Primary HIV-1 and Infectious Molecular Clones Are Differentially Susceptible to Broadly Neutralizing Antibodies
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Monoclonal Antibodies
2.3. Media and Cell Culture
2.4. HIV-1 Preparation and Purification
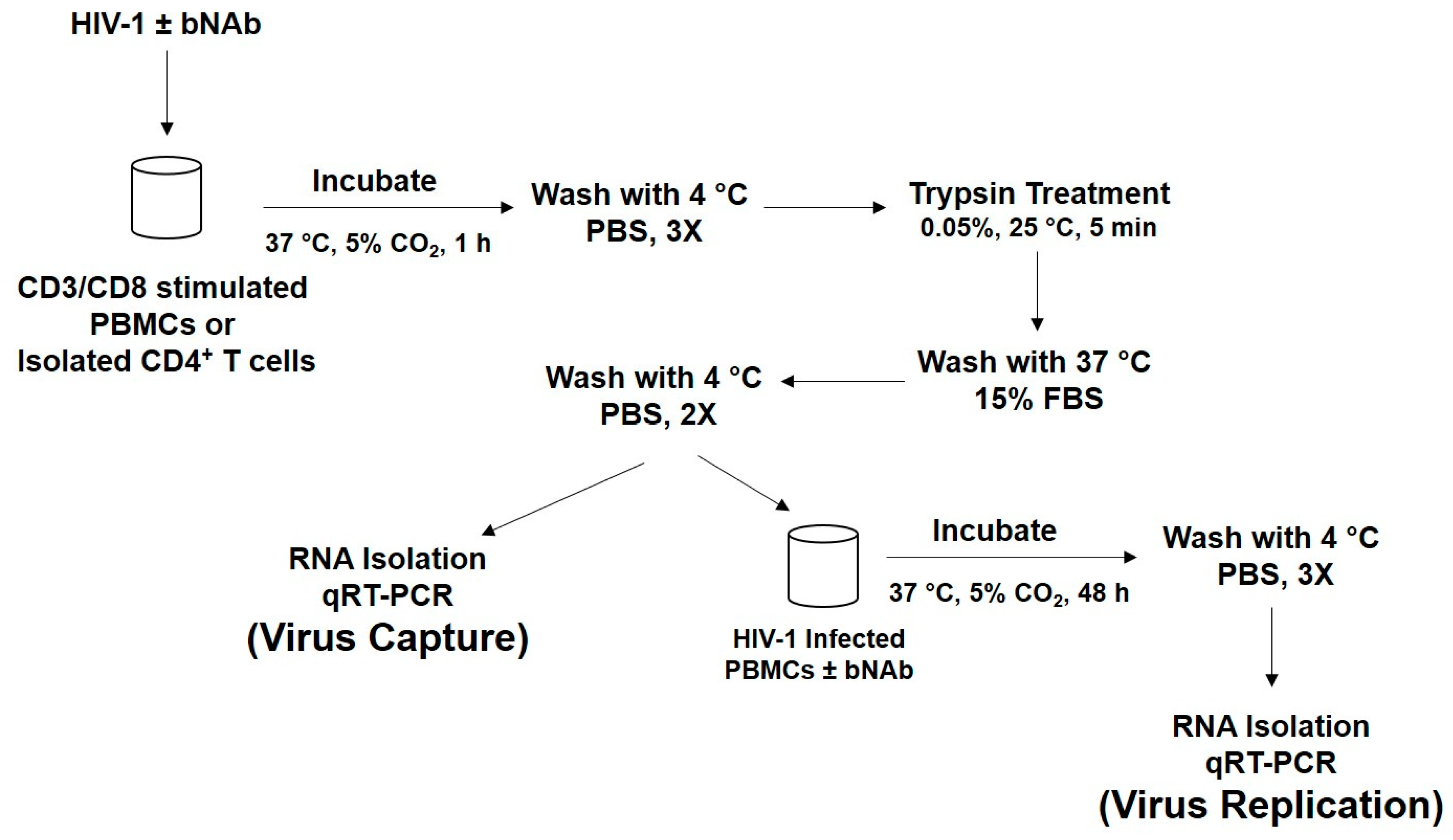
2.5. HIV-1 Capture Assay Using PBMCs
2.6. HIV-1 Replication Experiments
2.7. Isolation of CD4+ T Cells
2.8. RNA Extraction and qRT-PCR
2.9. Statistical Analysis
3. Results
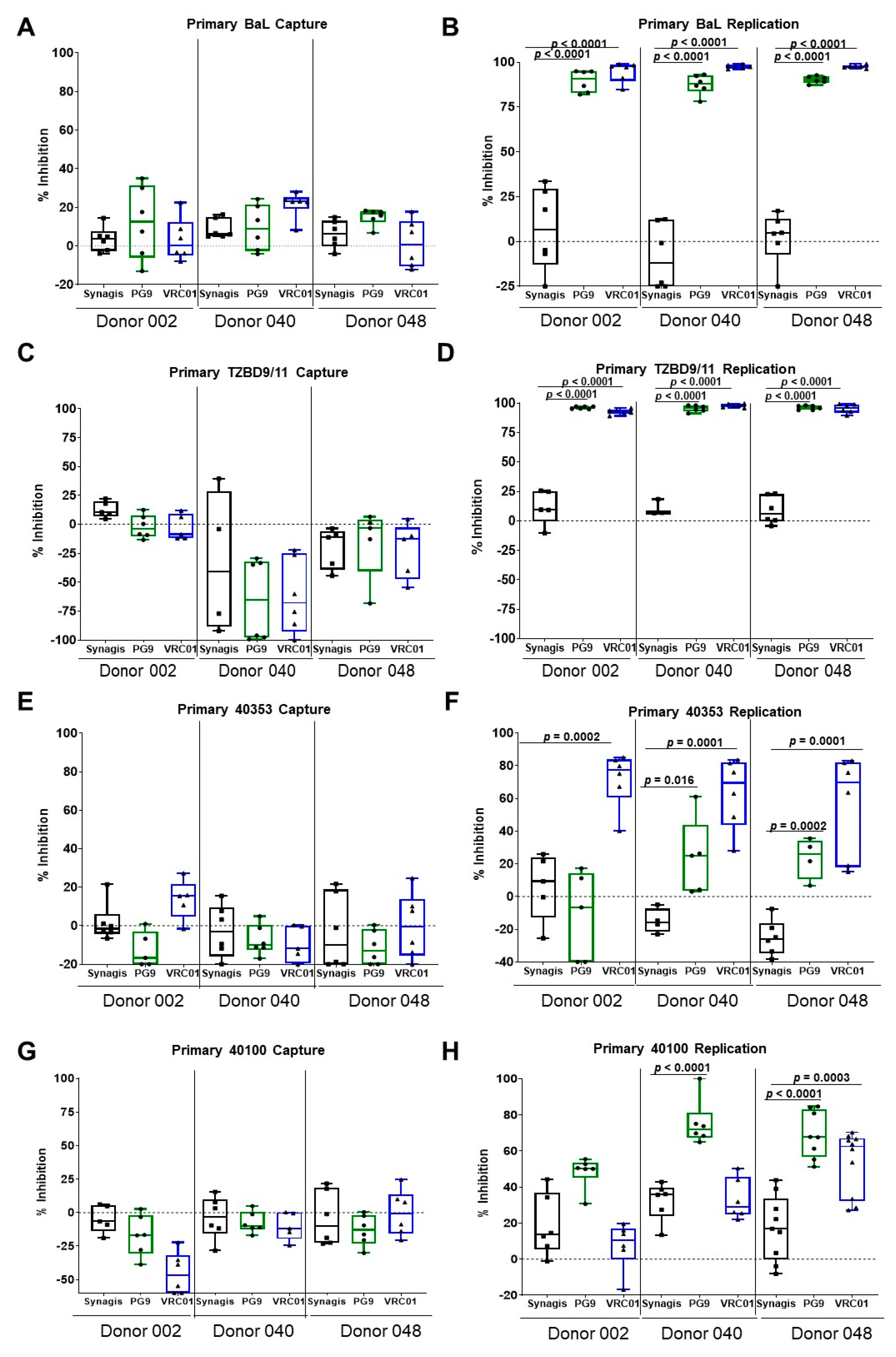
3.1. Capture and Replication of Primary HIV-1 in Human PBMCs
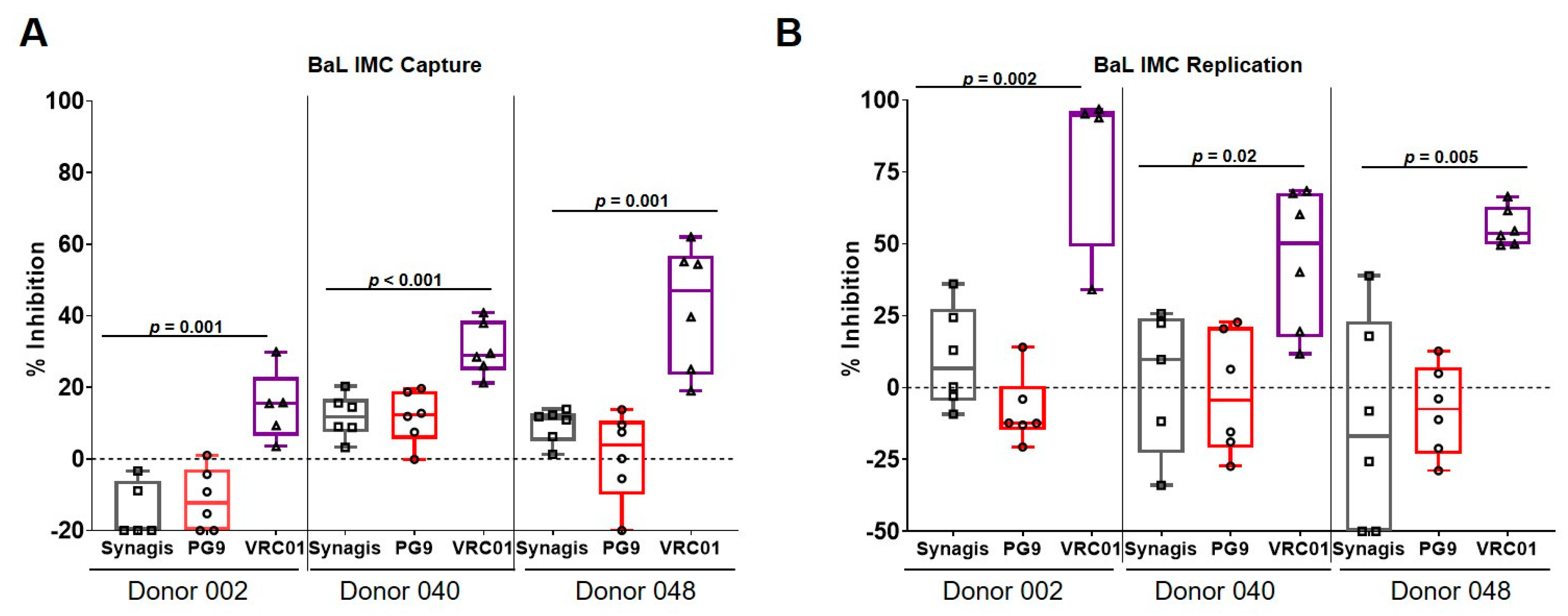
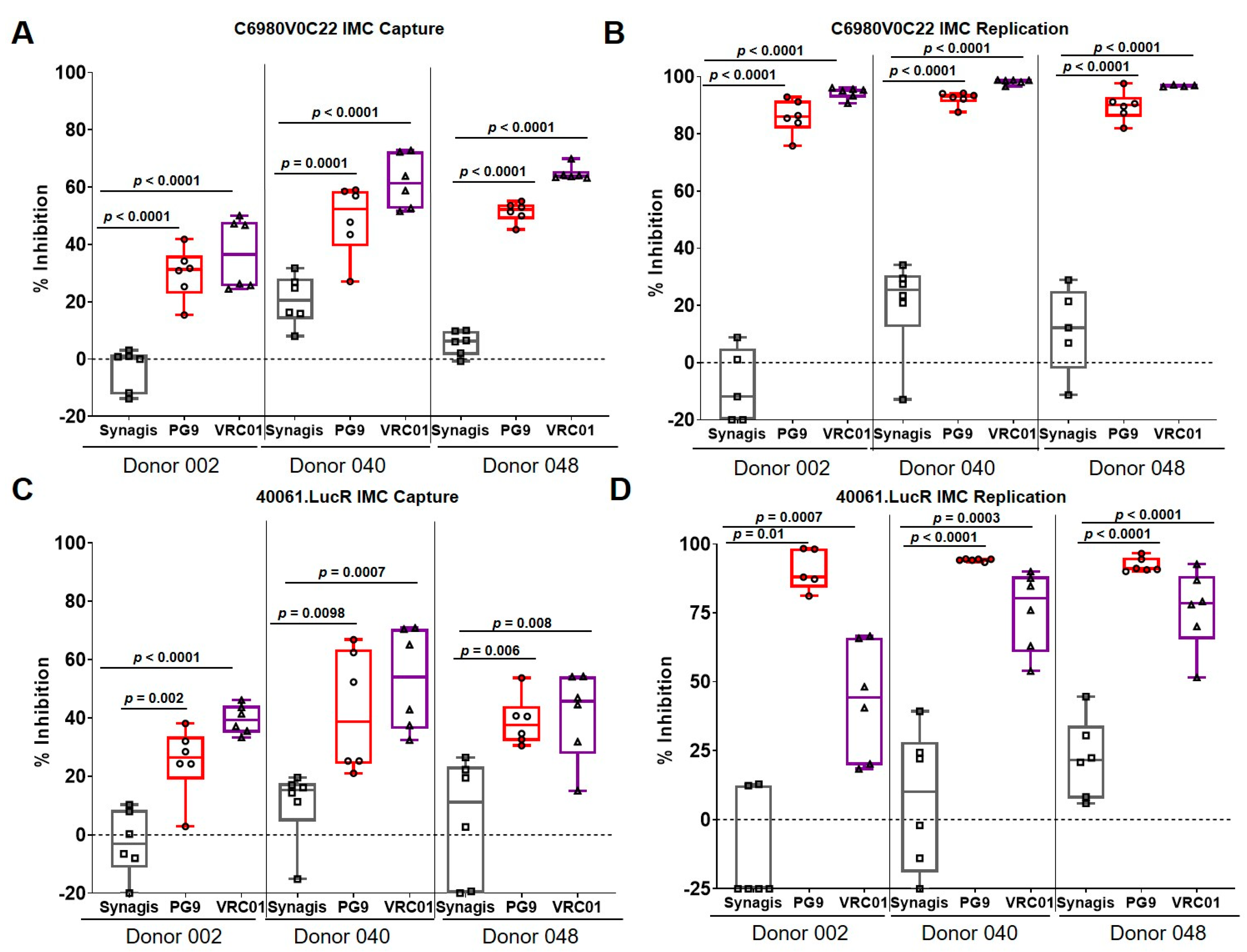
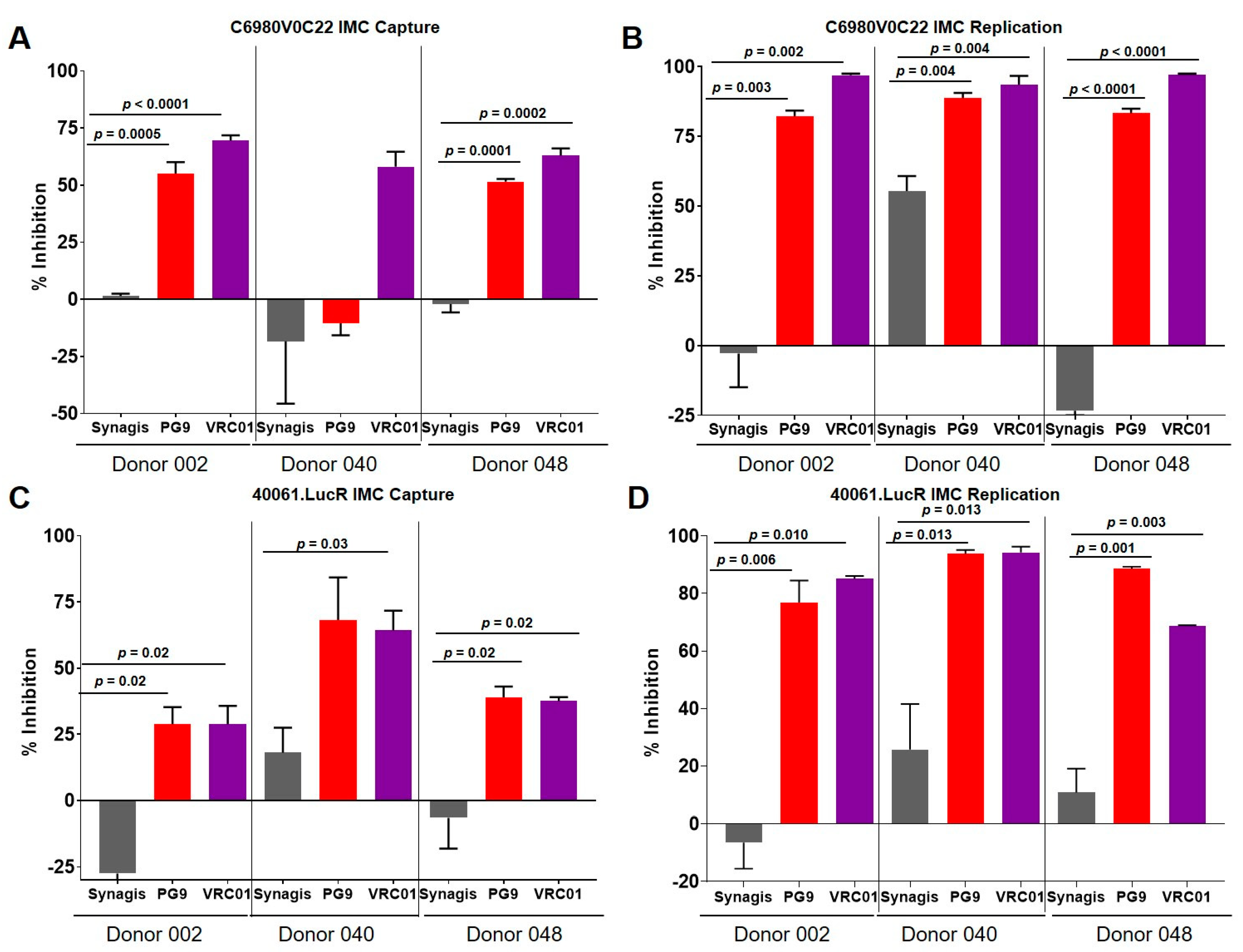
3.2. Capture and Replication of Infectious Molecular Clones in Human PBMCs
3.3. Virus Capture and Replication of IMCs with Isolated CD4+ T Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Montefiori, D.C. Evaluating Neutralizing Antibodies against Hiv, Siv, and Shiv in Luciferase Reporter Gene Assays. Curr. Protoc. Immunol. 2005. [CrossRef] [PubMed]
- Chuang, G.-Y.; Acharya, P.; Schmidt, S.D.; Yang, Y.; Louder, M.K.; Zhou, T.; Kwon, Y.D.; Pancera, M.; Bailer, R.T.; Doria-Rose, N.A.; et al. Residue-Level Prediction of Hiv-1 Antibody Epitopes Based on Neutralization of Diverse Viral Strains. J. Virol. 2013, 87, 10047–10058. [Google Scholar] [CrossRef]
