Abstract
Helminth parasites release extracellular vesicles which interact with the surrounding host tissues, mediating host–parasite communication and other fundamental processes of parasitism. As such, vesicle proteins present attractive targets for the development of novel intervention strategies to control these parasites and the diseases they cause. Herein, we describe the first proteomic analysis by LC-MS/MS of two types of extracellular vesicles (exosome-like, 120 k pellet vesicles and microvesicle-like, 15 k pellet vesicles) from adult Schistosoma haematobium worms. A total of 57 and 330 proteins were identified in the 120 k pellet vesicles and larger 15 k pellet vesicles, respectively, and some of the most abundant molecules included homologues of known helminth vaccine and diagnostic candidates such as Sm-TSP2, Sm23, glutathione S-transferase, saponins and aminopeptidases. Tetraspanins were highly represented in the analysis and found in both vesicle types. Vaccination of mice with recombinant versions of three of these tetraspanins induced protection in a heterologous challenge (S. mansoni) model of infection, resulting in significant reductions (averaged across two independent trials) in liver (47%, 38% and 41%) and intestinal (47%, 45% and 41%) egg burdens. These findings offer insight into the mechanisms by which anti-tetraspanin antibodies confer protection and highlight the potential that extracellular vesicle surface proteins offer as anti-helminth vaccines.
1. Introduction
Schistosomiasis is the second most important parasitic disease, only after malaria, in terms of social, economic, and public health impact [1]. Schistosoma haematobium, the causative agent of urogenital schistosomiasis, is highly prevalent in 53 Middle East and African countries [1] and it is also sporadically seen in India [2] and France [3]. Urogenital schistosomiasis affects more than 90 million people, mostly in sub-Saharan Africa where 180 million inhabitants are at risk, and is responsible for 150,000 deaths per year [4]. Furthermore, the most common complications associated with this disease include schistosomal hematuria, bladder wall pathology, hydronephrosis, and dysuria [4].
The current control programs against schistosomiasis are aimed at reducing the morbidity caused by the parasite by regularly treating infected populations with praziquantel [5]. Despite the efforts made to control this devastating disease, schistosomiasis is still spreading to new geographical areas [3,6]. Furthermore, praziquantel has shown reduced efficacy in field studies [7] and is not effective against the immature stages of the parasite [8,9]. Hence, a vaccine that reduces disease severity and/or reduces transmission is needed to control and eliminate schistosomiasis [10,11]. Despite efforts over decades, there is no licensed vaccine [1,12]. Even though various vaccine candidates have advanced into clinical trials targeting Schistosoma mansoni, the causative agent of intestinal schistosomiasis, the only vaccine candidate against S. haematobium to have progressed into clinical trial is a glutathione S-transferase recombinant protein, Sh28GST. However, results from a phase 3 trial conducted from 2009 to 2012 in S. haematobium infected children did not report any significant efficacy due to the vaccine [13]. Therefore, it is important to continue identifying new target antigens in the effort to develop a vaccine against S. haematobium and the other species of schistosomes [10,12].
S. haematobium adult worms live in the perivesicular veins where they can survive for years using evasion strategies to remain undetected by the host immune system. One of the main strategies the parasites employ to hijack the immune effector response is to release a suite of immunomodulatory excretory/secretory (ES) products. ES products comprise different proteins, glycans, lipids, and nucleic acids [14], and have been the focus of different studies aiming at understanding the molecular basis of host-parasite interactions and the subsequent use of this information to develop novel therapeutics and diagnostics [15,16,17,18]. Recently, it has been documented that the ES products from different helminths (including schistosomes) contain extracellular vesicles (EV)s [19,20,21,22]. EVs are membrane-bound organelles released by cells that can act as mediators of intercellular communication by transferring molecular signals mediated by proteins, lipids, metabolites, mRNAs, microRNAs, and other non-coding RNA species [23,24]. In addition to the transmission of information between cells within the same organism, recent studies have shown that EVs secreted by parasitic helminths are taken up by host cells within the parasite’s niche tissue and provide a means of inter-species communication [19,25,26,27,28,29,30,31,32,33,34,35,36,37,38]. For instance, EVs from trematodes and nematodes can be internalized by host cells whereupon they suppress effector immune responses [37,39,40], or in contrast, some helminth EVs contribute to pathogenesis by promoting cell proliferation and inflammatory cytokine production [29].
