Current State of Global African Swine Fever Vaccine Development under the Prevalence and Transmission of ASF in China
Abstract
1. Introduction
2. Epidemic Situation of ASF in China
3. Factors of ASF Prevalence in China
3.1. Strong Environmental Resistance of ASFV
3.2. Role of Viral Circulation in the Occurrence of ASF
3.3. Role of Human Activity in the Transmission of ASFV
3.4. Analysis on Breeding Pattern of Diseased Pig Farms
4. Chinese Model of ASF Control Strategy
- (1)
- Introduction control. Introduced breeding pigs must be isolated for more than 30 days and tested clinically and laboratory to ensure that ASFV was negative.
- (2)
- Transport vehicle control. Establishing a standardized cleaning and disinfection system and specifications for transport vehicles, and separate and specific trucks are recommended to transport the pig and material.
- (3)
- Control of items entering pigsties. Strict disinfection treatment of items entering pig farms by fumigation, ozone and other methods.
- (4)
- Personnel control. Personnel inside the pig farm should reduce their outgoing activities and isolation measures for admission personnel should be implement strictly.
- (5)
- Feed control. Eliminate potential sources of contamination of feed.
- (6)
- Environmental control. The surrounding environment of pig farms should be regularly monitored, detected and evaluated.
5. Current State of ASF Vaccine Development
5.1. Inactivated Vaccines
5.2. Live Attenuated Vaccines (LAVs)
5.2.1. Conventional LAVs
5.2.2. Recombinant LAVs/Gene-Deleted Vaccines
5.3. Subunit and DNA Vaccines
5.4. Virus Vectored Vaccines
5.5. Combined Vaccination Strategy
6. Discussion and Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ASF | African swine fever |
| ASFV | African swine fever virus |
| OIE | The World Organization for Animal Health |
| MGF | Multigene families |
| MARA | Ministry of Agriculture and Rural Affairs of the People’s Republic of China |
| CSF | Classical Swine Fever |
| HRP | High-risk periods |
| TADs | Transboundary animal diseases |
| DIVA | Differentiating infected from vaccinated animals |
| LAVs | Live attenuated vaccines |
| FACS | Fluorescence-activated cells sorting |
| PBMs | Peripheral blood monocytes |
| WSL-R | Spontaneously immortalized wild boar cell line |
| COS-7/COS-1 | A monkey cell line transformed with the large antigen of SV40 |
| PAMs | Pulmonary alveolar macrophage |
| CTL | Cytotoxic T lymphocyte |
| SLA-II | Swine leukocyte antigen II |
| ORFs | Open reading frames |
| ADE | Antibody-dependent enhancement |
| MVA | Modified vaccinia virus Ankara |
| VACV | Vaccinia virus |
| r Ad | Recombinant adenoviruses |
References
- Gaudreault, N.N.; Madden, D.W.; Wilson, W.C.; Trujillo, J.D.; Richt, J.A. African Swine Fever Virus: An Emerging DNA Arbovirus. Front. Vet. Sci. 2020, 7, 215. [Google Scholar] [CrossRef] [PubMed]
- Pietschmann, J.; Guinat, C.; Beer, M.; Pronin, V.; Tauscher, K.; Petrov, A.; Keil, G.; Blome, S. Course and transmission characteristics of oral low-dose infection of domestic pigs and European wild boar with a Caucasian African swine fever virus isolate. Arch. Virol. 2015, 160, 1657–1667. [Google Scholar] [CrossRef] [PubMed]
- Dixon, L.K.; Chapman, D.A.; Netherton, C.L.; Upton, C. African swine fever virus replication and genomics. Virus Res. 2013, 173, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Alejo, A.; Matamoros, T.; Guerra, M.; Andrés, G. A Proteomic Atlas of the African Swine Fever Virus Particle. J. Virol. 2018, 92, e01293-18. [Google Scholar] [CrossRef] [PubMed]
- Dixon, L.K.; Sun, H.; Roberts, H. African swine fever. Antivir. Res. 2019, 165, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Galindo, I.; Alonso, C. African Swine Fever Virus: A Review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef]
- Dixon, L.K.; Islam, M.; Nash, R.; Reis, A.L. African swine fever virus evasion of host defences. Virus Res. 2019, 266, 25–33. [Google Scholar] [CrossRef]
- Malogolovkin, A.; Burmakina, G.; Titov, I.; Sereda, A.; Gogin, A.; Baryshnikova, E.; Kolbasov, D. Comparative Analysis of African Swine Fever Virus Genotypes and Serogroups. Emerg. Infect. Dis. 2015, 21, 312–315. [Google Scholar] [CrossRef]
- Rowlands, R.J.; Michaud, V.; Heath, L.; Hutchings, G.; Oura, C.; Vosloo, W.; Dwarka, R.; Onashvili, T.; Albina, E.; Dixon, L.K. African Swine Fever Virus Isolate, Georgia, 2007. Emerg. Infect. Dis. 2008, 14, 1870–1874. [Google Scholar] [CrossRef]
- Costard, S.; Wieland, B.; De Glanville, W.; Jori, F.; Rowlands, R.; Vosloo, W.; Roger, F.; Pfeiffer, D.U.; Dixon, L.K. African swine fever: How can global spread be prevented? Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 2683–2696. [Google Scholar] [CrossRef]
- Wen, X.; He, X.; Zhang, X.; Zhang, X.; Liu, L.; Guan, Y.; Zhang, Y.; Bu, Z. Genome sequences derived from pig and dried blood pig feed samples provide important insights into the transmission of African swine fever virus in China in 2018. Emerg. Microbes Infect. 2019, 8, 303–306. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Liu, R.; Zhang, X.; Li, F.; Wang, J.; Zhang, J.; Liu, X.; Wang, L.; Zhang, J.; Wu, X.; et al. Replication and virulence in pigs of the first African swine fever virus isolated in China. Emerg. Microbes Infect. 2019, 8, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, N.; Luo, Y.; Liu, Y.; Miao, F.; Chen, T.; Zhang, S.; Cao, P.; Li, X.; Tian, K.; et al. Emergence of African Swine Fever in China, 2018. Transbound. Emerg. Dis. 2018, 65, 1482–1484. [Google Scholar] [CrossRef] [PubMed]
- Gaudreault, N.N.; Richt, J.A. Subunit Vaccine Approaches for African Swine Fever Virus. Vaccines (Basel) 2019, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- OIE, Reports on ASF: OIE-World Organisation for Animal Health. Available online: https://www.oie.int/en/animal-health-in-the-world/information-on-aquatic-and-terrestrial-animal-diseases/african-swine-fever/reports-on-asf/ (accessed on 22 August 2020).
- OIE, Global Situation of ASF. Available online: https://www.oie.int/fileadmin/Home/eng/Animal_Health_in_the_World/docs/pdf/Disease_cards/ASF/Report_47_Global_situation_ASF.pdf (accessed on 22 August 2020).
- Liu, J.; Liu, B.; Shan, B.; Wei, S.; An, T.; Shen, G.; Chen, Z. Prevalence of African Swine Fever in China, 2018–2019. J. Med. Virol. 2019, 92, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- MARA, Information on the Epidemic of ASF. Available online: http://www.moa.gov.cn/ztzl/fzzwfk/yqxx/ (accessed on 21 August 2020).