- Falkowska, E.; Le, K.M.; Ramos, A.; Doores, K.J.; Lee, J.H.; Blattner, C.; Ramirez, A.; Derking, R.; van Gils, M.J.; Liang, C.H.; et al. Broadly Neutralizing Hiv Antibodies Define a Glycan-Dependent Epitope on the Prefusion Conformation of Gp41 on Cleaved Envelope Trimers. Immunity 2014, 40, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Georgiev, I.S.; Gordon Joyce, M.; Zhou, T.; Kwong, P.D. Elicitation of Hiv-1-Neutralizing Antibodies against the Cd4-Binding Site. Curr. Opin. HIV AIDS 2013, 8, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Kang, B.H.; Pancera, M.; Lee, J.H.; Tong, T.; Feng, Y.; Imamichi, H.; Georgiev, I.S.; Chuang, G.Y.; Druz, A.; et al. Broad and Potent Hiv-1 Neutralization by a Human Antibody That Binds the Gp41-Gp120 Interface. Nature 2014, 515, 138–142. [Google Scholar] [CrossRef]
- Huang, J.; Ofek, G.; Laub, L.; Louder, M.K.; Doria-Rose, N.A.; Longo, N.S.; Imamichi, H.; Bailer, R.T.; Chakrabarti, B.K.; Sharma, S.K.; et al. Broad and Potent Neutralization of Hiv-1 by a Gp41-Specific Human Antibody. Nature 2012, 491, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Program, N.C.S.; Lynch, R.M.; Zhou, T.; Gao, F.; Alam, S.M.; Boyd, S.D.; Fire, A.Z.; Roskin, K.M.; Schramm, C.A.; et al. Co-Evolution of a Broadly Neutralizing Hiv-1 Antibody and Founder Virus. Nature 2013, 496, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.M.; Huber, M.; Doores, K.J.; Falkowska, E.; Pejchal, R.; Julien, J.-P.; Wang, S.-K.; Ramos, A.; Chan-Hui, P.-Y.; Moyle, M.; et al. Broad Neutralization Coverage of Hiv by Multiple Highly Potent Antibodies. Nature 2011, 477, 466–470. [Google Scholar] [CrossRef]
