Characteristics of a Novel Target Antigen against Myeloma Cells for Immunotherapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Reagents
2.3. Detection of CXorf48 Gene Expression by RT–PCR
2.4. Detection of Cancer/Testis Antigen (CTA) Gene Expression by RT-PCR
2.5. Detection of CXorf48 Protein Expression by Immunocytochemical Staining
2.6. Treatment with Demethylating Agent
2.7. Generation of CXorf48-Specific CTLs from Human PBMNC
2.8. Detection of IFN-γ Secretion from CTLs by Enzyme-Linked Immunospot (ELISpot) Assay
2.9. Detection of Cytotoxicity of CTLs by Cytotoxicity Assay
2.10. Dextramer Staining
2.11. Statistical Analysis
3. Results
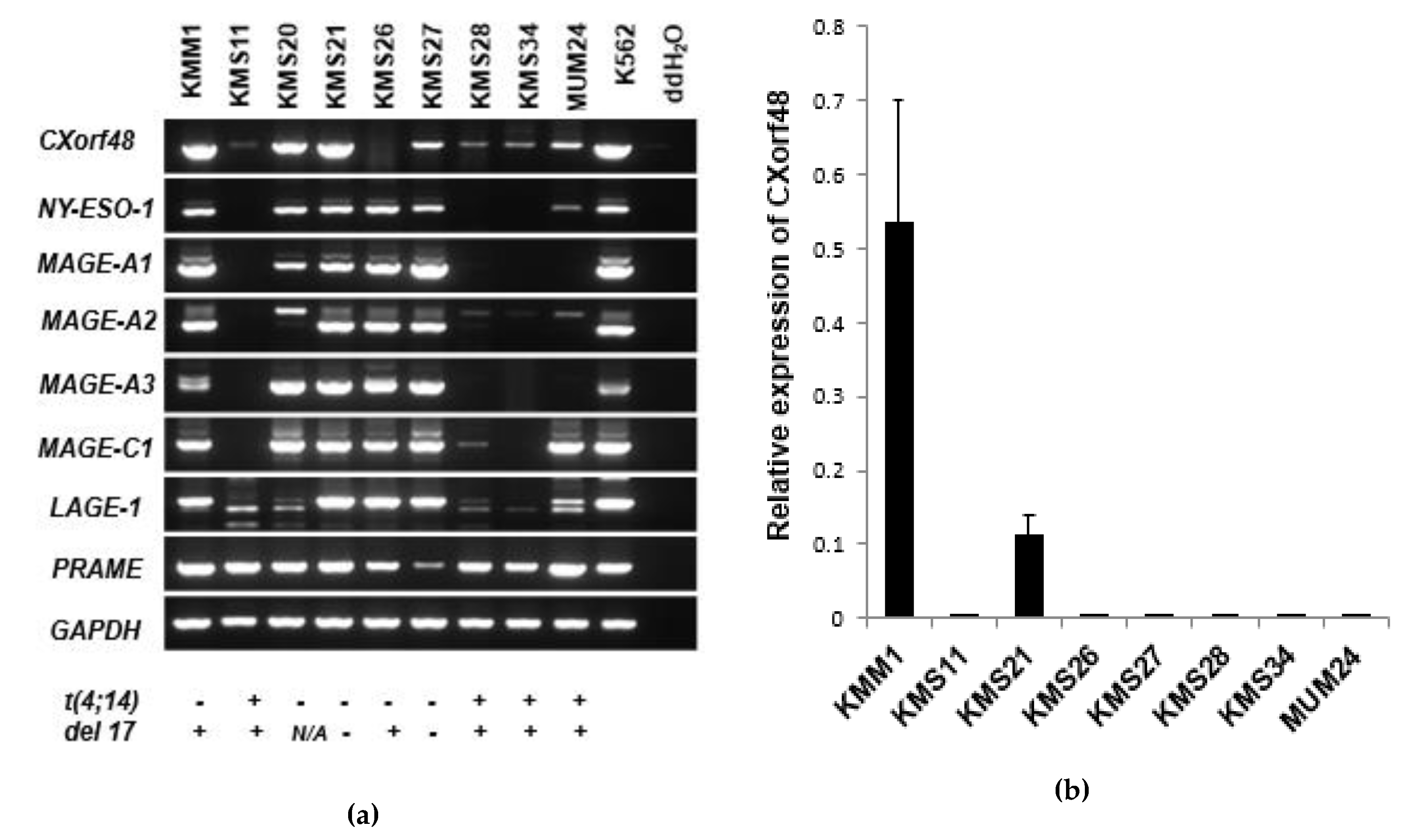
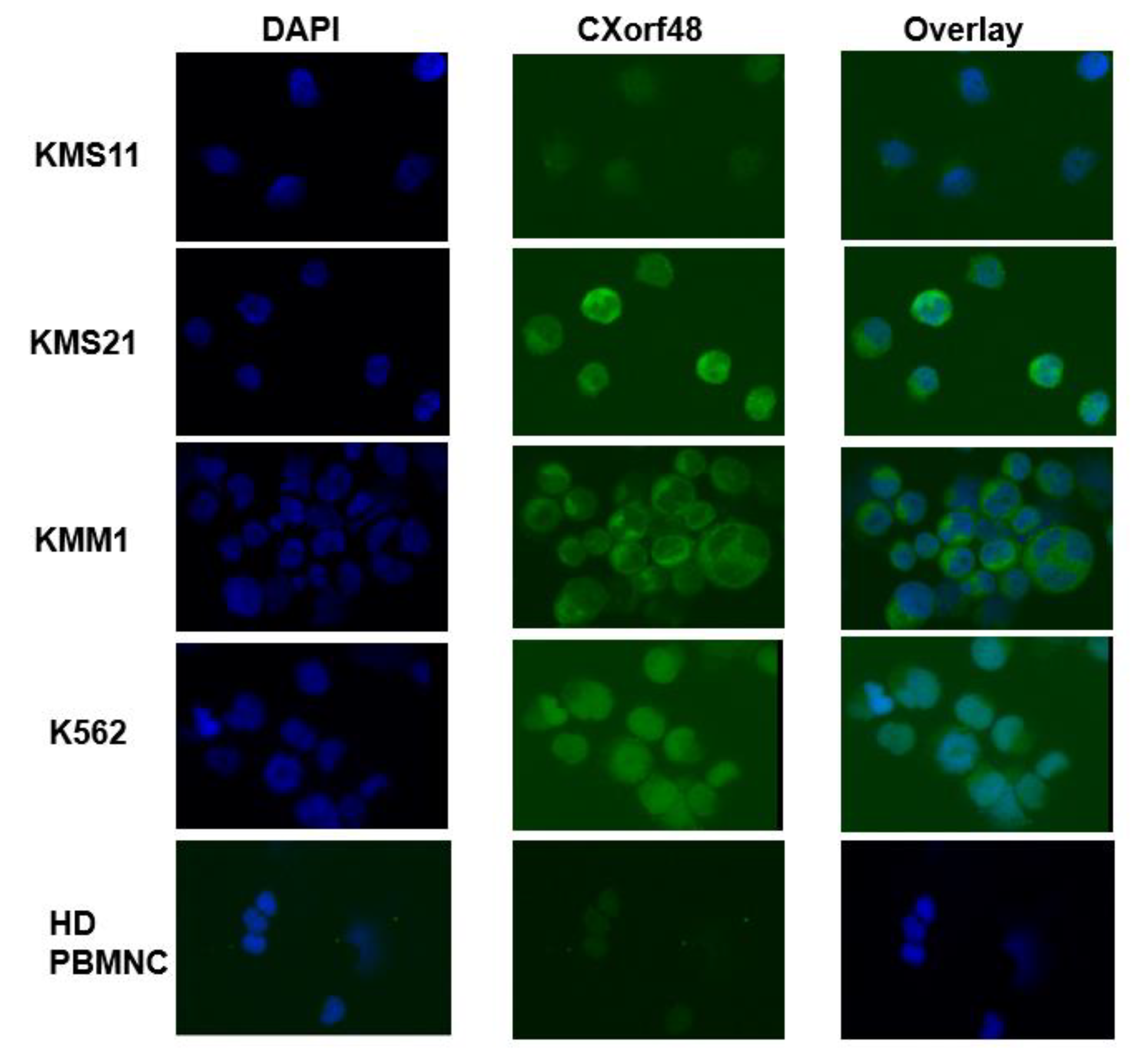
3.1. CXorf48 Is Expressed in Myeloma Cells
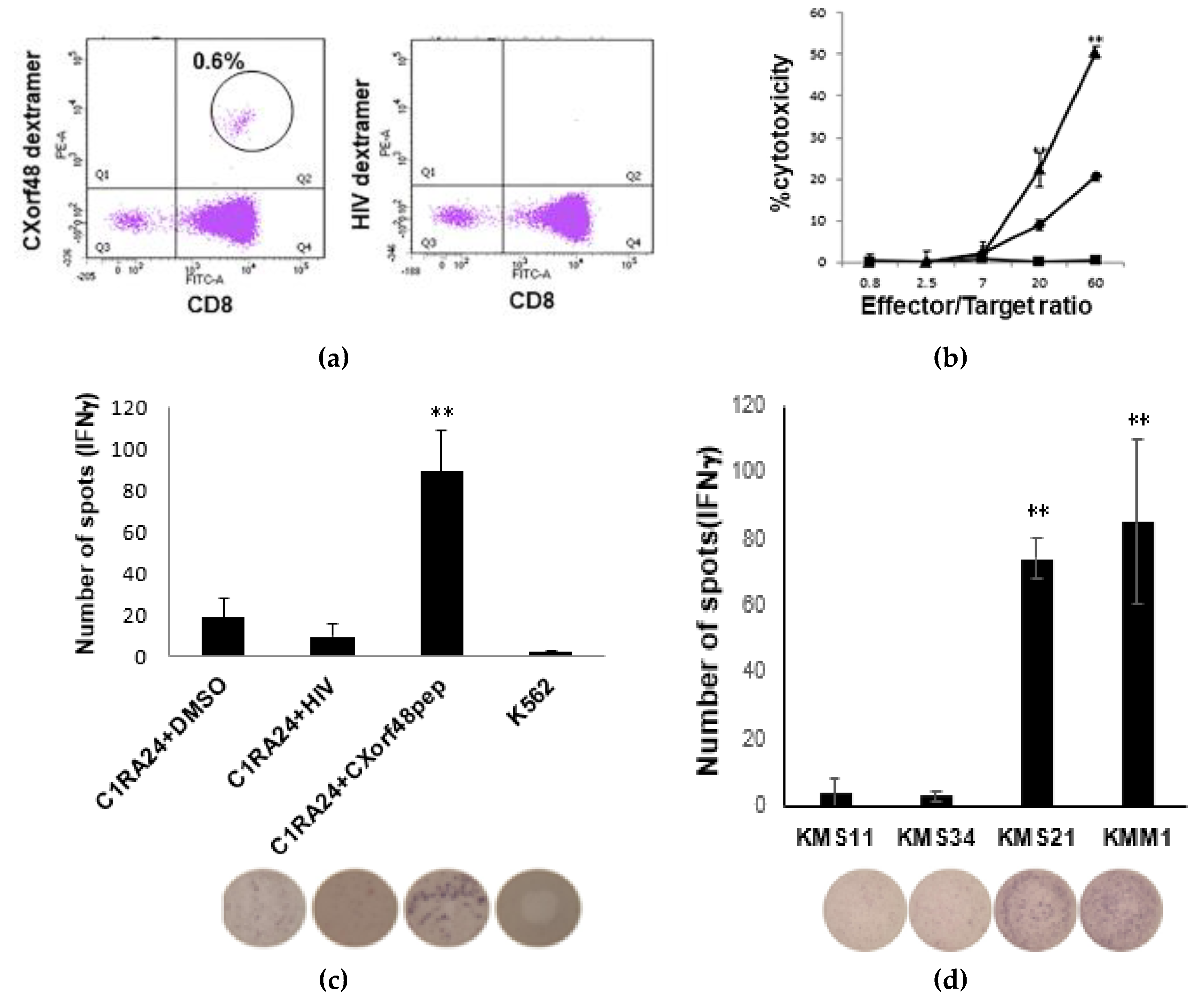
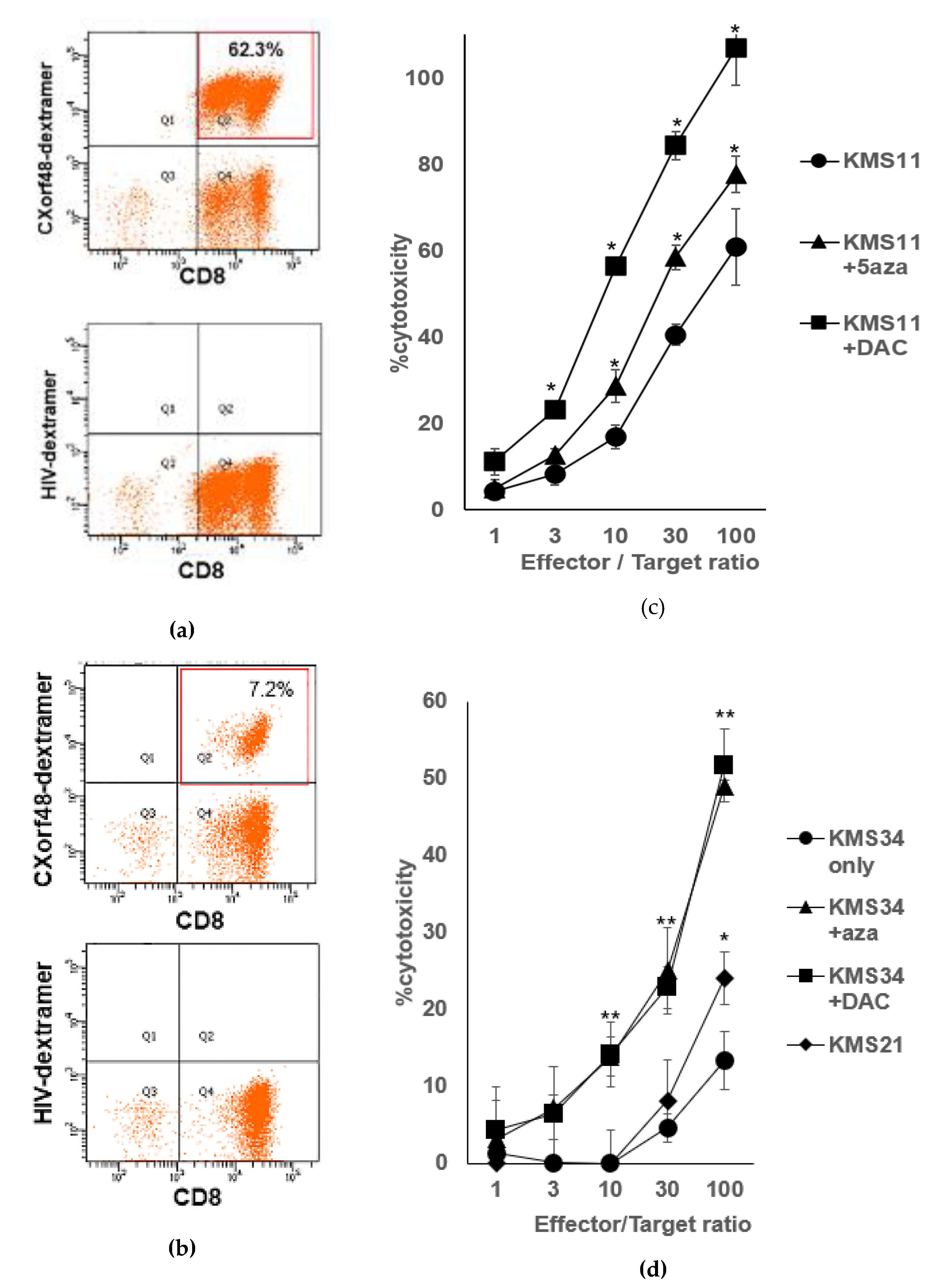
3.2. CXorf48-Specific CTLs Recognized Myeloma Cells with High Expression of CXorf48
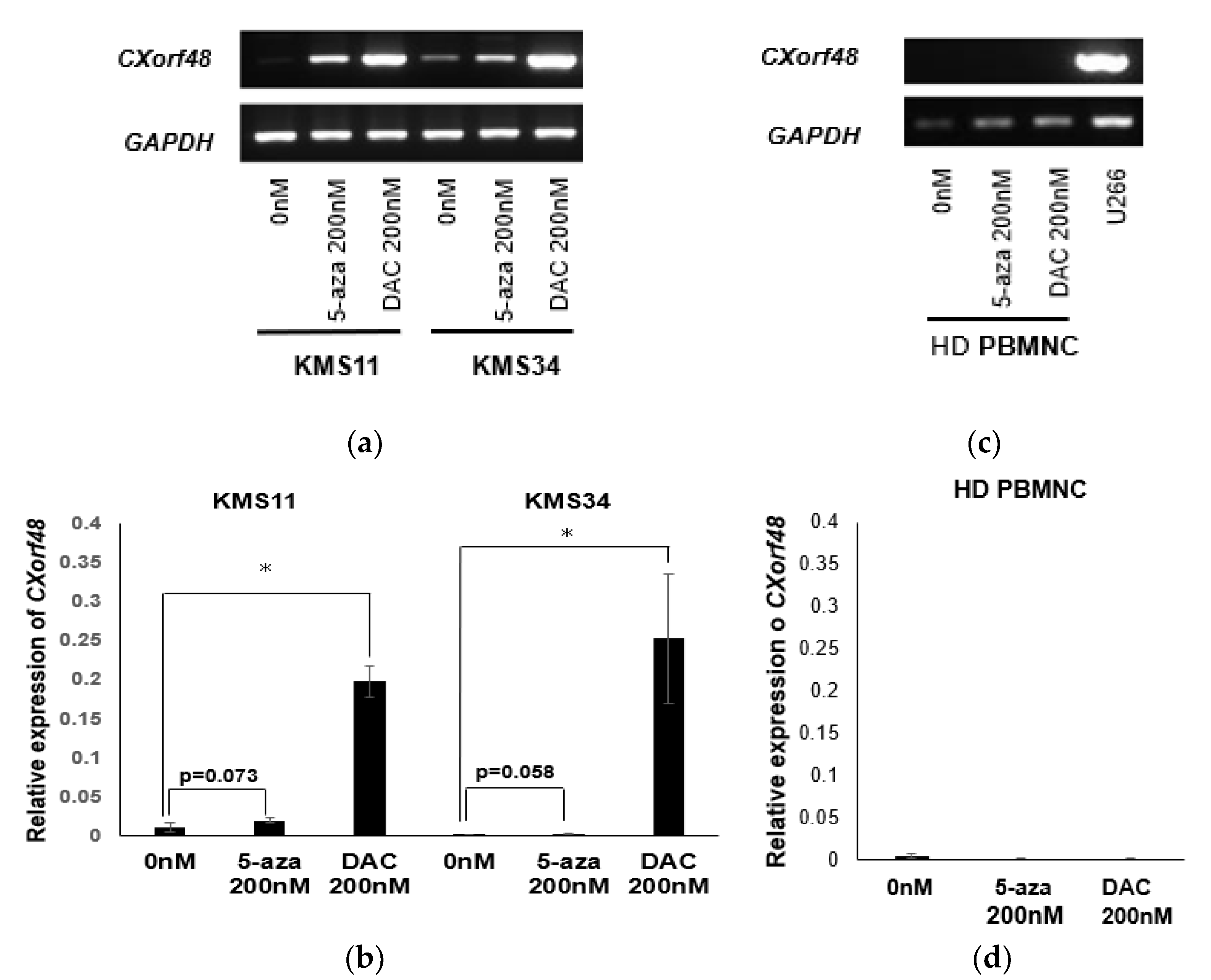
3.3. Up-regulation of CXorf48 Expression by Demethylating Agents
3.4. CXorf48-Specific CTL Recognized DMA-Treated Myeloma Cells with Low CXorf48 Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rajkumar, S.V.; Kumar, S. Multiple Myeloma: Diagnosis and Treatment. Mayo Clin. Proc. 2016, 91, 101–119. [Google Scholar] [CrossRef] [PubMed]
