SARS-CoV-2 Epitope Mapping on Microarrays Highlights Strong Immune-Response to N Protein Region
Abstract
1. Introduction
2. Materials and Methods
2.1. Serum Samples
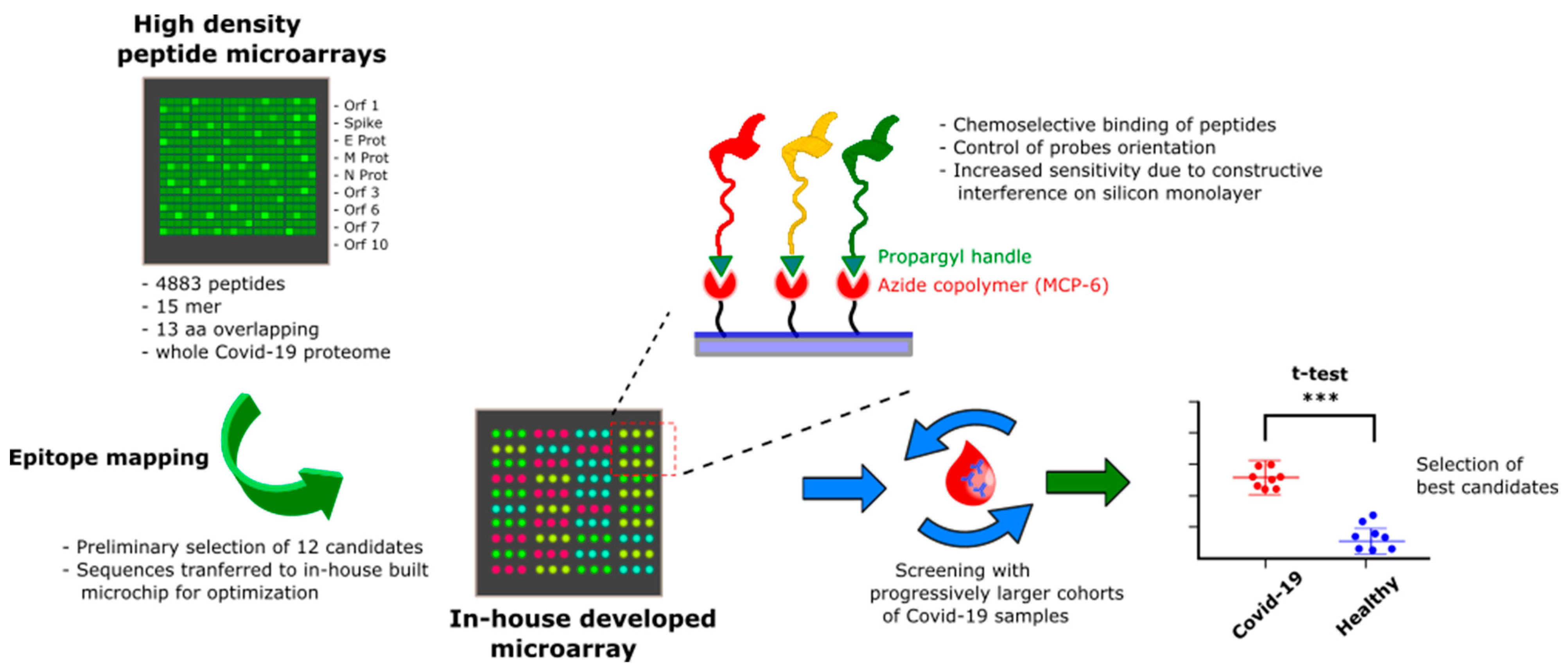
2.2. Proteome-Wide Microarray
2.3. Peptide Synthesis
2.4. Peptide Microarray
2.5. Bioassays
3. Results and Discussion
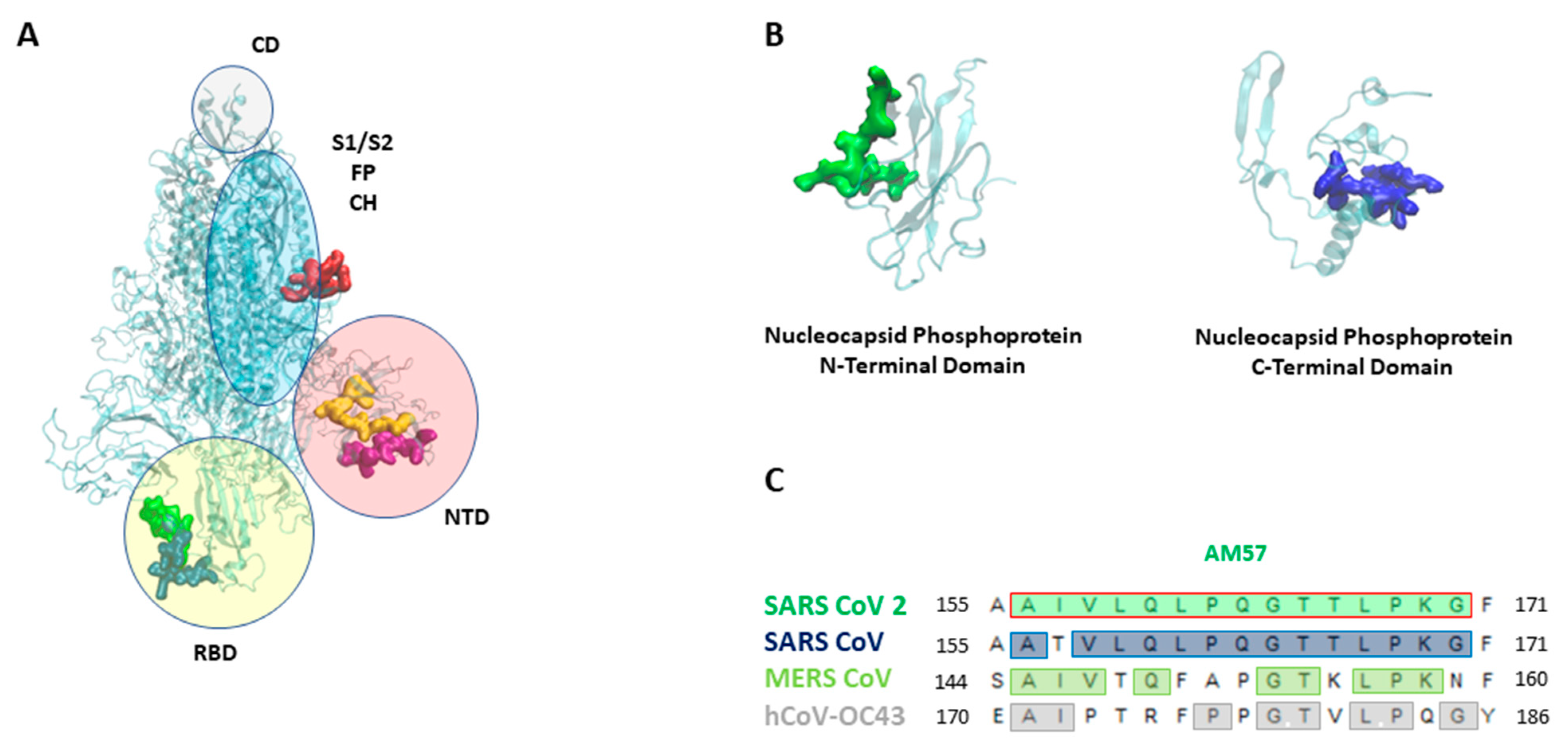
3.1. SARS-CoV-2 Whole-Proteome Epitope Mapping
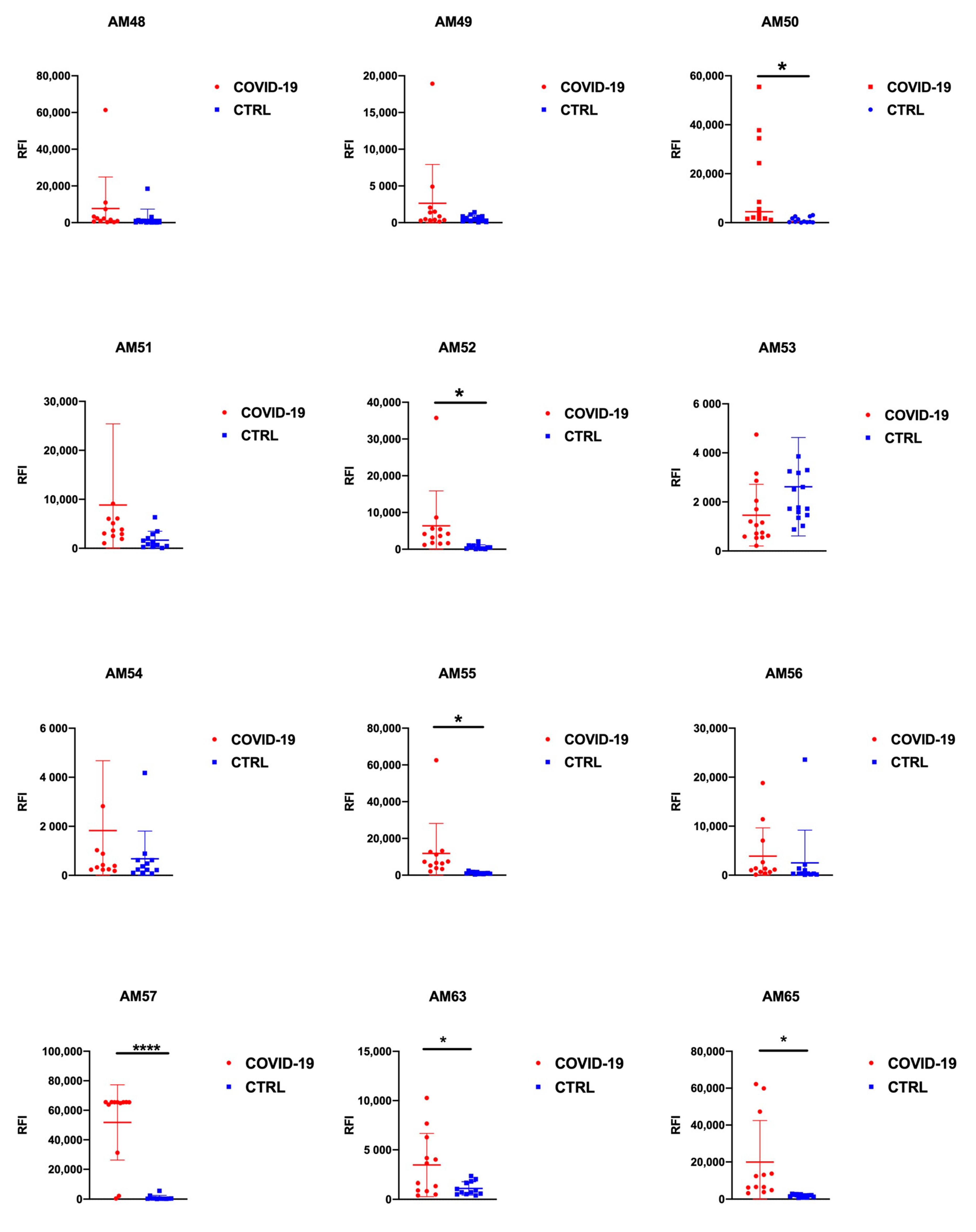
3.2. Epitopes Validation Screening
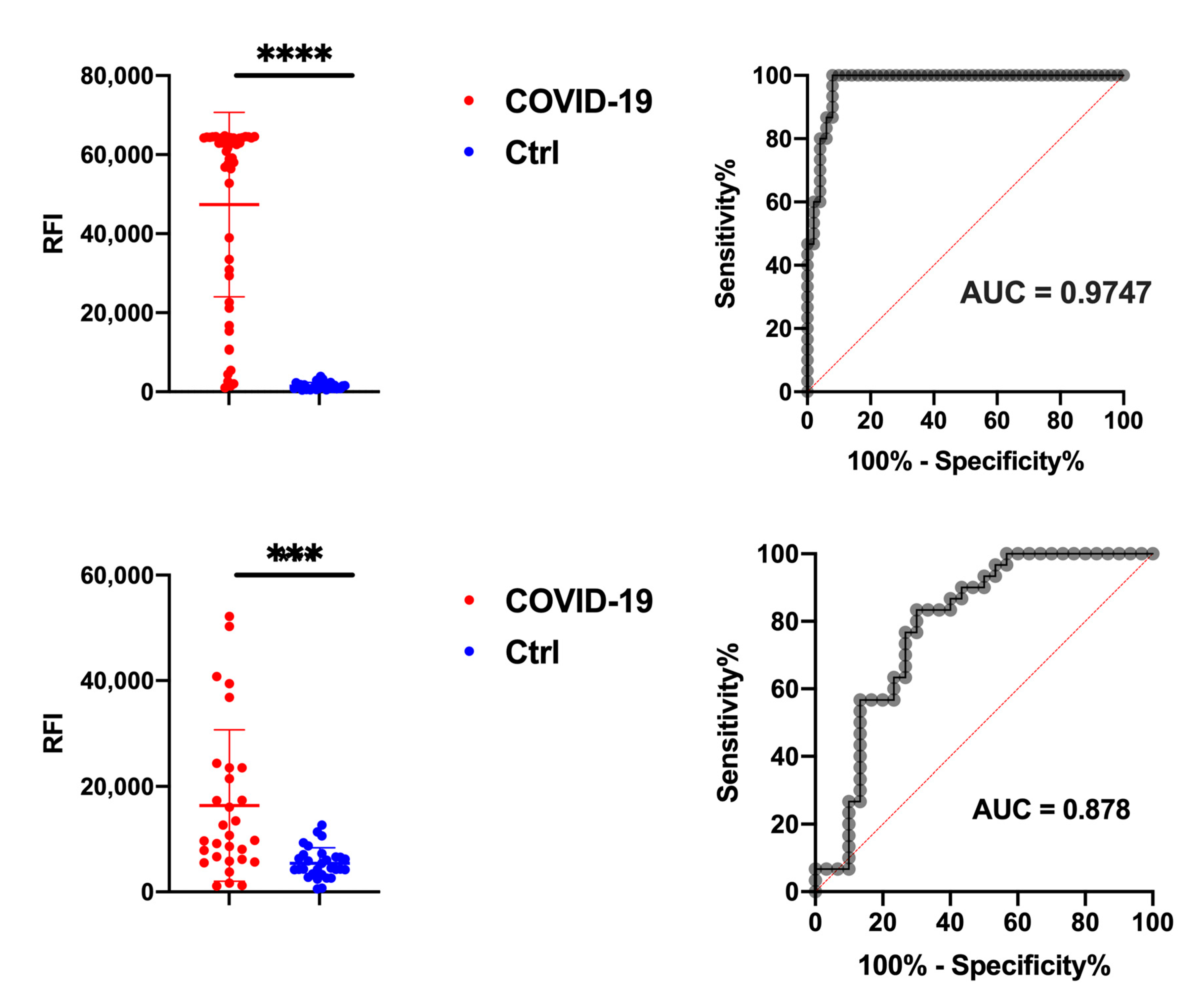
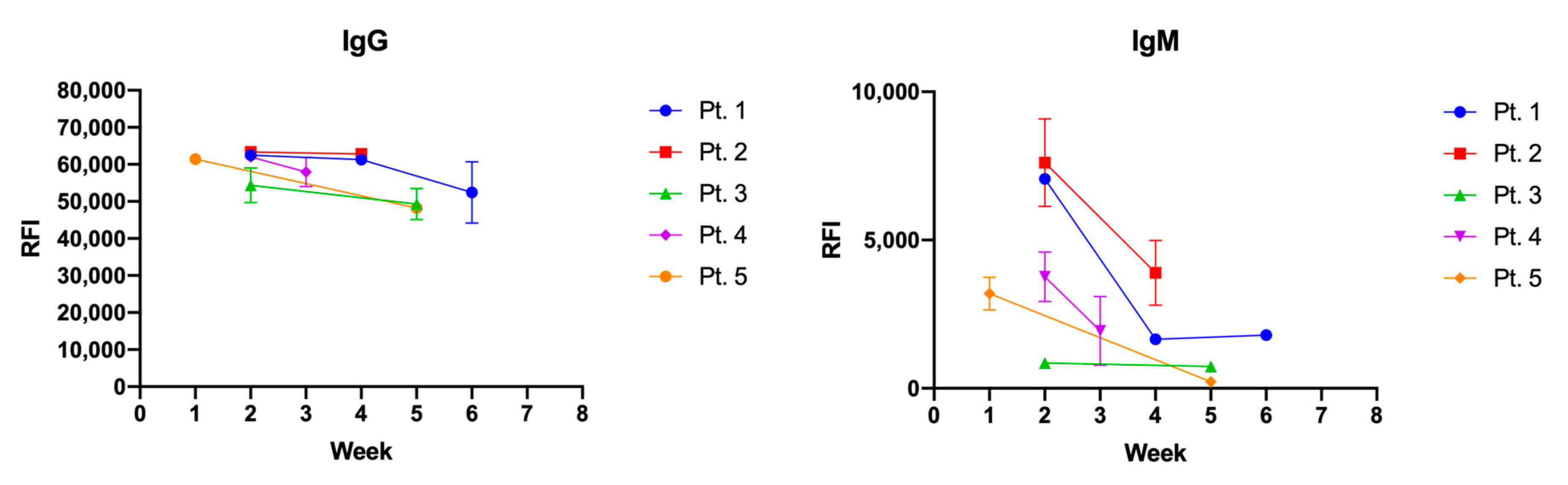
3.3. N Protein Epitope 155-171 (AM57) Shows Promising Diagnostic Potential
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Carter, L.J.; Garner, L.V.; Smoot, J.W.; Li, Y.; Zhou, Q.; Saveson, C.J.; Sasso, J.M.; Gregg, A.C.; Soares, D.J.; Beskid, T.R.; et al. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020, 6, 591–605. [Google Scholar] [CrossRef]
- Phelan, A.L. COVID-19 immunity passports and vaccination certificates: Scientific, equitable, and legal challenges. Lancet 2020, 395, 1595–1598. [Google Scholar] [CrossRef]
- Long, Q.-X.; Liu, B.-Z.; Deng, H.-J.; Wu, G.-C.; Deng, K.; Chen, Y.-K.; Liao, P.; Qiu, J.-F.; Lin, Y.; Cai, X.