary, S.; Palmer, K.E.; Pogue, G.P. Chemical Conjugate TMV-Peptide Bivalent Fusion Vaccines Improve Cellular Immunity and Tumor Protection. Bioconjug. Chem. 2006, 17, 1330–1338. [Google Scholar] [CrossRef]
- Ward, B.J.; Makarkov, A.; Séguin, A.; Pillet, S.; Trépanier, S.; Dhaliwall, J.; Libman, M.D.; Vesikari, T.; Landry, N. Efficacy, Immunogenicity, and Safety of a Plant-Derived, Quadrivalent, Virus-like Particle Influenza Vaccine in Adults (18-64 Years) and Older Adults (≥65 Years): Two Multicentre, Randomised Phase 3 Trials. Lancet Lond. Engl. 2020, 396, 1491–1503. [Google Scholar] [CrossRef]
- McCormick, A.A.; Palmer, K.E. Genetically Engineered Tobacco Mosaic Virus as Nanoparticle Vaccines. Expert Rev. Vaccines 2008, 7, 33–41. [Google Scholar] [CrossRef]
- Kemnade, J.O.; Seethammagari, M.; Collinson-Pautz, M.; Kaur, H.; Spencer, D.M.; McCormick, A.A. Tobacco Mosaic Virus Efficiently Targets DC Uptake, Activation and Antigen-Specific T Cell Responses in Vivo. Vaccine 2014, 32, 4228–4233. [Google Scholar] [CrossRef]
- McCormick, A.A.; Corbo, T.A.; Wykoff-Clary, S.; Nguyen, L.V.; Smith, M.L.; Palmer, K.E.; Pogue, G.P. TMV-Peptide Fusion Vaccines Induce Cell-Mediated Immune Responses and Tumor Protection in Two Murine Models. Vaccine 2006, 24, 6414–6423. [Google Scholar] [CrossRef]
- Smith, M.L.; Lindbo, J.A.; Dillard-Telm, S.; Brosio, P.M.; Lasnik, A.B.; McCormick, A.A.; Nguyen, L.V.; Palmer, K.E. Modified Tobacco Mosaic Virus Particles as Scaffolds for Display of Protein Antigens for Vaccine Applications. Virology 2006, 348, 475–488. [Google Scholar] [CrossRef]
- Banik, S.; Mansour, A.A.; Suresh, R.V.; Wykoff-Clary, S.; Malik, M.; McCormick, A.A.; Bakshi, C.S. Development of a Multivalent Subunit Vaccine against Tularemia Using Tobacco Mosaic Virus (TMV) Based Delivery System. PLoS ONE 2015, 10, e0130858. [Google Scholar] [CrossRef]
- Arnaboldi, P.M.; Sambir, M.; D’Arco, C.; Peters, L.A.; Seegers, J.F.M.L.; Mayer, L.; McCormick, A.A.; Dattwyler, R.J. Intranasal Delivery of a Protein Subunit Vaccine Using a Tobacco Mosaic Virus Platform Protects against Pneumonic Plague. Vaccine 2016, 34, 5768–5776. [Google Scholar] [CrossRef]
- Mallajosyula, J.K.; Hiatt, E.; Hume, S.; Johnson, A.; Jeevan, T.; Chikwamba, R.; Pogue, G.P.; Bratcher, B.; Haydon, H.; Webby, R.J.; et al. Single-Dose Monomeric HA Subunit Vaccine Generates Full Protection from Influenza Challenge. Hum. Vaccines Immunother. 2014, 10, 586–595. [Google Scholar] [CrossRef]
- Mansour, A.A.; Banik, S.; Suresh, R.V.; Kaur, H.; Malik, M.; McCormick, A.A.; Bakshi, C.S. An Improved Tobacco Mosaic Virus (TMV)-Conjugated Multiantigen Subunit Vaccine Against Respiratory Tularemia. Front. Microbiol. 2018, 9, 1195. [Google Scholar] [CrossRef]