- Kazandjian, D. Multiple myeloma epidemiology and survival: A unique malignancy. Semin. Oncol. 2016, 43, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Pinto, V.; Bergantim, R.; Caires, H.R.; Seca, H.; Guimarães, J.E.; Vasconcelos, M.H. Multiple Myeloma: Available Therapies and Causes of Drug Resistance. Cancers 2020, 12, 407. [Google Scholar] [CrossRef] [PubMed]
- Abramson, H.N. Monoclonal Antibodies for the Treatment of Multiple Myeloma: An Update. Int. J. Mol. Sci. 2018, 19, 3924. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, B.; Hari, P.N.; Usmani, S.Z.; Hamadani, M. Chimeric antigen receptor T cell therapy in multiple myeloma: Promise and challenges. Bone Marrow Transplant. 2020. [Google Scholar] [CrossRef] [PubMed]
- Mailankody, S.; Korde, N.; Lesokhin, A.M.; Lendvai, N.; Hassoun, H.; Stetler-Stevenson, M.; Landgren, O. Minimal residual disease in multiple myeloma: Bringing the bench to the bedside. Nat. Rev. Clin. Oncol. 2015, 12, 286–295. [Google Scholar] [CrossRef]
- Munshi, N.C.; Avet-Loiseau, H.; Rawstron, A.C.; Owen, R.G.; Child, J.A.; Thakurta, A.; Sherrington, P.; Samur, M.K.; Georgieva, A.; Anderson, K.C.; et al. Association of Minimal Residual Disease With Superior Survival Outcomes in Patients With Multiple Myeloma: A Meta-analysis. JAMA Oncol. 2017, 3, 28–35. [Google Scholar] [CrossRef]
- Oka, Y.; Tsuboi, A.; Nakata, J.; Nishida, S.; Hosen, N.; Kumanogoh, A.; Oji, Y.; Sugiyama, H. Wilms’ Tumor Gene 1 (WT1) Peptide Vaccine Therapy for Hematological Malignancies: From CTL Epitope Identification to Recent Progress in Clinical Studies Including a Cure-Oriented Strategy. Oncol. Res. Treat. 2017, 40, 682–690. [Google Scholar] [CrossRef]
- Ghobrial, I.; Cruz, C.H.; Garfall, A.; Shah, N.; Munshi, N.; Kaufman, J.; Boise, L.H.; Morgan, G.; Adalsteinsson, V.A.; Manier, S.; et al. Immunotherapy in Multiple Myeloma: Accelerating on the Path to the Patient. Clin. Lymphoma Myeloma Leuk. 2019, 19, 332–344. [Google Scholar] [CrossRef]
- Lu, C.; Meng, S.; Jin, Y.; Zhang, W.; Li, Z.; Wang, F.; Wang-Johanning, F.; Wei, Y.; Liu, H.; Tu, H.; et al. A novel multi-epitope vaccine from MMSA-1 and DKK1 for multiple myeloma immunotherapy. Br. J. Haematol. 2017, 178, 413–426. [Google Scholar] [CrossRef]