- Walker, L.M.; Phogat, S.K.; Chan-Hui, P.-Y.; Wagner, D.; Phung, P.; Goss, J.L.; Wrin, T.; Simek, M.D.; Fling, S.; Mitcham, J.L.; et al. Broad and Potent Neutralizing Antibodies from an African Donor Reveal a New Hiv-1 Vaccine Target. Science 2009, 326, 285–289. [Google Scholar] [CrossRef]
- Wu, X.; Yang, Z.-Y.; Li, Y.; Hogerkorp, C.-M.; Schief, W.R.; Seaman, M.S.; Zhou, T.; Schmidt, S.D.; Wu, L.; Xu, L.; et al. Rational Design of Envelope Identifies Broadly Neutralizing Human Monoclonal Antibodies to Hiv-1. Science 2010, 329, 856–861. [Google Scholar] [CrossRef]
- Brown, B.K.; Wieczorek, L.; Sanders-Buell, E.; Borges, A.R.; Robb, M.L.; Birx, D.L.; Michael, N.L.; McCutchan, F.E.; Polonis, V.R. Cross-Clade Neutralization Patterns among Hiv-1 Strains from the Six Major Clades of the Pandemic Evaluated and Compared in Two Different Models. Virology 2008, 375, 529–538. [Google Scholar] [CrossRef]
- Kim, J.; Jobe, O.; Peachman, K.K.; Michael, N.L.; Robb, M.L.; Rao, M.; Rao, V.B. Quantitative Analyses Reveal Distinct Sensitivities of the Capture of Hiv-1 Primary Viruses and Pseudoviruses to Broadly Neutralizing Antibodies. Virology 2017, 508, 188–198. [Google Scholar] [CrossRef]