- Palmer, K.E.; Benko, A.; Doucette, S.A.; Cameron, T.I.; Foster, T.; Hanley, K.M.; McCormick, A.A.; McCulloch, M.; Pogue, G.P.; Smith, M.L.; et al. Protection of Rabbits against Cutaneous Papillomavirus Infection Using Recombinant Tobacco Mosaic Virus Containing L2 Capsid Epitopes. Vaccine 2006, 24, 5516–5525. [Google Scholar] [CrossRef]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the Receptor-Binding Domain (RBD) of 2019 Novel Coronavirus: Implication for Development of RBD Protein as a Viral Attachment Inhibitor and Vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef]
- Crawford, K.H.D.; Eguia, R.; Dingens, A.S.; Loes, A.N.; Malone, K.D.; Wolf, C.R.; Chu, H.Y.; Tortorici, M.A.; Veesler, D.; Murphy, M.; et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses 2020, 12, 513. [Google Scholar] [CrossRef] [PubMed]
- Kam, Y.W.; Kien, F.; Roberts, A.; Cheung, Y.C.; Lamirande, E.W.; Vogel, L.; Chu, S.L.; Tse, J.; Guarner, J.; Zaki, S.R.; et al. Antibodies against Trimeric S Glycoprotein Protect Hamsters against SARS-CoV Challenge despite Their Capacity to Mediate FcgammaRII-Dependent Entry into B Cells in Vitro. Vaccine 2007, 25, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Jaume, M.; Yip, M.S.; Cheung, C.Y.; Leung, H.L.; Li, P.H.; Kien, F.; Dutry, I.; Callendret, B.; Escriou, N.; Altmeyer, R.; et al. Anti-Severe Acute Respiratory Syndrome Coronavirus Spike Antibodies Trigger Infection of Human Immune Cells via a PH- and Cysteine Protease-Independent FcγR Pathway. J. Virol. 2011, 85, 10582–10597. [Google Scholar] [CrossRef] [PubMed]
- Jaume, M.; Yip, M.S.; Kam, Y.W.; Cheung, C.Y.; Kien, F.; Roberts, A.; Li, P.H.; Dutry, I.; Escriou, N.; Daeron, M.; et al. SARS CoV Subunit Vaccine: Antibody-Mediated Neutralisation and Enhancement. Hong Kong Med. J. 2012, 18 (Suppl. 2), 31–36. [Google Scholar]
- Luo, F.; Liao, F.-L.; Wang, H.; Tang, H.-B.; Yang, Z.-Q.; Hou, W. Evaluation of Antibody-Dependent Enhancement of SARS-CoV Infection in Rhesus Macaques Immunized with an Inactivated SARS-CoV Vaccine. Virol. Sin. 2018, 33, 201–204. [Google Scholar] [CrossRef]
- Perlman, S.; Dandekar, A.A. Immunopathogenesis of Coronavirus Infections: Implications for SARS. Nat. Rev. Immunol. 2005, 5, 917–927. [Google Scholar] [CrossRef]
- Pewe, L.; Zhou, H.; Netland, J.; Tangudu, C.; Olivares, H.; Shi, L.; Look, D.; Gallagher, T.; Perlman, S. A Severe Acute Respiratory Syndrome-Associated Coronavirus-Specific Protein Enhances Virulence of an Attenuated Murine Coronavirus. J. Virol. 2005, 79, 11335–11342. [Google Scholar] [CrossRef][Green Version]
- Channappanavar, R.; Perlman, S. Pathogenic Human Coronavirus Infections: Causes and Consequences of Cytokine Storm and Immunopathology. Semin. Immunopathol. 2017, 39, 529–539. [Google Scholar] [CrossRef]
- Tseng, C.-T.; Sbrana, E.; Iwata-Yoshikawa, N.; Newman, P.C.; Garron, T.; Atmar, R.L.; Peters, C.J.; Couch, R.B. Immunization with SARS Coronavirus Vaccines Leads to Pulmonary Immunopathology on Challenge with the SARS Virus. PloS ONE 2012, 7, e35421. [Google Scholar] [CrossRef]