EVs from helminths also contain vaccine candidate antigens. For example, EVs from S. mansoni contain molecules that have known vaccine efficacy in animal models of schistosomiasis [27], and vaccination of mice with helminth EVs stimulates the production of protective immune responses that significantly reduce fecal egg counts, worm burdens, and symptom severity and mortality induced by infection after parasite challenge [25,41,42,43,44]. Moreover, antibodies produced against recombinant forms of Opisthorchis viverrini EV surface proteins hinders the uptake of EVs by cholangiocytes and suppresses the immune response that fuels pathogenesis [29,42].
In addition to other molecules, helminth EVs contain tetraspanins (TSPs), which have been shown to be effective vaccine candidates against Schistosoma spp. [21,27]. The TSPs Sm23, Sm-TSP-1, and Sm-TSP-2, all found in the membrane of S. mansoni EVs [20,27], have displayed partial efficacy in when used as adjuvanted subunit vaccines [45,46,47], and Sm-TSP-2 has successfully completed phase I clinical trials [48]. TSPs from other helminth species, including S. japonicum and O. viverrini, have also been shown to be efficacious vaccine candidates in animal challenge models of infection [42,49].
Although there are reports describing EVs and the vaccine efficacy of EV-derived molecules from other schistosomes, no studies have been conducted on S. haematobium. Herein, we have characterized for the first time the proteomic composition of small ( 120 k pellet) and large (15k pellet) subclasses of EVs from this parasite, and selected three of the EV surface TSPs for assessment as subunit vaccines in a heterologous challenge mouse challenge model of schistosomiasis.
2. Materials and Methods
2.1. Ethics Statement
All experimental procedures performed on animals in this study were approved by the James Cook University (JCU) animal ethics committee (A2391). All experiments were performed in accordance with the 2007 Australian Code of Practice for the Care and Use of Animals for Scientific Purposes and the 2001 Queensland Animal Care and Protection Act.
2.2. Parasite Material and Experimental Animals
Bulinus truncatus snails infected with S. haematobium (Egyptian strain) were provided by the Biomedical Research Institute, MD, USA, and maintained in aquaria in a 27 °C incubator. Male BALB/c mice were purchased from the Animal Resource Centre, Canningvale, Western Australia, and maintained at the Australian Institute of Tropical Health and Medicine (AITHM) animal facility in cages under controlled temperature and light with free access to pelleted food and water.
To obtain cercariae, snails were removed from the tank with a pair of forceps and washed several times with water to remove debris and rotifers, transferred to a Petri dish and incubated without water at 27 °C in the dark for 2 h. Water was then added and the snails were placed under light for 1.5 h at 28 °C. Cercariae were concentrated using a 20 µm pore size sieve and, finally, each BALB/c mouse (6 week-old) was infected with 1000 cercariae by tail penetration [50].
2.3. Adult Worm Culture, ES Collection, and EV Purification
S. haematobium adult worms were obtained by perfusion of mice at 16 weeks post-infection and parasites were washed several times with serum-free modified Basch media supplemented with 4 × antibiotic/antimycotic (SFB) [20] and then incubated in SFB (50 pairs/5 mL) at 37 °C in with 5% CO2 for 2 weeks. ES products were harvested daily, differentially centrifuged at 4 °C (500× g, 2000× g and 4000× g for 30 min each) to remove large parasite material such as eggs and tegumental debris and stored at −80 °C until use.