- Tao, D.; Sun, D.; Liu, Y.; Wei, S.; Yang, Z.; An, T.; Shan, F.; Chen, Z.; Liu, J. One Year of African Swine Fever Outbreak in China. Acta Trop. 2020, 211, 105602. [Google Scholar] [CrossRef]
- Ma, Y.; Han, Y.; Jin, X.; Li, Y.; Qiu, H. Research progress, difficulties and breakthrough points of African swine fever vaccine. China J. Prev. Vet. Med. 2020, 1–6. (In Chinese). Available online: http://tow.cnki.net/kcms/detail/detail.aspx?filename=ZGXQ20200608007&dbcode=CRJT_CJFD&dbname=CAPJLAST&v= (accessed on 15 September 2020).
- National Bureau of China, Statistics of the Number of Live Pigs and Pork Production in China in Q1 in 2020. Available online: https://data.stats.gov.cn/easyquery.htm?cn=C01 (accessed on 21 August 2020).
- Gong, L.; Xu, R.; Wang, Z.; Deng, Q.; Wang, H.; Zhang, G. African swine fever recovery in China. Vet. Med. Sci. 2020. [Google Scholar] [CrossRef]
- China Daily.com.cn, ASFV May Affect Pork for Several Years. Available online: https://www.chinadaily.com.cn (accessed on 14 August 2020).
- Mazur-Panasiuk, N.; Woźniakowski, G. Natural inactivation of African swine fever virus in tissues: Influence of temperature and environmental conditions on virus survival. Vet. Microbiol. 2020, 242, 108609. [Google Scholar] [CrossRef]
- Olesen, A.S.; Lohse, L.; Boklund, A.; Halasa, T.; Gallardo, C.; Pejsak, Z.; Belsham, G.J.; Rasmussen, T.B.; Bøtner, T.A. Transmission of African swine fever virus from infected pigs by direct contact and aerosol routes. Vet. Microbiol. 2017, 211, 92–102. [Google Scholar] [CrossRef]
- Olesen, A.S.; Lohse, L.; Boklund, A.; Halasa, T.; Belsham, G.J.; Rasmussen, T.B.; Bøtner, T.A. Short time window for transmissibility of African swine fever virus from a contaminated environment. Transbound. Emerg. Dis. 2018, 65, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Eblé, P.L.; Hagenaars, T.; Weesendorp, E.; Quak, S.; Moonen-Leusen, H.W.; Loeffen, W.L.A. Transmission of African Swine Fever Virus via carrier (survivor) pigs does occur. Vet. Microbiol. 2019, 237, 108345. [Google Scholar] [CrossRef] [PubMed]
- Cwynar, P.; Stojkov, J.; Wlazlak, K. African Swine Fever Status in Europe. Viruses 2019, 11, 310. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ren, Z.; Wang, Q.; Ge, S.; Liu, Y.; Liu, C.; Liu, F.; Hu, Y.; Li, J.; Bao, J.; et al. Infection of African swine fever in wild boar, China, 2018. Transbound. Emerg. Dis. 2019, 66, 1395–1398. [Google Scholar] [CrossRef]
- Vergne, T.; Chen-Fu, C.; Li, S.; Cappelle, J.; Edwards, J.; Martin, V.; Pfeiffer, D.U.; Fusheng, G.; Roger, F.L. Pig empire under infectious threat: Risk of African swine fever introduction into the People’s Republic of China. Vet. Rec. 2017, 181, 117. [Google Scholar] [CrossRef]
- Probst, C.; Gethmann, J.; Amler, S.; Globig, A.; Knoll, B.; Conraths, F.J. The potential role of scavengers in spreading African swine fever among wild boar. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Costard, S.; Mur, L.; Lubroth, J.; Sánchez-Vizcaíno, J.M.; Pfeiffer, D.U. Epidemiology of African swine fever virus. Virus Res. 2013, 173, 191–197. [Google Scholar] [CrossRef]
- De Oliveira, R.P.; Hutet, E.; Duhayon, M.; Guionnet, J.-M.; Paboeuf, F.; Vial, L.; Le Potier, M. Successful Infection of Domestic Pigs by Ingestion of the European Soft Tick O. Erraticus That Fed on African Swine Fever Virus Infected Pig. Viruses 2020, 12, 300. [Google Scholar] [CrossRef]
- MARA, Press Conference on African Swine Fever. Available online: http://www.moa.gov.cn/ztzl/fzzwfk/gzdt/201903/t20190301_6173098.htm (accessed on 14 August 2020).
- Gao, X.; Liu, T.; Liu, Y.; Xiao, J.; Wang, H. Transmission of African swine fever in China Through Legal Trade of Live Pigs. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef]
- Liu, R.; Sun, Y.; Chai, Y.; Li, S.; Li, S.; Wang, L.; Su, J.; Yu, S.; Yan, J.; Gao, G.F.; et al. The structural basis of African swine fever virus pA104R binding to DNA and its inhibition by stilbene derivatives. Proc. Natl. Acad. Sci. USA 2020, 117, 11000–11009. [Google Scholar] [CrossRef]
- Li, Y.; Salman, M.; Shen, C.; Yang, H.; Wang, Y.; Jiang, Z.; Edwards, J.; Huang, B. African Swine Fever in a commercial pig farm: Outbreak investigation and an approach for identifying the source of infection. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Fan, Z.; Tian, H.; Sun, Y.; Feng, F.; Zhou, H.; Guo, K. Epidemiological profile and prevention and control strategy of African swine fever. Chin. J. Vet. Med. 2019, 39, 1027–1034. [Google Scholar] [CrossRef]
- MARA, Emergency Response to Epidemics. Available online: http://www.moa.gov.cn/gk/yjgl_1/yqfb (accessed on 14 August 2020).