- Du, L.; Zhao, G.; Chan, C.C.S.; Sun, S.; Chen, M.; Liu, Z.; Guo, H.; He, Y.; Zhou, Y.; Zheng, B.-J.; et al. Recombinant Receptor-Binding Domain of SARS-CoV Spike Protein Expressed in Mammalian, Insect and E. Coli Cells Elicits Potent Neutralizing Antibody and Protective Immunity. Virology 2009, 393, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Zhao, G.; He, Y.; Guo, Y.; Zheng, B.-J.; Jiang, S.; Zhou, Y. Receptor-Binding Domain of SARS-CoV Spike Protein Induces Long-Term Protective Immunity in an Animal Model. Vaccine 2007, 25, 2832–2838. [Google Scholar] [CrossRef] [PubMed]
- Mekhaiel, D.N.A.; Czajkowsky, D.M.; Andersen, J.T.; Shi, J.; El-Faham, M.; Doenhoff, M.; McIntosh, R.S.; Sandlie, I.; He, J.; Hu, J.; et al. Polymeric Human Fc-Fusion Proteins with Modified Effector Functions. Sci. Rep. 2011, 1, 124. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, F.; Shen, C.; Peng, W.; Li, D.; Zhao, C.; Li, Z.; Li, S.; Bi, Y.; Yang, Y.; et al. A Noncompeting Pair of Human Neutralizing Antibodies Block COVID-19 Virus Binding to Its Receptor ACE2. Science 2020, 368, 1274–1278. [Google Scholar] [CrossRef]
- Ward, B.J.; Gobeil, P.; Séguin, A.; Atkins, J.; Boulay, I.; Charbonneau, P.-Y.; Couture, M.; D’Aoust, M.-A.; Dhaliwall, J.; Finkle, C.; et al. Phase 1 Randomized Trial of a Plant-Derived Virus-like Particle Vaccine for COVID-19. Nat. Med. 2021, 27, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Fiala, B.; Schäfer, A.; Wrenn, S.; Pham, M.N.; Murphy, M.; Tse, L.V.; Shehata, L.; O’Connor, M.A.; Chen, C.; et al. Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2. Cell 2020, 183, 1367–1382.e17. [Google Scholar] [CrossRef]
- Burger, M.I. Ludwig Delivering Super-Cooled COVID-19 Vaccine a Daunting Challenge for Some Countries. Reuters 2020. Available online: https://www.reuters.com/article/us-health-coronavirus-logistics-idUSKBN25S417 (accessed on 13 June 2021).
- Huang, W.-C.; Zhou, S.; He, X.; Chiem, K.; Mabrouk, M.T.; Nissly, R.H.; Bird, I.M.; Strauss, M.; Sambhara, S.; Ortega, J.; et al. SARS-CoV-2 RBD Neutralizing Antibody Induction Is Enhanced by Particulate Vaccination. Adv. Mater. Deerfield Beach Fla 2020, 32, e2005637. [Google Scholar] [CrossRef]
- Olotu, A.; Lusingu, J.; Leach, A.; Lievens, M.; Vekemans, J.; Msham, S.; Lang, T.; Gould, J.; Dubois, M.-C.; Jongert, E.; et al. Efficacy of RTS,S/AS01E Malaria Vaccine and Exploratory Analysis on Anti-Circumsporozoite Antibody Titres and Protection in Children Aged 5-17 Months in Kenya and Tanzania: A Randomised Controlled Trial. Lancet Infect. Dis. 2011, 11, 102–109. [Google Scholar] [CrossRef]
- Lusingu, J.; Olotu, A.; Leach, A.; Lievens, M.; Vekemans, J.; Olivier, A.; Benns, S.; Olomi, R.; Msham, S.; Lang, T.; et al. Safety of the Malaria Vaccine Candidate, RTS,S/AS01E in 5 to 17 Month Old Kenyan and Tanzanian Children. PLoS ONE 2010, 5, e14090. [Google Scholar] [CrossRef]