- Matsushita, M.; Ozawa, K.; Suzuki, T.; Nakamura, M.; Nakano, N.; Kanchi, S.; Ichikawa, D.; Matsuki, E.; Sakurai, M.; Karigane, D.; et al. CXorf48 is a potential therapeutic target for achieving treatment-free remission in CML patients. Blood Cancer J. 2017, 7, e601. [Google Scholar] [CrossRef] [PubMed]
- Hattori, Y.; Du, W.; Yamada, T.; Ichikawa, D.; Matsunami, S.; Matsushita, M. A myeloma cell line established from a patient refractory to thalidomide therapy revealed high-risk cytogenetic abnormalities and produced vascular endothelial growth factor. Blood Cancer J. 2013, 3, e115. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Matsushita, M.; Otsuka, Y.; Tsutsumida, N.; Tanaka, C.; Uchiumi, A.; Ozawa, K.; Suzuki, T.; Ichikawa, D.; Aburatani, H.; Okamoto, S.; et al. Identification of Novel HLA-A*24:02-Restricted Epitope Derived from a Homeobox Protein Expressed in Hematological Malignancies. PLoS ONE 2016, 11, e0146371. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Bartlett, D.L.; Gorry, M.C.; O’Malley, M.E.; Guo, Z.S. Three epigenetic drugs up-regulate homeobox gene Rhox5 in cancer cells through overlapping and distinct molecular mechanisms. Mol. Pharmacol. 2009, 76, 1072–1081. [Google Scholar] [CrossRef]
- Cruz, C.R.; Gerdemann, U.; Leen, A.M.; Shafer, J.A.; Ku, S.; Tzou, B.; Horton, T.M.; Sheehan, A.; Copeland, A.; Younes, A.; et al. Improving T-cell therapy for relapsed EBV-negative Hodgkin lymphoma by targeting upregulated MAGE-A4. Clin. Cancer Res. 2011, 17, 7058–7066. [Google Scholar] [CrossRef]