-F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Xiang, F.; Wang, X.; He, X.; Peng, Z.; Yang, B.; Zhang, J.; Zhou, Q.; Ye, H.; Ma, Y.; Li, H.; et al. Antibody detection and dynamic characteristics in patients with COVID-19. In Clinical Infectious Diseases an Official Publication of the Infectious Diseases Society of America; Infectious Diseases Society of America: Arlington, VA, USA, 2020. [Google Scholar] [CrossRef]
- Okba, N.; Müller, M.A.; Li, W.; Wang, C.; GeurtsvanKessel, C.H.; Corman, V.M.; Lamers, M.M.; Sikkema, R.S.; de Bruin, E.; Chandler, F.D.; et al. SARS-CoV-2 specific antibody responses in COVID-19 patients. Emerg. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Amrun, S.N.; Lee, C.Y.P.; Lee, B.; Fong, S.W.; Young, B.E.; Chee, R.S.L.; Ng, L.F.; Yeo, N.K.-W.; Torres-Ruesta, A.; Carissimo, G.; et al. Linear B-cell epitopes in the spike and nucleocapsid proteins as markers of SARS-CoV-2 exposure and disease severity. EBioMedicine 2020. [Google Scholar] [CrossRef] [PubMed]
- Che, X.Y.; Qiu, L.W.; Liao, Z.Y.; Wang, Y.; Wen, K.; Pan, Y.X.; Hao, W.; Mei, Y.B.; Cheng, V.C.C.; Yuen, K.Y. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J. Infect. Dis. 2005. [Google Scholar] [CrossRef][Green Version]
- Zhang, B.; Hu, Y.F.; Chen, L.L.; Yau, T.; Tong, Y.G.; Hu, J.C.; Cai, J.P.; Chan, K.H.; Dou, Y.; Deng, J.; et al. Mining of epitopes on spike protein of SARS-CoV-2 from COVID-19 patients. Cell Res. 2020, 30, 702–704. [Google Scholar] [CrossRef]