EVs were isolated using established methods previously described for S. mansoni [20]. Stored supernatants were thawed on ice, concentrated at 4 °C using a 10 kDa spin concentrator (Merck Millipore, USA) and centrifuged for 1 h at 15,000× g at 4 °C. The resultant pellet (containing 15 k vesicles) was washed with 1 mL of PBS, centrifuged at 15,000× g for 1 h at 4 °C, resuspended in 200 μL PBS and stored at −80 °C. The supernatant was ultracentrifuged at 120,000× g for 3 h at 4 °C using an MLS-50 rotor (Beckman Coulter, Brea, CA, USA) to collect 120 k pellet vesicles. The resultant pellet was resuspended in 70 μL of PBS and subjected to Optiprep® density gradient (ODG) separation. The ODG was prepared by diluting a 60% Iodixanol solution (Optiprep®, Sigma-Aldrich, St. Louis, MO, USA) with 0.25 M sucrose in 10 mM Tris-HCl pH 7.2 to make 40%, 20%, 10%, and 5% iodixanol solutions, and 1.0 mL of these solutions was layered in decreasing density in an ultracentrifuge tube. The resuspended 120 k pellet vesicles were added to the top layer and ultracentrifuged at 120,000× g for 18 h at 4 °C. A control tube was similarly prepared using PBS instead of the 120 k pellet vesicle sample to measure the density of the different fractions recovered from the gradient. Fractions obtained from the ODG were diluted with 8 mL of PBS containing 1 × EDTA-free protease inhibitor cocktail (Santa Cruz, CA, USA), and concentrated using a 10 kDa spin concentrator to remove the excess Optiprep® solution.
The density of different fractions obtained from the ODG (12 each for the sample and control) was calculated by measuring the absorbance of each fraction at 340 nm using a POLARstar Omega (BMG Labtech, Cary, NC, USA) spectrophotometer and interpolating the absorbance in a standard curve as previously shown [27]. The protein concentration of all fractions was quantified using the Quick Start™ Bradford Protein Assay Kit (Bio-Rad Laboratories, Inc., Life Science Research, Hercules, CA, USA) following the manufacturer’s instructions.
2.4. Determination of the Size and Concentration of EVs
The size distribution and particle concentration of the different fractions recovered after ODG, as well as the 15 k vesicle fraction, was measured using tunable resistive pulse sensing (TRPS) using a qNano instrument (Izon, Christchurch, New Zealand) following an established protocol [34]. A Nanopore NP150 and a NP400 (Izon, New Zealand) were used to measure each fraction containing 120 k pellet vesicles and the 15 k pellet vesicle fraction, respectively. Thirty-five μL of measurement electrolyte (Izon, New Zealand) was added to the upper fluid well and maximum pressure was applied; the shielding lid was clicked five to 10 times to wet the Nanopore. Then, 75 μL of measurement electrolyte was added to the lower fluid well, maximum pressure and an appropriate voltage (0.1 V) was applied, and Nanopore current was checked for stability. Thirty-five and 75 μL of filtered coating solution (Izon, New Zealand) was loaded in the upper and lower fluid well, respectively, and maximum pressure was applied for 10 min followed by maximum vacuum for another 10 min. The coating solution was flushed out of the upper and lower fluid wells two to three times with measurement electrolyte, maximum pressure was applied for 10 min and the voltage was increased until the current reached between 120 and 140 nA and the baseline current was stable. Then, 35 μL of calibration particles (CP200 carboxylated polystyrene calibration particles; Izon, New Zealand) was loaded to the upper fluid well at a 1:200 dilution when calibrating for 120 k pellet vesicle fractions and 1:1,500 when calibrating for the 15 k pellet vesicle fraction, incubated for 2 min at maximum pressure and the stretch was reduced and the calibration particles were measured at two different pressures (P10 and P5). Then, the 120 k and 15 k pellet vesicle fractions were diluted 1:5, applied to the Nanopore and measured similarly to the calibration particles. The size and concentration of particles were determined using the software provided by Izon (version 3.2).