- Jobe, O.; Peachman, K.K.; Matyas, G.R.; Asher, L.V.; Alving, C.R.; Rao, M. An Anti-Phosphoinositide-Specific Monoclonal Antibody That Neutralizes Hiv-1 Infection of Human Monocyte-Derived Macrophages. Virology 2012, 430, 110–119. [Google Scholar] [CrossRef]
- Chackerian, B.; Long, E.M.; Luciw, P.A.; Overbaugh, J. Human Immunodeficiency Virus Type 1 Coreceptors Participate in Postentry Stages in the Virus Replication Cycle and Function in Simian Immunodeficiency Virus Infection. J. Virol. 1997, 71, 3932–3939. [Google Scholar] [CrossRef]
- Robb, M.L.; Eller, L.A.; Kibuuka, H.; Rono, K.; Maganga, L.; Nitayaphan, S.; Kroon, E.; Sawe, F.K.; Sinei, S.; Sriplienchan, S.; et al. Prospective Study of Acute Hiv-1 Infection in Adults in East Africa and Thailand. N. Engl. J. Med. 2016, 374, 2120–2130. [Google Scholar] [CrossRef]
- Landais, E.; Murrell, B.; Briney, B.; Murrell, S.; Rantalainen, K.; Berndsen, Z.T.; Ramos, A.; Wickramasinghe, L.; Smith, M.L.; Eren, K.; et al. Hiv Envelope Glycoform Heterogeneity and Localized Diversity Govern the Initiation and Maturation of a V2 Apex Broadly Neutralizing Antibody Lineage. Immunity 2017, 47, 990–1003.e9. [Google Scholar] [CrossRef]
- Wieczorek, L.; Krebs, S.J.; Kalyanaraman, V.; Whitney, S.; Tovanabutra, S.; Moscoso, C.G.; Sanders-Buell, E.; Williams, C.; Slike, B.; Molnar, S.; et al. Comparable Antigenicity and Immunogenicity of Oligomeric Forms of a Novel, Acute Hiv-1 Subtype C Gp145 Envelope for Use in Preclinical and Clinical Vaccine Research. J. Virol. 2015, 89, 7478–7493. [Google Scholar] [CrossRef][Green Version]