- Li, Y.; Lai, D.Y.; Zhang, H.N.; Jiang, H.W.; Tian, X.L.; Ma, M.L.; Tao, S.C.; Qi, H.; Meng, Q.-F.; Guo, S.-J.; et al. Linear epitopes of SARS-CoV-2 spike protein elicit neutralizing antibodies in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 1095–1097. [Google Scholar] [CrossRef]
- Poh, C.M.; Carissimo, G.; Wang, B.; Amrun, S.N.; Lee, C.Y.; Chee, R.S.; Fong, S.W.; Yeo, N.K.; Lee, W.H.; Torres-Ruesta, A.; et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun. 2020, 11, 2806. [Google Scholar] [CrossRef]
- Farrera-Soler, L.; Daguer, J.; Barluenga, S.; Vadas, O.; Cohen, P.; Pagano, S.; Yerly, S.; Kaiser, L.; Vuilleumier, N.; Winssinger, N. Identification of immunodominant linear epitopes from SARS-CoV-2 patient plasma. PLoS ONE 2020, 15, e0238089. [Google Scholar] [CrossRef]
- Shrock, E.; Fujimura, E.; Kula, T.; Timms, R.T.; Lee, I.H.; Leng, Y.; Elledge, S.J.; Robinson, M.L.; Sie, B.M.; Li, M.Z.; et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, X.; Zhang, X.; Hou, X.; Liang, T.; Wang, D.; Teng, F.; Dai, J.; Duan, H.; Guo, S.; et al. SARS-CoV-2 proteome microarray for mapping COVID-19 antibody interactions at amino acid resolution. ACS Cent. Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, D.; Chiari, M.; Gori, A.; Cretich, M. Towards precision medicine: The role and potential of protein and peptide microarrays. Analyst 2019, 144, 5353–5367. [Google Scholar] [CrossRef] [PubMed]