- Yang, H. Swine Disease Prevention and Control Strategy in the Context of African Swine Fever. Vet. Orientat. 2020, 4–5. (In Chinese) [Google Scholar]
- Chen, W.; Zhao, D.; He, X.; Liu, R.; Wang, Z.; Zhang, X.; Li, F.; Shan, D.; Chen, H.; Zhang, J.; et al. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Sci. China Life Sci. 2020, 63, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Goatley, L.C.; Reis, A.L.; Portugal, R.; Goldswain, H.; Shimmon, G.L.; Hargreaves, Z.; Ho, C.-S.; Montoya, M.; Sanchez-Cordon, P.; Taylor, G.; et al. A Pool of Eight Virally Vectored African Swine Fever Antigens Protect Pigs against Fatal Disease. Vaccines (Basel) 2020, 8, 234. [Google Scholar] [CrossRef]
- Stone, S.S.; Hess, W.R. Antibody response to inactivated preparations of African swine fever virus in pigs. Am. J. Vet.Res. 1967, 28, 475–481. [Google Scholar]
- Blome, S.; Gabriel, C.; Beer, M. Modern adjuvants do not enhance the efficacy of an inactivated African swine fever virus vaccine preparation. Vaccine 2014, 32, 3879–3882. [Google Scholar] [CrossRef]
- Tlaxca, J.L.; Ellis, S.; Remmele, R.L. Live attenuated and inactivated viral vaccine formulation and nasal delivery: Potential and challenges. Adv. Drug Deliv. Rev. 2015, 93, 56–78. [Google Scholar] [CrossRef]
- Krug, P.W.; Holinka, L.G.; O’Donnell, V.; Reese, B.; Sanford, B.; Fernandez-Sainz, I.; Gladue, D.P.; Arzt, J.; Rodriguez, L.; Risatti, G.R.; et al. The Progressive Adaptation of a Georgian Isolate of African Swine Fever Virus to Vero Cells Leads to a Gradual Attenuation of Virulence in Swine Corresponding to Major Modifications of the Viral Genome. J. Virol. 2014, 89, 2324–2332. [Google Scholar] [CrossRef]
- Leitão, A.; Cartaxeiro, C.; Coelho, R.; Cruz, B.; Parkhouse, R.M.E.; Portugal, F.C.; Vigário, J.D.; Martins, C.L.V. The non-haemadsorbing African swine fever virus isolate ASFV/NH/P68 provides a model for defining the protective anti-virus immune response. J. Gen. Virol. 2001, 82, 513–523. [Google Scholar] [CrossRef]
- King, K.; Chapman, D.; Argilaguet, J.M.; Fishbourne, E.; Hutet, E.; Cariolet, R.; Hutchings, G.; Oura, C.A.L.; Netherton, C.L.; Moffat, K.; et al. Protection of European domestic pigs from virulent African isolates of African swine fever virus by experimental immunisation. Vaccine 2011, 29, 4593–4600. [Google Scholar] [CrossRef] [PubMed]
- Netherton, C.L.; Goatley, L.C.; Reis, A.L.; Portugal, R.; Nash, R.H.; Morgan, S.B.; Gault, L.; Nieto, R.; Norlin, V.; Gallardo, C.; et al. Identification and Immunogenicity of African Swine Fever Virus Antigens. Front. Immunol. 2019, 10, 1318. [Google Scholar] [CrossRef]
- Reis, A.L.; Goatley, L.C.; Jabbar, T.; Lopez, E.; Rathakrishnan, A.; Dixon, L.K. Deletion of the Gene for the Type I Interferon Inhibitor I329L from the Attenuated African Swine Fever Virus OURT88/3 Strain Reduces Protection Induced in Pigs. Vaccines (Basel) 2020, 8, 262. [Google Scholar] [CrossRef]
- Granja, A.G.; Sabina, P.; Salas, M.L.; Fresno, M.; Revilla, Y. Regulation of Inducible Nitric Oxide Synthase Expression by Viral A238L-Mediated Inhibition of p65/RelA Acetylation and p300 Transactivation. J. Virol. 2006, 80, 10487–10496. [Google Scholar] [CrossRef] [PubMed]
- Barber, C.; Netherton, C.; Goatley, L.; Moon, A.; Goodbourn, S.; Dixon, L.K. Identification of residues within the African swine fever virus DP71L protein required for dephosphorylation of translation initiation factor eIF2α and inhibiting activation of pro-apoptotic CHOP. Virology 2017, 504, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Moon, A.; Childs, K.; Goodbourn, S.; Dixon, L.K. The African Swine Fever Virus DP71L Protein Recruits the Protein Phosphatase 1 Catalytic Subunit to Dephosphorylate eIF2α and Inhibits CHOP Induction but Is Dispensable for These Activities during Virus Infection. J. Virol. 2010, 84, 10681–10689. [Google Scholar] [CrossRef]
- Dixon, L.K.; Sanchez-Cordon, P.; Galindo, I.; Alonso, C. Investigations of Pro- and Anti-Apoptotic Factors Affecting African Swine Fever Virus Replication and Pathogenesis. Viruses 2017, 9, 241. [Google Scholar] [CrossRef]
- Sanchez-Cordon, P.; Jabbar, T.; Berrezaie, M.; Chapman, D.; Reis, A.; Sastre, P.; Rueda, P.; Goatley, L.; Dixon, L.K. Evaluation of protection induced by immunisation of domestic pigs with deletion mutant African swine fever virus BeninΔMGF by different doses and routes. Vaccine 2017, 36, 707–715. [Google Scholar] [CrossRef]
- O’Donnell, V.; Holinka, L.G.; Sanford, B.; Krug, P.W.; Carlson, J.; Pacheco, J.M.; Reese, B.; Risatti, G.R.; Gladue, D.P.; Borca, M.V. African swine fever virus Georgia isolate harboring deletions of 9GL and MGF360/505 genes is highly attenuated in swine but does not confer protection against parental virus challenge. Virus Res. 2016, 221, 8–14. [Google Scholar] [CrossRef]