2.5. In-Gel Trypsin Digestion of EVs
Fractions containing 120 k pellet vesicles of sufficient size and concentration, and the 15 k pellet vesicle fraction, were resuspended in 1 × loading buffer (10% glycerol, 80 mM Tris-HCl, 2% SDS, 0.01% bromophenol blue and 1.25% beta-mercaptoethanol, pH 6.8), boiled at 95 °C for 5 min and electrophoresed in a 15% SDS-PAGE gel at 100 V. The gel was stained with 0.03% Coomassie Brilliant Blue (40% methanol, 10% acetic acid, and 50% water) for 30 min at room temperature (RT) with gentle shaking and destained using destaining buffer 1 (60% water, 10% acetic acid, and 30% methanol) for 1 h at RT with gentle shaking. Each lane was sliced into three pieces with a surgical blade and placed into a fresh Eppendorf tube. Then, slices were further destained three times using destaining buffer 2 (50% acetonitrile (ACN), 20% ammonium bicarbonate, and 30% milliQ water) by adding 200 μL of buffer to the gel slice, and incubating them at 37 °C for 45 min. Supernatants were discarded and, finally, gel slices were dried in a vacuum concentrator (LabGear, QLD 4064, Australia) on low/high medium heat (<45 °C). Next, 100 µL of reduction buffer (20 mM dithiothreitol, 25 mM ammonium bicarbonate) was added to each dried slice, incubated at 65 °C for 1 h, and supernatants were discarded. Alkylation was achieved by adding 100 µL of alkylation buffer (50 mM iodoacetamide, 25 mM ammonium bicarbonate) to each gel slice, which were further incubated in darkness for 40 min at RT. Gel slices were washed with 200 µL of wash buffer (25 mM ammonium bicarbonate) and incubated at 37 °C for 15 min twice after the gel slices were dried in a speedivac. For trypsin digestion, a total of 2 µg of trypsin (Sigma-Aldrich, USA) was added to each gel slice and incubated for 5 min at RT. Finally, 50 µL of trypsin reaction buffer (40 mM ammonium bicarbonate, 9% ACN) was added to gel slices and incubated overnight at 37 °C. Peptides were extracted in 50% acetonitrile with 0.1% trifluoroacetic acid. The last step was performed three times to maximize peptide recovery. All peptides were finally dried in a vacuum concentrator. Samples were then resuspended in 10 μL of 0.1% trifluoroacetic acid and tryptic peptides were desalted using a Zip-Tip® column (Merck Millipore, Burlington, MA, USA) pipette tip according to the manufacturer’s protocol and dried in a vacuum concentrator before analysis using liquid chromatography-tandem mass spectrometry (LC-MS/MS).
2.6. LC-MS/MS Analysis, Database Search, and Bioinformatic Analysis
Each 120 k pellet and 15 k pellet vesicle preparation was reconstituted in 10 μL of 5% formic acid and injected onto a 50 mm 300 μm C18 trap column (Agilent Technologies, Santa Clara, CA, USA). The samples were then desalted for 5 min at 30 μL/min using 0.1% formic acid and the peptides were then eluted onto an analytical nano-HPLC column (150 mm × 75 μm 300SBC18, 3.5 μm, Agilent Technologies, USA) at a flow rate of 300 nL/min. Peptides were separated using a 95 min gradient of 1–40% buffer B (90/10 ACN/0.1% formic acid) followed by a steeper gradient from 40 to 80% buffer B in 5 min. A 5600 ABSciex mass spectrometer operated in information-dependent acquisition mode, in which a 1 s TOF-MS scan from 350–1400 m/z was used, and for product ion ms/ms 80–1400 m/z ions observed in the TOF-MS scan exceeding a threshold of 100 counts and a charge state of +2 to +5 were set to trigger the acquisition of product ion. Analyst 1.6.1 (ABSCIEX, Framingham, MA, USA) software was used for data acquisition.