- Carmona, S.J.; Nielsen, M.; Schafer-Nielsen, C.; Mucci, J.; Altcheh, J.; Balouz, V.; Tekiel, V.; Frasch, A.C.; Campetella, O.; Buscaglia, C.A.; et al. Towards high-throughput immunomics for infectious diseases: Use of next-generation peptide microarrays for rapid discovery and mapping of antigenic determinants. Mol. Cell. Proteom. 2015, 14, 1871–1884. [Google Scholar] [CrossRef] [PubMed]
- Cretich, M.; Gori, A.; D’Annessa, I.; Chiari, M.; Colombo, G. Peptides for infectious diseases: From probe design to diagnostic microarrays. Antibodies 2019, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Gori, A.; Sola, L.; Gagni, P.; Bruni, G.; Liprino, M.; Peri, C.; Colombo, G.; Cretich, M.; Chiari, M. Screening complex biological samples with peptide microarrays: The favorable impact of probe orientation via chemoselective immobilization strategies on clickable polymeric coatings. Bioconjugate Chem. 2016, 27, 2669–2677. [Google Scholar] [CrossRef] [PubMed]
- Sola, L.; Gagni, P.; D’Annessa, I.; Capelli, R.; Bertino, C.; Romanato, A.; Damin, F.; Bergamaschi, G.; Marchisio, E.; Cuzzocrea, A.; et al. Enhancing antibody serodiagnosis using a controlled peptide coimmobilization strategy. ACS Infect. Dis. 2018, 4, 998–1006. [Google Scholar] [CrossRef]