- O’Donnell, V.; Holinka, L.G.; Gladue, D.P.; Sanford, B.; Krug, P.W.; Lu, X.; Arzt, J.; Reese, B.; Carrillo, C.; Risatti, G.R.; et al. African Swine Fever Virus Georgia Isolate Harboring Deletions of MGF360 and MGF505 Genes Is Attenuated in Swine and Confers Protection against Challenge with Virulent Parental Virus. J. Virol. 2015, 89, 6048–6056. [Google Scholar] [CrossRef]
- Sanna, G.; Giudici, S.D.; Bacciu, D.; Angioi, P.P.; Giammarioli, M.; Oggiano, A.; De Mia, G.M. Improved Strategy for Molecular Characterization of African Swine Fever Viruses from Sardinia, Based on Analysis of p30, CD2V and I73R / I329L Variable Regions. Transbound. Emerg. Dis. 2016, 64, 1280–1286. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, V.L.; Almeida, S.C.P.; Soares, H.R.; Crespo, A.; Marshall-Clarke, S.; Parkhouse, R.M.E. A novel TLR3 inhibitor encoded by African swine fever virus (ASFV). Arch. Virol. 2011, 156, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Banjara, S.; Shimmon, G.L.; Dixon, L.K.; Netherton, C.L.; Hinds, M.G.; Kvansakul, M. Crystal Structure of African Swine Fever Virus A179L with the Autophagy Regulator Beclin. Viruses 2019, 11, 789. [Google Scholar] [CrossRef] [PubMed]
- Banjara, S.; Caria, S.; Dixon, L.K.; Hinds, M.G.; Kvansakul, M. Structural Insight into African Swine Fever Virus A179L-Mediated Inhibition of Apoptosis. J. Virol. 2017, 91, e02228-16. [Google Scholar] [CrossRef]
- Hurtado, C.; Bustos, M.J.; Granja, A.G.; De León, P.; Sabina, P.; López-Viñas, E.; Gómez-Puertas, P.; Revilla, Y.; Carrascosa, A.L. The African swine fever virus lectin EP153R modulates the surface membrane expression of MHC class I antigens. Arch. Virol. 2010, 156, 219–234. [Google Scholar] [CrossRef]
- Hurtado, C.; Granja, A.G.; Bustos, M.J.; Nogal, M.L.; De Buitrago, G.G.; De Yébenes, V.G.; Salas, M.L.; Revilla, Y.; Carrascosa, A.L. The C-type lectin homologue gene (EP153R) of African swine fever virus inhibits apoptosis both in virus infection and in heterologous expression. Virology 2004, 326, 160–170. [Google Scholar] [CrossRef]
- Galindo, I.; Almazán, F.; Bustos, M.J.; Viñuela, E.; Carrascosa, A.L. African Swine Fever Virus EP153R Open Reading Frame Encodes a Glycoprotein Involved in the Hemadsorption of Infected Cells. Virology 2000, 266, 340–351. [Google Scholar] [CrossRef]
- Petrovan, V.; Murgia, M.V.; Wu, P.; Lowe, A.D.; Jia, W.; Rowland, R.R.R. Epitope mapping of African swine fever virus (ASFV) structural protein, p54. Virus Res. 2020, 279, 197871. [Google Scholar] [CrossRef]
- Hernáez, B.; Díaz-Gil, G.; García-Gallo, M.; Quetglas, J.I.; Rodríguez-Crespo, I.; Dixon, L.K.; Escribano, J.M.; Alonso, C. The African swine fever virus dynein-binding protein p54 induces infected cell apoptosis. FEBS Lett. 2004, 569, 224–228. [Google Scholar] [CrossRef]
- Mima, K.A.; Katorkina, E.I.; Katorkin, S.A.; Tsybanov, S.Z.; Malogolovkin, A.S. In silico prediction of B- and T-cell epitopes in the CD2v protein of african swine fever virus (African swine fever virus, Asfivirus, Asfarviridae). Vopr. Virusol. 2020, 65, 103–112. [Google Scholar] [CrossRef]
- Mazloum, A.; Zhukov, I.U.; Aronova, E.B.; Igolkin, A.S.; Vlasova, N.N. ASF virus replication features in the presence of recombinant proteins CD2v, pX69R and pE248R. Vopr. Virusol. 2019, 64, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Burmakina, G.; Malogolovkin, A.; Tulman, E.R.; Xu, W.; Delhon, G.; Kolbasov, D.; Rock, D.L. Identification of T-cell epitopes in African swine fever virus CD2v and C-type lectin proteins. J. Gen. Virol. 2019, 100, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Borca, M.; O’Donnell, V.; Holinka, L.G.; Ramírez-Medina, E.; Clark, B.A.; Vuono, E.A.; Berggren, K.A.; Alfano, M.; Carey, L.B.; Richt, J.A.; et al. The L83L ORF of African swine fever virus strain Georgia encodes for a non-essential gene that interacts with the host protein IL-1β. Virus Res. 2018, 249, 116–123. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, V.; Risatti, G.R.; Holinka, L.G.; Krug, P.W.; Carlson, J.; Velazquez-Salinas, L.; Azzinaro, P.A.; Gladue, D.P.; Borca, M. Simultaneous Deletion of the 9GL and UK Genes from the African Swine Fever Virus Georgia 2007 Isolate Offers Increased Safety and Protection against Homologous Challenge. J. Virol. 2016, 91, 1. [Google Scholar] [CrossRef]
- Zsak, L.; Caler, E.; Lu, Z.; Kutish, G.F.; Neilan, J.G.; Rock, D.L. A Nonessential African Swine Fever Virus Gene UK Is a Significant Virulence Determinant in Domestic Swine. J. Virol. 1998, 72, 1028–1035. [Google Scholar] [CrossRef]
- Woźniakowski, G.; Mazur-Panasiuk, N.; Walczak, M.; Juszkiewicz, M.; Frant, M.; Niemczuk, K. Attempts at the development of a recombinant African swine fever virus strain with abrogated EP402R, 9GL, and A238L gene structure using the CRISPR/Cas9 system. J. Vet. Res. 2020, 64, 197–205. [Google Scholar] [CrossRef]
- O’Donnell, V.; Holinka, L.G.; Krug, P.W.; Gladue, D.P.; Carlson, J.; Sanford, B.; Alfano, M.; Kramer, E.; Lu, Z.; Arzt, J.; et al. African Swine Fever Virus Georgia 2007 with a Deletion of Virulence-Associated Gene9GL(B119L), when Administered at Low Doses, Leads to Virus Attenuation in Swine and Induces an Effective Protection against Homologous Challenge. J. Virol. 2015, 89, 8556–8566. [Google Scholar] [CrossRef]