For database search and protein identification, a database was built using a concatenated target/decoy version of the S. haematobium predicted proteome [51,52] sequences; Bioproject PRJNA78265 downloaded from Parasite WormBase (www.parasite.wormbase.org) and concatenated to the common repository of adventitious proteins (cRAP, https://www.thegpm.org/crap/), as well as to the Sh-TSP-2 protein (Genbank QCO69687.1). A database search was performed using a combination of four search engines—X! Tandem version X! Tandem Vengeance (2015.12.15.2) [53], MS-GF+ version Release (v2018.04.09) [54], OMSSA, and Tide [55] using SearchGUI version v3.3.3 [56]. The identification settings were as follows: Trypsin, Specific, with a maximum of two missed cleavages 10.0 ppm as MS1 and 0.2 Da as MS2 tolerances; fixed modifications: Carbamidomethylation of C (+57.021464 Da), variable modifications: Deamidation of N (+0.984016 Da), Deamidation of Q (+0.984016 Da), Oxidation of M (+15.994915 Da). Peptides and proteins were inferred from the spectrum identification results using PeptideShaker version 1.16.38 [56]. Peptide Spectrum Matches (PSMs), peptides, and proteins were validated at a 1.0% False Discovery Rate (FDR) estimated using the decoy hit distribution. Only proteins having at least two unique peptides were considered as identified. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019462 and 10.6019/PXD019462 (Username: reviewer46995@ebi.ac.uk Password: 03jNpGFk).
Protein family (Pfam) domains were classified using HMMER v3.1b1 [57] and protein gene ontology (GO) categories were classified using Blast2GO v5.2 [58]. ReviGO was used to visualize GO terms using semantic similarity-based scatterplots [59]. TMHMM [60] and SignalP 4.1 [61] software was used to predict transmembrane domains and putative signal peptides, respectively.
2.7. Cloning and Expression of Sh-TSP2 and MS3_09198 in Pichia Pastoris
DNA sequences encoding the large extracellular loop (LEL) regions of Sh-TSP2 and MS3_09198 (predicted using TMPRED) were codon optimized based on yeast codon usage preference and synthesized by GenScript (Piscataway, NJ, USA). The synthesized coding DNAs with a 6-His-tag expressed at the C-terminus were cloned into the pPinkα-HC expression vector (Thermofisher, Waltham, MA, USA) using XhoI/KpnI restriction sites. The recombinant plasmids with correct insert confirmed by double-strand DNA sequencing were linearized with AflII, and then transformed by electroporation into PichiaPink™ Strain 4 with endogenous proteinases A and B knocked out to prevent degradation of expressed proteins. The transformants were selected on PAD (Pichia Adenine Dropout) agar plates and the expression yield of picked colonies was evaluated in 10 mL BMMY medium with 0.5% methanol for 72 h. The clones with the highest expression yield were used to express recombinant Sh-TSP2 and MS3_09198 in 2L BMMY under induction of 0.5% methanol at 30 °C with 250 rpm shaking for 72 h. The culture medium containing the secreted proteins was harvested by centrifugation (5000× g for 20 min at RT) and filtered through a 0.22 µM membrane filter (Millipore). Recombinant proteins were purified by immobilized metal affinity chromatography (IMAC) using a prepacked 5 mL His-Trap HP column (GE Healthcare, Chicago, IL, USA). The purified recombinant proteins were buffer-exchanged in 1 × PBS, pH 7.4. The purity of the proteins was analyzed by SDS-PAGE and the concentration was measured by OD280. The purified recombinant proteins were adjusted to 1 mg/mL, aliquoted and stored at −80 °C.