- Casalino, L.; Gaieb, Z.; Goldsmith, J.A.; Hjorth, C.K.; Dommer, A.C.; Harbison, A.M.; Fogarty, C.A.; Barros, E.P.; Taylor, B.C.; McLellan, J.S.; et al. Beyond shielding: The roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 2020. [Google Scholar] [CrossRef]
- Gori, A.; Cretich, M.; Vanna, R.; Sola, L.; Gagni, P.; Bruni, G.; Chiari, M.; Liprino, M.; Gramatica, F.; Burastero, S. Multiple epitope presentation and surface density control enabled by chemoselective immobilization lead to enhanced performance in IgE-binding fingerprinting on peptide microarrays. Anal. Chim. Acta 2017, 983, 189–197. [Google Scholar] [CrossRef]
- Odinolfi, M.T.; Romanato, A.; Bergamaschi, G.; Strada, A.; Sola, L.; Girella, A.; Cretich, M.; Milanese, C.; Chiari, M.; Gori, A. Clickable cellulosic surfaces for peptide-based bioassays. Talanta 2019, 205, 120152. [Google Scholar] [CrossRef]
- Cretich, M.; di Carlo, G.; Longhi, R.; Gotti, C.; Spinella, N.; Coffa, S.; Galati, C.; Renna, L.; Chiari, M. High sensitivity protein assays on microarray silicon slides. Anal. Chem. 2009, 81, 5197–5203. [Google Scholar] [CrossRef] [PubMed]
- Cretich, M.; Galati, C.; Renna, L.; Condorelli, G.G.; Gagni, P.; Chiari, M. Characterization of a new fluorescence-enhancing substrate for microarrays with femtomolar sensitivity. Sens. Actuators B Chem. 2014, 192, 15–22. [Google Scholar] [CrossRef]