- Li, L.; Goedegebuure, S.P.; Gillanders, W.E. Preclinical and clinical development of neoantigen vaccines. Ann. Oncol. 2017, 28, xii11–xii17. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Paul, B.; Kang, S.; Zheng, Z.; Kang, Y. The challenges of checkpoint inhibition in the treatment of multiple myeloma. Cell Immunol. 2018, 334, 87–98. [Google Scholar] [CrossRef]
- Gjerstorff, M.F.; Andersen, M.H.; Ditzel, H.J. Oncogenic cancer/testis antigens: Prime candidates for immunotherapy. Oncotarget 2015, 6, 15772–15787. [Google Scholar] [CrossRef]
- Tsuboi, A.; Oka, Y.; Nakajima, H.; Fukuda, Y.; Elisseeva, O.A.; Yoshihara, S.; Hosen, N.; Ogata, A.; Kito, K.; Fujiki, F.; et al. Wilms tumor gene WT1 peptide-based immunotherapy induced a minimal response in a patient with advanced therapy-resistant multiple myeloma. Int. J. Hematol. 2007, 86, 414–417. [Google Scholar] [CrossRef]
- Dawood, R.M.; Moustafa, R.I.; Abdelhafez, T.H.; El-Shenawy, R.; El-Abd, Y.; Bader El Din, N.G.; Dubuisson, J.; El Awady, M.K. A multiepitope peptide vaccine against HCV stimulates neutralizing humoral and persistent cellular responses in mice. BMC Infect Dis. 2019, 19, 932. [Google Scholar] [CrossRef] [PubMed]