- Lewis, T.; Zsak, L.; Burrage, T.G.; Lu, Z.; Kutish, G.F.; Neilan, J.G.; Rock, D.L. An African Swine Fever Virus ERV1-ALRHomologue, 9GL, Affects Virion Maturation and Viral Growth in Macrophages and Viral Virulence in Swine. J. Virol. 2000, 74, 1275–1285. [Google Scholar] [CrossRef]
- Sanford, B.; Holinka, L.G.; O’Donnell, V.; Krug, P.W.; Carlson, J.; Alfano, M.; Carrillo, C.; Wu, P.; Lowe, A.; Risatti, G.; et al. Deletion of the thymidine kinase gene induces complete attenuation of the Georgia isolate of African swine fever virus. Virus Res. 2016, 213, 165–171. [Google Scholar] [CrossRef]
- Reis, A.L.; Goatley, L.C.; Jabbar, T.; Sanchez-Cordon, P.; Netherton, C.L.; Chapman, D.A.G.; Dixon, L.K. Deletion of the African Swine Fever Virus Gene DP148R Does Not Reduce Virus Replication in Culture but Reduces Virus Virulence in Pigs and Induces High Levels of Protection against Challenge. J. Virol. 2017, 91, e01428-17. [Google Scholar] [CrossRef]
- Rathakrishnan, A.; Moffat, K.; Reis, A.L.; Dixon, L.K. Production of Recombinant African Swine Fever Viruses: Speeding Up the Process. Viruses 2020, 12, 615. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Sánchez, E.G.; Pérez-Núñez, D.; Nogal, M.; De León, P.; Carrascosa, A.L.; Nieto, R.; Soler, A.; Arias, M.L.; Revilla, Y. African swine fever virus (ASFV) protection mediated by NH/P68 and NH/P68 recombinant live-attenuated viruses. Vaccine 2018, 36, 2694–2704. [Google Scholar] [CrossRef] [PubMed]
- Monteagudo, P.L.; Lacasta, A.; López, E.; Bosch, L.; Collado, J.; Pina-Pedrero, S.; Correa-Fiz, F.; Accensi, F.; Navas, M.J.; Vidal, E.; et al. BA71ΔCD2: A New Recombinant Live Attenuated African Swine Fever Virus with Cross-Protective Capabilities. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Borca, M.V.; O’Donnell, V.; Holinka, L.G.; Risatti, G.R.; Ramirez-Medina, E.; Vuono, E.A.; Shi, J.; Pruitt, S.; Rai, A.; Silva, E.; et al. Deletion of CD2-like gene from the genome of African swine fever virus strain Georgia does not attenuate virulence in swine. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef]
- Ramirez-Medina, E.; Vuono, E.; O’Donnell, V.; Holinka, L.G.; Silva, E.; Rai, A.; Pruitt, S.; Carrillo, C.; Gladue, D.P.; Borca, M.V. Differential Effect of the Deletion of African Swine Fever Virus Virulence-Associated Genes in the Induction of Attenuation of the Highly Virulent Georgia Strain. Viruses 2019, 11, 599. [Google Scholar] [CrossRef]
- Reis, A.L.; Abrams, C.C.; Goatley, L.C.; Netherton, C.; Chapman, D.G.; Sanchez-Cordon, P.; Dixon, L.K. Deletion of African swine fever virus interferon inhibitors from the genome of a virulent isolate reduces virulence in domestic pigs and induces a protective response. Vaccine 2016, 34, 4698–4705. [Google Scholar] [CrossRef]
- Carlson, J.; O’Donnell, V.; Alfano, M.; Velazquez-Salinas, L.; Holinka, L.G.; Krug, P.W.; Gladue, D.P.; Higgs, S.; Borca, M.V. Association of the Host Immune Response with Protection Using a Live Attenuated African Swine Fever Virus Model. Viruses 2016, 8, 291. [Google Scholar] [CrossRef]
- Gómez-Puertas, P.; Rodríguez, F.; Oviedo, J.M.; Ramiro-Ibáñez, F.; Ruiz-Gonzalvo, F.; Alonso, C.; Escribano, J.M. Neutralizing antibodies to different proteins of African swine fever virus inhibit both virus attachment and internalization. J. Virol. 1996, 70, 5689–5694. [Google Scholar] [CrossRef]
- Escribano, J.M.; Galindo, I.; Alonso, C. Antibody-mediated neutralization of African swine fever virus: Myths and facts. Virus Res. 2013, 173, 101–109. [Google Scholar] [CrossRef]
- Ruiz-Gonzalvo, F.; Rodriguez, F.; Escribano, J.M. Functional and Immunological Properties of the Baculovirus-Expressed Hemagglutinin of African Swine Fever Virus. Virology 1996, 218, 285–289. [Google Scholar] [CrossRef]
- Gómez-Puertas, P.; Rodríguez, F.; Oviedo, J.M.; Brun, A.; Alonso, C.; Escribano, J.M. The African Swine Fever Virus Proteins p54 and p30 Are Involved in Two Distinct Steps of Virus Attachment and Both Contribute to the Antibody-Mediated Protective Immune Response. Virology 1998, 243, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Barderas, M.G.; Rodríguez, F.; Gómez-Puertas, P.; Avilés, M.; Beitia, F.J.; Alonso, C.; Escribano, J.M. Antigenic and immunogenic properties of a chimera of two immunodominant African swine fever virus proteins. Arch. Virol. 2001, 146, 1681–1691. [Google Scholar] [CrossRef] [PubMed]
- Neilan, J.G.; Zsak, L.; Lu, Z.; Burrage, T.G.; Kutish, G.F.; Rock, D.L. Neutralizing antibodies to African swine fever virus proteins p30, p54, and p72 are not sufficient for antibody-mediated protection. Virology 2004, 319, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Bosch-Camós, L.; López, E.; Rodriguez, F. African swine fever vaccines: A promising work still in progress. Porc. Health Manag. 2020, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Argilaguet, J.M.; Pérez-Martin, E.; Gallardo, C.; Salguero, F.J.; Borrego, B.; Lacasta, A.; Accensi, F.; Díaz, I.; Nofrarias, M.; Pujols, J.; et al. Enhancing DNA immunization by targeting ASFV antigens to SLA-II bearing cells. Vaccine 2011, 29, 5379–5385. [Google Scholar] [CrossRef]