- Lehmann, M.J.; Sherer, N.M.; Marks, C.B.; Pypaert, M.; Mothes, W. Actin- and Myosin-Driven Movement of Viruses Along Filopodia Precedes Their Entry into Cells. J. Cell Biol. 2005, 170, 317–325. [Google Scholar] [CrossRef]
- Sherer, N.M.; Lehmann, M.J.; Jimenez-Soto, L.F.; Horensavitz, C.; Pypaert, M.; Mothes, W. Retroviruses Can Establish Filopodial Bridges for Efficient Cell-to-Cell Transmission. Nat. Cell Biol. 2007, 9, 310–315. [Google Scholar] [CrossRef]
- Endres, T.; Lampe, M.; Briggs, J.A.G.; Kräusslich, H.-G.; Bräuchle, C.; Müller, B.; Lamb, D.C. Hiv-1-Cellular Interactions Analyzed by Single Virus Tracing. Eur. Biophys. J. 2008, 37, 1291–1301. [Google Scholar] [CrossRef]
- Burckhardt, C.J.; Greber, U.F. Virus Movements on the Plasma Membrane Support Infection and Transmission between Cells. PLoS Pathog. 2009, 5, e1000621. [Google Scholar] [CrossRef]
- Sherer, N.M.; Jin, J.; Mothes, W. Directional Spread of Surface-Associated Retroviruses Regulated by Differential Virus-Cell Interactions. J. Virol. 2010, 84, 3248–3258. [Google Scholar] [CrossRef]
- Kukura, P.; Ewers, H.; Müller, C.; Renn, A.; Helenius, A.; Sandoghdar, V. High-Speed Nanoscopic Tracking of the Position and Orientation of a Single Virus. Nat. Methods 2009, 6, 923–927. [Google Scholar] [CrossRef]