- Zamunér, F.T.; Karia, B.T.; de Oliveira, C.Z.; Santos, C.R.; Carvalho, A.L.; Vettore, A.L. A Comprehensive Expression Analysis of Cancer Testis Antigens in Head and Neck Squamous Cell Carcinoma Revels MAGEA3/6 as a Marker for Recurrence. Mol. Cancer Ther. 2015, 14, 828–834. [Google Scholar] [CrossRef] [PubMed]
- Santra, M.; Zhan, F.; Tian, E.; Barlogie, B.; Shaughnessy, J. A subset of multiple myeloma harboring the t(4;14)(p16;q32) translocation lacks FGFR3 expression but maintains an IGH/MMSET fusion transcript. Blood 2003, 101, 2374–2376. [Google Scholar] [CrossRef]
- Chesi, M.; Nardini, E.; Brents, L.A.; Schröck, E.; Ried, T.; Kuehl, W.M.; Bergsagel, P.L. Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat. Genet. 1997, 16, 260–264. [Google Scholar] [CrossRef]
- Matsushita, M.; Yamazaki, R.; Ikeda, H.; Kawakami, Y. Preferentially expressed antigen of melanoma (PRAME) in the development of diagnostic and therapeutic methods for hematological malignancies. Leuk. Lymphoma 2003, 44, 439–444. [Google Scholar] [CrossRef]