- Argilaguet, J.M.; Pérez-Martin, E.; Nofrarías, M.; Gallardo, C.; Accensi, F.; Lacasta, A.; Mora, M.; Ballester, M.; Galindo-Cardiel, I.; Lopez-Soria, S.; et al. DNA Vaccination Partially Protects against African Swine Fever Virus Lethal Challenge in the Absence of Antibodies. PLoS ONE 2012, 7, e40942. [Google Scholar] [CrossRef]
- Lacasta, A.; Ballester, M.; Monteagudo, P.L.; Rodríguez, J.M.; Salas, M.L.; Accensi, F.; Pina-Pedrero, S.; Bensaid, A.; Argilaguet, J.; Lopez-Soria, S.; et al. Expression Library Immunization Can Confer Protection against Lethal Challenge with African Swine Fever Virus. J. Virol. 2014, 88, 13322–13332. [Google Scholar] [CrossRef]
- Ivanov, V.; Efremov, E.E.; Novikov, B.V.; Balyshev, V.M.; Tsibanov, S.; Kalinovsky, T.; Kolbasov, D.V.; Niedzwiecki, A.; Rath, M. Vaccination with viral protein-mimicking peptides postpones mortality in domestic pigs infected by African swine fever virus. Mol. Med. Rep. 2011, 4, 395–401. [Google Scholar] [CrossRef]
- Lokhandwala, S.; Waghela, S.D.; Bray, J.; Sangewar, N.; Charendoff, C.; Martin, C.L.; Hassan, W.S.; Koynarski, T.; Gabbert, L.; Burrage, T.G.; et al. Adenovirus-vectored novel African Swine Fever Virus antigens elicit robust immune responses in swine. PLoS ONE 2017, 12, e0177007. [Google Scholar] [CrossRef]
- Argilaguet, J.M.; Pérez-Martin, E.; López, S.; Goethe, M.; Escribano, J.M.; Giesow, K.; Keil, G.M.; Rodríguez, F.; Lopez-Soria, S. BacMam immunization partially protects pigs against sublethal challenge with African swine fever virus. Antivir. Res. 2013, 98, 61–65. [Google Scholar] [CrossRef]
- Lokhandwala, S.; Waghela, S.D.; Bray, J.; Martin, C.L.; Sangewar, N.; Charendoff, C.; Shetti, R.; Ashley, C.; Chen, C.-H.; Berghman, L.R.; et al. Induction of Robust Immune Responses in Swine by Using a Cocktail of Adenovirus-Vectored African Swine Fever Virus Antigens. Clin. Vaccine Immunol. 2016, 23, 888–900. [Google Scholar] [CrossRef] [PubMed]
- Lokhandwala, S.; Petrovan, V.; Popescu, L.; Sangewar, N.; Elijah, C.; Stoian, A.; Olcha, M.; Ennen, L.; Bray, J.; Bishop, R.P.; et al. Adenovirus-vectored African Swine Fever Virus antigen cocktails are immunogenic but not protective against intranasal challenge with Georgia 2007/1 isolate. Vet. Microbiol. 2019, 235, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Murgia, M.V.; Mogler, M.; Certoma, A.; Green, D.; Monaghan, P.; Williams, D.T.; Rowland, R.R.R.; Gaudreault, N.N. Evaluation of an African swine fever (ASF) vaccine strategy incorporating priming with an alphavirus-expressed antigen followed by boosting with attenuated ASF virus. Arch. Virol. 2018, 164, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Hübner, A.; Keil, G.M.; Kabuuka, T.; Mettenleiter, T.C.; Fuchs, W. Efficient transgene insertion in a pseudorabies virus vector by CRISPR/Cas9 and marker rescue-enforced recombination. J. Virol. Methods 2018, 262, 38–47. [Google Scholar] [CrossRef]
- Pérez-Núñez, D.; Sunwoo, S.-Y.; Sánchez, E.G.; Haley, N.; García-Belmonte, R.; Nogal, M.; Morozov, I.; Madden, D.; Gaudreault, N.N.; Mur, L.; et al. Evaluation of a viral DNA-protein immunization strategy against African swine fever in domestic pigs. Vet. Immunol. Immunopathol. 2019, 208, 34–43. [Google Scholar] [CrossRef]
- Sunwoo, S.Y.; Pérez-Núñez, D.; Morozov, I.; Sánchez, E.G.; Gaudreault, N.N.; Trujillo, J.D.; Mur, L.; Nogal, M.; Madden, D.W.; Urbaniak, K.; et al. DNA-Protein Vaccination Strategy Does Not Protect from Challenge with African Swine Fever Virus Armenia 2007 Strain. Vaccines (Basel) 2019, 7, 12. [Google Scholar] [CrossRef]
- Lopera-Madrid, J.; Osorio, J.E.; Grethe, J.S.; Xiang, Z.; Adams, L.G.; Laughlin, R.C.; Mwangi, W.; Subramanya, S.; Neilan, J.; Brake, D.; et al. Safety and immunogenicity of mammalian cell derived and Modified Vaccinia Ankara vectored African swine fever subunit antigens in swine. Vet. Immunol. Immunopathol. 2017, 185, 20–33. [Google Scholar] [CrossRef]
- Jancovich, J.K.; Chapman, D.; Hansen, D.T.; Robida, M.D.; Loskutov, A.; Craciunescu, F.; Borovkov, A.; Kibler, K.; Goatley, L.; King, K.; et al. Immunization of Pigs by DNA Prime and Recombinant Vaccinia Virus Boost To Identify and Rank African Swine Fever Virus Immunogenic and Protective Proteins. J. Virol. 2018, 92, e02219-17. [Google Scholar] [CrossRef]
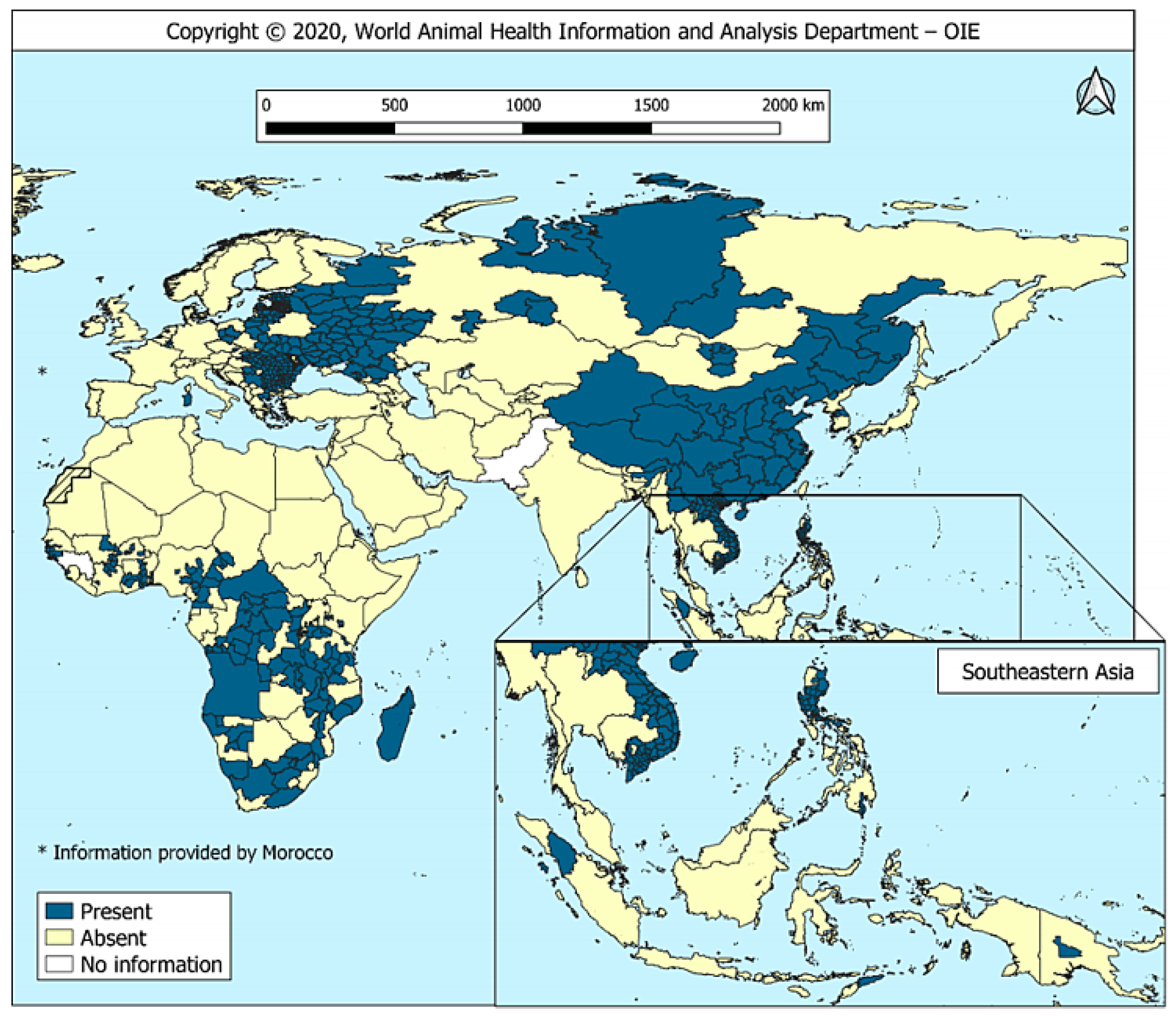
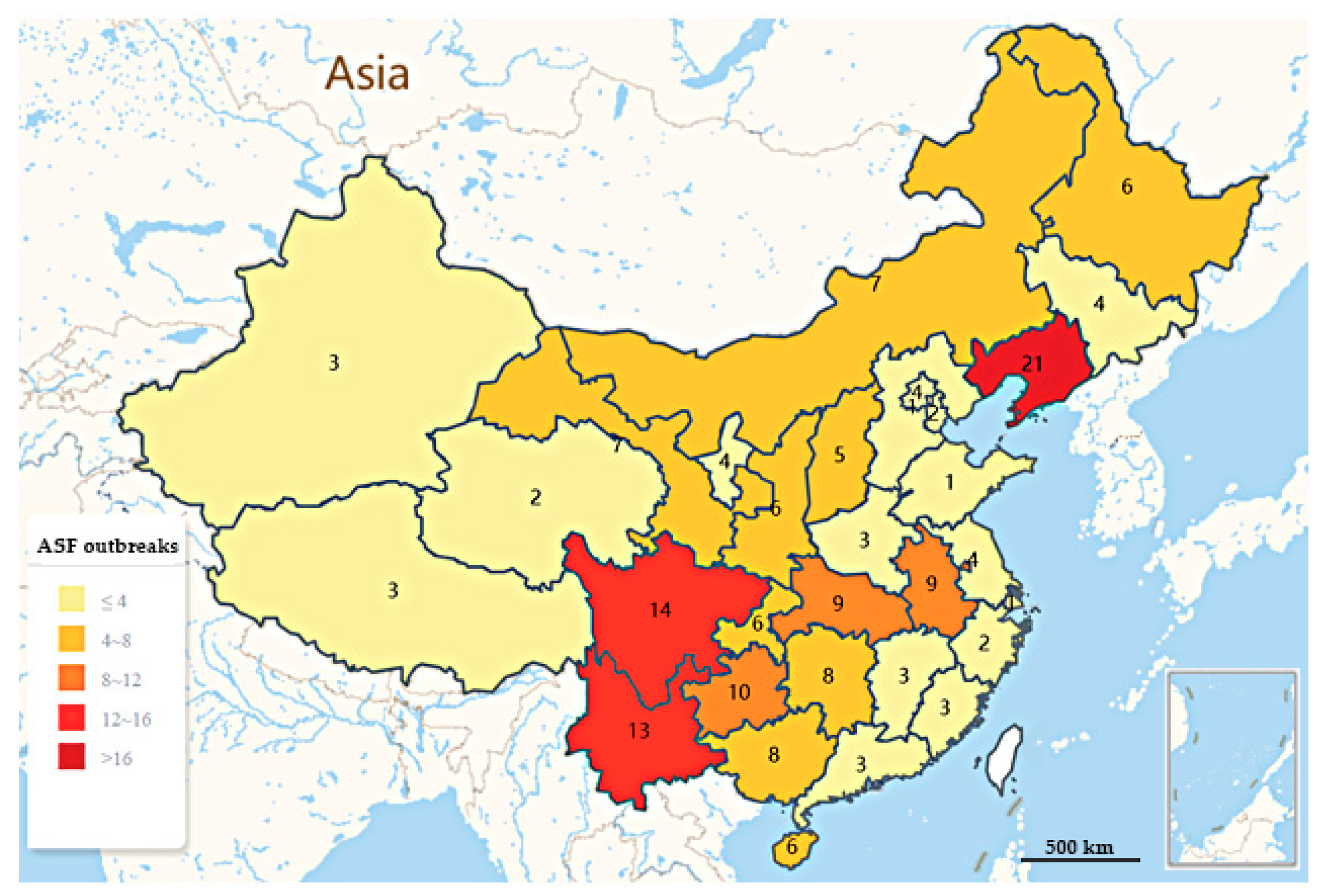
| Region | Provinces | Outbreaks | No. of Susceptible | No. of Incidence | No. of Death | Total Outbreaks | Outbreaks-ASF, % |
|---|---|---|---|---|---|---|---|
| Southwest | Sichuan | 14 | 1873 | 308 | 247 | 46 | 25.84% (46/178) |
| Guizhou | 10 | 1763 | 259 | 215 | |||
| Yunnan | 13 | 2861 | 1155 | 921 | |||
| Tibet | 3 | / | / | 55 | |||