- Chand, S.; Messina, E.L.; AlSalmi, W.; Ananthaswamy, N.; Gao, G.; Uritskiy, G.; Padilla-Sanchez, V.; Mahalingam, M.; Peachman, K.K.; Robb, M.L.; et al. Glycosylation and Oligomeric State of Envelope Protein Might Influence Hiv-1 Virion Capture by Alpha4beta7 Integrin. Virology 2017, 508, 199–212. [Google Scholar] [CrossRef]
- Blumenthal, R.; Durell, S.R.; Viard, M. Hiv Entry and Envelope Glycoprotein-Mediated Fusion. J. Biol. Chem. 2012, 287, 40841–40849. [Google Scholar] [CrossRef]
- Herold, N.; Anders-Osswein, M.; Glass, B.; Eckhardt, M.; Müller, B.; Kräusslich, H.-G.; Anders-Ößwein, M. Hiv-1 Entry in Supt1-R5, Cem-Ss, and Primary Cd4+ T Cells Occurs at the Plasma Membrane and Does Not Require Endocytosis. J. Virol. 2014, 88, 13956–13970. [Google Scholar] [CrossRef]
- Fackler, O.T.; Peterlin, B. Endocytic Entry of Hiv-1. Curr. Biol. 2000, 10, 1005–1008. [Google Scholar] [CrossRef]
- Schaeffer, E.; Soros, V.B.; Greene, W.C. Compensatory Link between Fusion and Endocytosis of Human Immunodeficiency Virus Type 1 in Human Cd4 T Lymphocytes. J. Virol. 2004, 78, 1375–1383. [Google Scholar] [CrossRef]
- Daecke, J.; Fackler, O.T.; Dittmar, M.T.; Kräusslich, H.-G. Involvement of Clathrin-Mediated Endocytosis in Human Immunodeficiency Virus Type 1 Entry. J. Virol. 2005, 79, 1581–1594. [Google Scholar] [CrossRef]