- Nguyen, L.P.; Galtier, N.; Nabholz, B. Gene expression, chromosome heterogeneity and the fast-X effect in mammals. Boil. Lett. 2015, 11, 20150010. [Google Scholar] [CrossRef] [PubMed]
- Pujol, J.L.; De Pas, T.; Rittmeyer, A.; Vallières, E.; Kubisa, B.; Levchenko, E.; Wiesemann, S.; Masters, G.A.; Shen, R.; Tjulandin, S.A.; et al. Safety and Immunogenicity of the PRAME Cancer Immunotherapeutic in Patients with Resected Non-Small Cell Lung Cancer: A Phase I Dose Escalation Study. J. Thorac. Oncol. 2016, 11, 2208–2217. [Google Scholar] [CrossRef]
- Azizi, A.; Ediriwickrema, A.; Dutta, R.; Patel, S.A.; Shomali, W.; Medeiros, B.; Iberri, D.; Gotlib, J.; Mannis, G.; Greenberg, P.; et al. Venetoclax and hypomethylating agent therapy in high risk myelodysplastic syndromes: A retrospective evaluation of a real-world experience. Leuk. Lymphoma 2020, 16, 1–8. [Google Scholar] [CrossRef]
- Li, N.; Liu, L.; Xiang, P.; Liang, L.; Wang, J.; Wang, Y.; Luo, S.; Song, Y.; Fang, B. Addition of low-dose decitabine to bortezomib and dexamethasone as second-line therapy in multiple myeloma. Br. J. Haematol. 2020, 189, e258–e262. [Google Scholar] [CrossRef]
- Zhou, J.; Shen, Q.; Lin, H.; Hu, L.; Li, G.; Zhang, X. Decitabine shows potent anti-myeloma activity by depleting monocytic myeloid-derived suppressor cells in the myeloma microenvironment. J. Cancer. Res. Clin. Oncol. 2019, 145, 329–336. [Google Scholar] [CrossRef]