| Chongqing | 6 | 770 | 24 | 78 | |||
| Northeast | Liaoning | 21 | 425.63 | 2276 | 2087 | 30 | 16.85% (30/178) |
| Jilin | 4 | 1458 | 196 | 204 | |||
| Heilongjiang | 6 | 746.49 | 5044 | 4158 | |||
| Central | Hubei | 9 | 2026 | 167 | 121 | 23 | 12.92% (23/178) |
| Hunan | 8 | 134.43 | 729 | 400 | |||
| Henan | 3 | 260 | 178 | 94 | |||
| Jiangxi | 3 | 463 | 75 | 63 | |||
| Northwest | Ningxia | 4 | 465 | 43 | 29 | 22 | 12.36% (22/178) |
| Xinjiang | 3 | 1124 | 204 | 146 | |||
| Qinghai | 2 | 101 | 46 | 31 | |||
| Shanxi | 6 | 119.06 | 459 | 266 | |||
| Gansu | 7 | 111.61 | 732 | 612 | |||
| Eastern China | Shandong | 1 | 4504 | 17 | 3 | 21 | 11.80% (21/178) |
| Jiangsu | 4 | 690.83 | 3087 | 1469 | |||
| Anhui | 9 | 110.18 | 586 | 358 | |||
| Zhejiang | 2 | / | 486 | 396 | |||
| Fujian | 3 | 222.47 | 147 | 123 | |||
| Shanghai | 1 | 314 | 50 | 11 | |||
| Northern China | Beijing | 4 | 140.50 | 138 | 129 | 19 | 10.67% (19/178) |
| Tianjin | 2 | 1000 | 292 | 256 | |||
| Hebei | 1 | 5600 | / | / | |||
| Shanxi | 5 | 8379 | 178 | 100 | |||
| Inner Mongolia | 7 | 995 | 348 | 311 | |||
| Southern China | Guangdong | 3 | 6167 | 1681 | 31 | 17 | 9.55% (17/178) |
| Guangxi | 8 | 278.39 | 129 | 966 | |||
| Hainan | 6 | 2162 | 432 | 223 |
| Pathway | Encoded Protein/Genes | Essential/Nonessential | Mechanism | Reference |
|---|---|---|---|---|
| Regulation of host protein expression | A238L | Nonessential | Inhibits NF-κ B and NFAT activation | [51] |
| NL(DP71L) | Nonessential | Inhibition of activation of peIF2α-ATF4-CHOP signaling pathway and its mediated apoptosis | [52,53] | |
| Interference with innate immune system | MGF360,MGF505/530 | Nonessential | Inhibition of transcription of type I IFN and other cytokines Increased survival of infected cells | [54,55,56,57] |
| I329L | Nonessential | Inhibits TLR signaling Inhibiting NF-κ B and IRF3 signaling pathways | [50,58,59] | |
| Regulation of apoptosis/autophagy | A179L | Nonessential | A member of the Bcl-2 family Inhibition of apoptosis in the early stage of infection Inhibition of cell autophagy | [60,61] |
| 4CL (A224L) | Nonessential | Inhibition of TNF-alpha-induced caspase 3 activation and apoptosis | [54] | |
| EP153R | Nonessential | C-type lectin, participate in the process of blood cell adsorption Regulating apoptosis and inhibiting MHC-1 expression | [62,63,64] | |
| P54 (E183L) | Nonessential | Participate in viral particle assembly and viral adhesion to host cells Induce apoptosis in the late stage of infection | [65,66] | |
| Others | CD2v(EP402R) | Nonessential | Mediate erythrocyte adsorption and promote virus transmission Interacts with cellular AP-1 protein and participates in intracellular transport of virus Inhibition of lymphocyte proliferation | [58,67,68,69] |
| L83L | Nonessential | Binding host protein IL-1beta to inhibit its antiviral effect | [70] | |
| UK(DP96R) | Nonessential | Negative regulation of type I IFN expression and NF-κ B signaling by inhibition of TBK1 and IKKβ | [71,72] | |
| 9GL(B119L) | Nonessential | Influencing virion maturation and viral growth in macrophages and viral virulence in swine | [56,71,73,74,75] | |
| TK(A240L) | Essential | Determines virulence | [76] | |
| DP148R | Nonessential | The function is elusive yet. Deletion of DP148R greatly reduces viral virulence | [77,78] |
| Source Strain | Genotype | Deletion Protein (Gene) | Cell | Virulence Changes | Protection | Reference |
|---|---|---|---|---|---|---|
| Benin 97/1 | I | DP148R, CD2v (EP402R), EP153R | PBMs, WSL-R 1 | [78] (2020) | ||
| OURT88/3 (attenuated) | I | I329L | PBMs | Reduced | [50] (2020) | |
| Georgia/2007 | II | PBMs | No attenuated | No | ||
| China HLJ/18 | II | MGF360/505 3, CD2v (EP402R) | PBMs | Attenuated | Homologous | [41] (2020) |
| China HLJ/18 | II | 9GL, UK | PBMs | Attenuated | No | |
| China HLJ/18 | II | MGF360/505 3 | PBMs | Attenuated | Homologous | |
| Georgia/2010 | II | CD2v(EP402R) | Primary macrophages | No attenuated | No | [81] (2020) |
| Georgia/2007 | II | 9GL (B119L), UK (DP96R), NL (DP71L) | Primary macrophages | Attenuated | No | [82] (2019) |
| NH/P68 (attenuated) | I | A238L | COS-7 2 | Highly attenuated | Homologous (I L60) No heterologous (II Arm07) | [79] (2018) |
| Benin 97/1 | I | MGF360/530/505 | PAMs | Attenuated | Homologous | [55] (2016), [83] (2018) |
| Benin 97/1 | I | DP148R | PAMs | Attenuated | Homologous | [77] (2017) |
| BA71 | I | CD2v (EP402R) | COS-1 2 | Highly attenuated | Homologous and heterologous (I E75, II Georgia 2007/1) | [80] (2017) |
| Georgia/2007 | II | 9GL, UK | Primary macrophages | Fully attenuated | Homologous | [71] (2017) |
| ASFVG/VP30 | II | TK | Primary macrophages, Vero | Attenuated | No | [76] (2016) |
| Pr4 | II | 9GL | Macrophages | Fully attenuated | Homologous | [84] (2016) |
| Georgia/2007 | II | 9GL, MGF360/505 3 | Primary macrophages | Highly attenuated | No | [56] (2016) |
| Georgia/2007 | II | MGF360/505 3 | Primary macrophages | Fully attenuated | Homologous | [57] (2015) |