- Miyauchi, K.; Kim, Y.; Latinovic, O.; Morozov, V.; Melikyan, G.B. Hiv Enters Cells Via Endocytosis and Dynamin-Dependent Fusion with Endosomes. Cell 2009, 137, 433–444. [Google Scholar] [CrossRef]
- Pritschet, K.; Donhauser, N.; Schuster, P.; Ries, M.; Haupt, S.; Kittan, N.A.; Korn, K.; Pöhlmann, S.; Holland, G.; Bannert, N.; et al. Cd4− and Dynamin-Dependent Endocytosis of Hiv-1 into Plasmacytoid Dendritic Cells. Virology 2012, 423, 152–164. [Google Scholar] [CrossRef]
- Van Wilgenburg, B.; Moore, M.D.; James, W.S.; Cowley, S.A. The Productive Entry Pathway of Hiv-1 in Macrophages Is Dependent on Endocytosis through Lipid Rafts Containing Cd4. PLoS ONE 2014, 9, e86071. [Google Scholar] [CrossRef]
- Maréchal, V.; Prevost, M.-C.; Petit, C.; Perret, E.; Heard, J.-M.; Schwartz, O. Human Immunodeficiency Virus Type 1 Entry into Macrophages Mediated by Macropinocytosis. J. Virol. 2001, 75, 11166–11177. [Google Scholar]
- Liu, J.; Ghneim, K.; Sok, D.; Bosche, W.J.; Li, Y.; Chipriano, E.; Berkemeier, B.; Oswald, K.; Borducchi, E.; Cabral, C.; et al. Antibody-Mediated Protection against Shiv Challenge Includes Systemic Clearance of Distal Virus. Science 2016, 353, 1045–1049. [Google Scholar] [CrossRef]
- Lu, M.; Ma, X.; Castillo-Menendez, L.R.; Gorman, J.; Alsahafi, N.; Ermel, U.; Terry, D.S.; Chambers, M.; Peng, D.; Zhang, B.; et al. Associating Hiv-1 Envelope Glycoprotein Structures with States on the Virus Observed by Smfret. Nature 2019, 568, 415–419. [Google Scholar] [CrossRef] [PubMed]