| Patients Data | Group A (Whole-Proteome Screening) | Group B (1st Microarray Screening) | Group C (2nd Microarray Screening) | Group D (Test Validation) | Group E (Longitudinal Samples) |
|---|---|---|---|---|---|
| No. patients | 7 | 12 | 28 | 50 | 5 |
| Mean age, IQR | 86 (79–95) | 78 (62–91) | 80 (72–86) | 81 (71–87) | 67 (64–72) |
| Females | 71% | 58% | 75% | 66% | 0% |
| Mild/moderate COVID-19 | 100% | 92% | 96% | 86% | 0% |
| Severe COVID-19 | 0% | 8% | 4% | 14% | 100% |
| Days since symptom onset, IQR | 28 (27–30) | 30 (25–35) | 38 (28–119) | 98 (28–120) | Time series (range 30–150 days) |
| Protein | Code | Sequence | p Value 1st Screening N = 12 | p Value 2nd Screening N = 28 |
|---|---|---|---|---|
| Orf1ab | AM48 | CASYQTQTNSPRRAR | 0.2958 | |
| S | AM49 | YQAGSTPCNGVEGFN | 0.1942 | |
| S | AM50 | FDNPVLPFNDGVYFA | 0.0180 | 0.0101 |
| Orf1ab | AM51 | LGVYDYLVSTQEFRY | 0.1499 | |
| Orf1ab | AM52 | QDGNAAISDYDYYRY | 0.0470 | 0.3314 |
| Orf1ab | AM54 | GLPGTILRTTNGDFL | 0.2074 | |
| S | AM55 | CASYQTQTNSPRRAR | 0.0359 | 0.2882 |
| Orf1ab | AM56 | IDLVPNQPYPNASFD | 0.5966 | |
| N | AM57 | AIVLQLPQGTTLPKG | <0.0001 | <0.0001 |
| S | AM63 | SNLKPFERDISTEIY | 0.0249 | 0.2640 |
| Orf1ab | AM65 | DNQDLNGNWYDFGDF | 0.0115 | 0.0317 |