| Georgia/2007 | II | 9GL | Primary macrophages | Attenuated | Homologous | [74] (2015) |
| Sequence Source | Gene/Protein | Vector/System | Adjuvant | Specific Antibodies | Neutralizing Antibody | Cellular Immunity | Protection | Reference |
|---|---|---|---|---|---|---|---|---|
| Protein-based subunit vaccines | ||||||||
| E75CV | HA (CD2v) | Baculovirus | Freund’s | Yes | No | Homologous protection (3/3) Dose dependent | [87] | |
| E75 | p54,p30 | Baculovirus | Freund’s | Yes | Yes | Partial protection (3/6), | [88] | |
| E75 | p54/p30 chimera | Baculovirus | Freund’s | Yes | Yes | Homologous protection (2/2) Mild clinical symptoms | [89] | |
| Pr4 | p54,p30,p72,p22 | Baculovirus | Freund’s | Yes | Yes | No (0/6) Delayed clinical disease Reduced viremia | [90] | |
| E70 | Group1:p158,p327,p14,p220 Group3:p30,p72 | Synthetic peptides | Freund’s | No | No; Group1&3 Increased average survival Reduced mean viral titers | [95] | ||
| DNA vaccines | ||||||||
| E75 | p54/p30 fusion | pCMV | No | No | No (0/4) | [92] | ||
| p54/p30/SLA-II fusion | pCMV | Yes | No | T cell response | No (0/4) Viremia enhancement | |||
| E75 | sHA/p54/p30 fusion | pCMV | Yes (p54;p30) | No | IFN-γ | No (0/6); | [93] | |
| sHA/p54/p30/Ub fusion | pCMV | No | Strong CTL IFN-γ | Partial protection (2/6) The absence of viremia | ||||
| E75 | 80 ORFs fragments/Ub fusion | DNA expression library | Yes | Yes | Partial protection (6/10) Reduced virus titers | [94] | ||
| Virus-vectored vaccines | ||||||||
| E75 | sHA 1/p54/p30fusion | BacMam | No | No | IFN-γ | Partial protection (4/6) The absence of viremia | [97] | |
| Georgia 2007/1 | p30,p54,pp62,p72 | Adenovirus | BioMize | Strong | IFN-γand CTL | [98] | ||
| Georgia 2007/1 | A151R,B119L,B602L,EP402R∆PRR,B438L,K205R,A104R | Adenovirus | BioMize; ZTS-01 | Strong | IFN-γ | [96] | ||
| Georgia 2007/1 | Ad-ASFV-I: A151R,B119L,B602L,EP402R∆PRR,B438L,K205R,A104R,pp62,p72 | Adenovirus | BioMize | Strong | IFN-γ | No Immune-response dependent enhancement of disease | [96] | |
| Ad-ASFV-II: p30,p54,pp62,p72,pp220 (p37-34-14,p150-I,p150-II) | BioMize | Higher | IFN-γ | Partial protection: (2/10) | ||||
| ZTS-01 | Lower | IFN-γ | Partial protection: (5/9) Lower clinical score The absence of viremia | |||||
| Combined vaccination strategy | ||||||||
| Georgia 2007/1 | p72, p54, p12 | HEK 293cell | TS6 | Yes | No | Less T Cell response | [104] | |
| p72, C-type Lectin (EP153R), CD2v | MVA 2 | TS6 | No | T Cell response | ||||
| p72, C-type Lectin (EP153R), CD2v | r VACV 3 prime + protein boost | TS6 | T Cell response IFN-γ | |||||
| Georgia 2007/1 | 47 antigens | DNA prime + r VACV boost | CpG oligo | Yes | No | T Cell response | No Reduced viral load Higher clinical scores | [105] |
| E70;Ba71V | DNA:CD2v,p30,p72,CP312R; Proteins: p15, p35, p54, p72, CD2v-E (s HA) | DNA+ Protein | ISA25 | Yes | 20%; 10% | Some | [102] | |
| Georgia 2007/1; Ba71V | DNA:CD2v, p72, p30, +/-p17; Proteins: p15, p35, p54, +/-p17 | DNA+ protein | ISA25 | Yes | No | Some | Challenge: Armenia 2007 No Disease enhancement | [103] |
| Ba71V | p30, p54, p72, s HA/p72 | Alphavirus RPs 4 | Yes | [100] | ||||
| p30 (Ba71V) + OURT88/3 | Alphavirus RP prime + LAV boost | Yes | Yes | |||||
| OUR T88/3 | A151R, p72, C129R, p30, p54, E146L, I215L, I73R, L8L, M448R, MGF110-4 L, MGF110-5 L | r Ad prime + MVA boost | Yes | Yes | Challenge: OUR T88/1 No Reduced and delayed clinical signs; Reduced viremia and viral load | [49] | ||
| OUR T88/3 Benin 97/1 | p72, p30, p54, E183L, E199L, EP153R, F317L, MGF505-5R | r Ad prime + MVA boost | No | Yes (Expect:B646L,E183L,EP153R) | Some (E183L,CP204L) | IFN-γ | Challenge: OUR T88/1 Exp.2 (6/6) Reduced Viremia Infectious virus persisted Exp.1 (2/6) | [42] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, K.; Liu, J.; Wang, L.; Fan, S.; Li, Z.; Li, Y.; Yi, L.; Ding, H.; Zhao, M.; Chen, J. Current State of Global African Swine Fever Vaccine Development under the Prevalence and Transmission of ASF in China. Vaccines 2020, 8, 531. https://doi.org/10.3390/vaccines8030531
Wu K, Liu J, Wang L, Fan S, Li Z, Li Y, Yi L, Ding H, Zhao M, Chen J. Current State of Global African Swine Fever Vaccine Development under the Prevalence and Transmission of ASF in China. Vaccines. 2020; 8(3):531. https://doi.org/10.3390/vaccines8030531
Chicago/Turabian StyleWu, Keke, Jiameng Liu, Lianxiang Wang, Shuangqi Fan, Zhaoyao Li, Yuwan Li, Lin Yi, Hongxing Ding, Mingqiu Zhao, and Jinding Chen. 2020. "Current State of Global African Swine Fever Vaccine Development under the Prevalence and Transmission of ASF in China" Vaccines 8, no. 3: 531. https://doi.org/10.3390/vaccines8030531
APA StyleWu, K., Liu, J., Wang, L., Fan, S., Li, Z., Li, Y., Yi, L., Ding, H., Zhao, M., & Chen, J. (2020). Current State of Global African Swine Fever Vaccine Development under the Prevalence and Transmission of ASF in China. Vaccines, 8(3), 531. https://doi.org/10.3390/vaccines8030531