- White, T.A.; Bartesaghi, A.; Borgnia, M.J.; Meyerson, J.R.; De La Cruz, M.J.V.; Bess, J.W.; Nandwani, R.; Hoxie, J.A.; Lifson, J.D.; Milne, J.L.S.; et al. Molecular Architectures of Trimeric Siv and Hiv-1 Envelope Glycoproteins on Intact Viruses: Strain-Dependent Variation in Quaternary Structure. PLoS Pathog. 2010, 6, e1001249. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Rao, V.B.; Rao, M. Primary HIV-1 and Infectious Molecular Clones Are Differentially Susceptible to Broadly Neutralizing Antibodies. Vaccines 2020, 8, 782. https://doi.org/10.3390/vaccines8040782
Kim J, Rao VB, Rao M. Primary HIV-1 and Infectious Molecular Clones Are Differentially Susceptible to Broadly Neutralizing Antibodies. Vaccines. 2020; 8(4):782. https://doi.org/10.3390/vaccines8040782
Chicago/Turabian StyleKim, Jiae, Venigalla B. Rao, and Mangala Rao. 2020. "Primary HIV-1 and Infectious Molecular Clones Are Differentially Susceptible to Broadly Neutralizing Antibodies" Vaccines 8, no. 4: 782. https://doi.org/10.3390/vaccines8040782
APA StyleKim, J., Rao, V. B., & Rao, M. (2020). Primary HIV-1 and Infectious Molecular Clones Are Differentially Susceptible to Broadly Neutralizing Antibodies. Vaccines, 8(4), 782. https://doi.org/10.3390/vaccines8040782