- Krueger, C.C.; Thoms, F.; Keller, E.; Leoratti, F.M.S.; Vogel, M.; Bachmann, M.F. RNA and Toll-Like Receptor 7 License the Generation of Superior Secondary Plasma Cells at Multiple Levels in a B Cell Intrinsic Fashion. Front. Immunol. 2019, 10, 736. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.O.; West, A.P.; Huey-Tubman, K.E.; Hoffmann, M.A.G.; Sharaf, N.G.; Hoffman, P.R.; Koranda, N.; Gristick, H.B.; Gaebler, C.; Muecksch, F.; et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell 2020, 182, 828–842.e16. [Google Scholar] [CrossRef]
- Brouwer, P.J.M.; Caniels, T.G.; van der Straten, K.; Snitselaar, J.L.; Aldon, Y.; Bangaru, S.; Torres, J.L.; Okba, N.M.A.; Claireaux, M.; Kerster, G.; et al. Potent Neutralizing Antibodies from COVID-19 Patients Define Multiple Targets of Vulnerability. Science 2020, 369, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, W.; Drabek, D.; Okba, N.M.A.; van Haperen, R.; Osterhaus, A.D.M.E.; van Kuppeveld, F.J.M.; Haagmans, B.L.; Grosveld, F.; Bosch, B.-J. A Human Monoclonal Antibody Blocking SARS-CoV-2 Infection. Nat. Commun. 2020, 11, 2251. [Google Scholar] [CrossRef]
- Jeyanathan, M.; Afkhami, S.; Smaill, F.; Miller, M.S.; Lichty, B.D.; Xing, Z. Immunological Considerations for COVID-19 Vaccine Strategies. Nat. Rev. Immunol. 2020, 20, 615–632. [Google Scholar] [CrossRef] [PubMed]
- Cole, G.A. Interferon-Gamma ELISPOT Assay for the Quantitative Measurement of Antigen-Specific Murine CD8+ T-Cells. Methods Mol. Biol. 2005, 302, 191–204. [Google Scholar] [CrossRef]
- Schoenborn, J.R.; Wilson, C.B. Regulation of Interferon-Gamma during Innate and Adaptive Immune Responses. Adv. Immunol. 2007, 96, 41–101. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Royal, J.M.; Simpson, C.A.; McCormick, A.A.; Phillips, A.; Hume, S.; Morton, J.; Shepherd, J.; Oh, Y.; Swope, K.; DeBeauchamp, J.L.; et al. Development of a SARS-CoV-2 Vaccine Candidate Using Plant-Based Manufacturing and a Tobacco Mosaic Virus-like Nano-Particle. Vaccines 2021, 9, 1347. https://doi.org/10.3390/vaccines9111347
Royal JM, Simpson CA, McCormick AA, Phillips A, Hume S, Morton J, Shepherd J, Oh Y, Swope K, DeBeauchamp JL, et al. Development of a SARS-CoV-2 Vaccine Candidate Using Plant-Based Manufacturing and a Tobacco Mosaic Virus-like Nano-Particle. Vaccines. 2021; 9(11):1347. https://doi.org/10.3390/vaccines9111347
Chicago/Turabian StyleRoyal, Joshua M., Carrie A. Simpson, Alison A. McCormick, Amanda Phillips, Steve Hume, Josh Morton, John Shepherd, Youngjun Oh, Kelsi Swope, Jennifer L. DeBeauchamp, and et al. 2021. "Development of a SARS-CoV-2 Vaccine Candidate Using Plant-Based Manufacturing and a Tobacco Mosaic Virus-like Nano-Particle" Vaccines 9, no. 11: 1347. https://doi.org/10.3390/vaccines9111347
APA StyleRoyal, J. M., Simpson, C. A., McCormick, A. A., Phillips, A., Hume, S., Morton, J., Shepherd, J., Oh, Y., Swope, K., DeBeauchamp, J. L., Webby, R. J., Cross, R. W., Borisevich, V., Geisbert, T. W., Demarco, J. K., Bratcher, B., Haydon, H., & Pogue, G. P. (2021). Development of a SARS-CoV-2 Vaccine Candidate Using Plant-Based Manufacturing and a Tobacco Mosaic Virus-like Nano-Particle. Vaccines, 9(11), 1347. https://doi.org/10.3390/vaccines9111347