- Grieve, S.; Wajnberg, G.; Lees, M.; Chacko, S.; Weir, J.; Crapoulet, N.; Reiman, T. TAZ functions as a tumor suppressor in multiple myeloma by downregulating MYC. Blood Adv. 2019, 3, 3613–3625. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsushita, M.; Saito, S.; Yokoe, S.; Ichikawa, D.; Hattori, Y. Characteristics of a Novel Target Antigen against Myeloma Cells for Immunotherapy. Vaccines 2020, 8, 579. https://doi.org/10.3390/vaccines8040579
Matsushita M, Saito S, Yokoe S, Ichikawa D, Hattori Y. Characteristics of a Novel Target Antigen against Myeloma Cells for Immunotherapy. Vaccines. 2020; 8(4):579. https://doi.org/10.3390/vaccines8040579
Chicago/Turabian StyleMatsushita, Maiko, Saku Saito, Shinya Yokoe, Daiju Ichikawa, and Yutaka Hattori. 2020. "Characteristics of a Novel Target Antigen against Myeloma Cells for Immunotherapy" Vaccines 8, no. 4: 579. https://doi.org/10.3390/vaccines8040579
APA StyleMatsushita, M., Saito, S., Yokoe, S., Ichikawa, D., & Hattori, Y. (2020). Characteristics of a Novel Target Antigen against Myeloma Cells for Immunotherapy. Vaccines, 8(4), 579. https://doi.org/10.3390/vaccines8040579