| N | AM66 | GNFGDQELIRQGTDY | 0.0771 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Musicò, A.; Frigerio, R.; Mussida, A.; Barzon, L.; Sinigaglia, A.; Riccetti, S.; Gobbi, F.; Piubelli, C.; Bergamaschi, G.; Chiari, M.; et al. SARS-CoV-2 Epitope Mapping on Microarrays Highlights Strong Immune-Response to N Protein Region. Vaccines 2021, 9, 35. https://doi.org/10.3390/vaccines9010035
Musicò A, Frigerio R, Mussida A, Barzon L, Sinigaglia A, Riccetti S, Gobbi F, Piubelli C, Bergamaschi G, Chiari M, et al. SARS-CoV-2 Epitope Mapping on Microarrays Highlights Strong Immune-Response to N Protein Region. Vaccines. 2021; 9(1):35. https://doi.org/10.3390/vaccines9010035
Chicago/Turabian StyleMusicò, Angelo, Roberto Frigerio, Alessandro Mussida, Luisa Barzon, Alessandro Sinigaglia, Silvia Riccetti, Federico Gobbi, Chiara Piubelli, Greta Bergamaschi, Marcella Chiari, and et al. 2021. "SARS-CoV-2 Epitope Mapping on Microarrays Highlights Strong Immune-Response to N Protein Region" Vaccines 9, no. 1: 35. https://doi.org/10.3390/vaccines9010035
APA StyleMusicò, A., Frigerio, R., Mussida, A., Barzon, L., Sinigaglia, A., Riccetti, S., Gobbi, F., Piubelli, C., Bergamaschi, G., Chiari, M., Gori, A., & Cretich, M. (2021). SARS-CoV-2 Epitope Mapping on Microarrays Highlights Strong Immune-Response to N Protein Region. Vaccines, 9(1), 35. https://doi.org/10.3390/vaccines9010035