2.8. Cloning and Expression of MS3_01370 in Escherichia Coli
Unlike Sh-TSP2 and MS3_09198, we were unsuccessful at expressing the LEL region of MS3_01370 in soluble form in P. pastoris, thus, the protein was expressed as a thioredoxin (TrX) fusion in E. coli. Primers incorporating NcoI (forward primer) and XhoI restriction enzyme sites (reverse primer) were used to amplify the LEL of MS3_01370 from S. haematobium cDNA and the amplicon was cloned into the pET32a expression vector (Novagen), in-frame with the N-terminal TrX tag. Protein expression was induced for 24 h in E. coli BL21(DE3) by the addition of 1 mM Isopropyl beta-D-1-thiogalactopyranoside (IPTG) using standard methods. The culture was harvested by centrifugation (8000× g for 20 min at 4 °C), re-suspended in 50 mL lysis buffer (50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 40 mM imidazole) and stored at −80 °C. The cell pellet was lysed by three freeze-thaw cycles at −80 °C and 42 °C followed by sonication on ice (10 × 5 sec pulses [70% amplitude] with 30 s rest periods between each pulse) with a Qsonica Sonicator. Insoluble material was pelleted by centrifugation at 20,000× g for 20 min at 4 °C. The supernatant was diluted 1:4 in lysis buffer and filtered through a 0.22 μm membrane (Millipore). MS3_01370 was purified by IMAC by loading onto a prepacked 1 mL His-Trap HP column (GE Healthcare) equilibrated with lysis buffer at a flow rate of 1 mL/min using an AKTA-pure-25 FPLC (GE Healthcare). After washing with 20 mL lysis buffer, bound His-tagged protein was eluted using the same buffer with a stepwise gradient of 50–250 mM imidazole (50 mM steps). Fractions containing MS3_01370 (as determined by SDS-PAGE) were pooled and concentrated using Amicon Ultra-15 centrifugal devices with a 3 kDa MWCO and quantified using the Pierce BCA Protein Assay kit. The final concentration of MS3_01370 was adjusted to 1 mg/mL and the protein was aliquoted and stored at −80 °C.
2.9. Vaccine Formulation and Immunization Schedule
Numbers of S. haematobium cercariae were insufficient to perform appropriately powered vaccination trials using an S. haematobium model of infection so vaccine experiments were instead performed using an S. mansoni challenge model [62]. Four groups of 10 BALB/c mice (6–8 weeks) were immunized intraperitoneally on day 1 with either recombinant Sh-TSP-2, MS3_09198, MS3_01370, or TrX control protein (50 μg/mouse), each formulated with an equal volume of Imject alum adjuvant (Thermofisher) and 5 μg of CpG ODN1826 (InvivoGen, San Diego, CA, USA). Immunizations were repeated on days 15 and 29 and each mouse was challenged (tail penetration) with 120 S. mansoni cercariae on day 43. Blood was sampled at day 42 to determine pre-challenge antibody titers. Two independent trials were performed.
2.10. Necropsy and Estimation of Parasite Burden
Mice were necropsied at day 91 (7 weeks post-infection) [62]. Blood was collected and worms harvested by vascular perfusion and counted. Livers were removed, weighed, and digested for 5 h with 5% KOH at 37 °C with shaking. Schistosome eggs from digested livers were concentrated by centrifugation at 1000× g for 10 min and re-suspended in 1 mL of 10% formalin. The number of eggs in a 5 μL aliquot was counted in triplicate and the number of eggs per gram (EPG) of liver tissue was calculated. Small intestines were removed and cleaned of debris before being weighed and digested as per the livers. Eggs were similarly concentrated and counted to calculate intestinal EPG.
2.11. Statistics
All statistics were performed using GraphPad Prism 7.0. The reductions in worm and egg numbers were analyzed using a Student’s t-test and results were expressed as the mean ± standard error of the mean. For antibody titers, the reactivity cut-off values were determined as the mean + 3SD of the naive serum.
3. Results
3.1. Density, Protein Concentration, Particle Concentration, and Purity of 120 k and 15 k Pellet Vesicles from Schistosoma haematobium
S. haematobium adult worm 120 k pellet vesicles were purified using an iodixanol gradient. The density of the 12 fractions obtained after gradient separation ranged from 1.039 to 1.4 g/mL (Supplementary Table S1). The protein and particle concentration of the fractions ranged from 1.6 to 25.35 μg/mL and 3.72 × 106 to 2.06 × 108 particles/mL, respectively, while the protein and particle concentration of the 15 k pellet vesicle fraction was 18.00 μg/mL and 1.48 × 107 particles/mL, respectively. The size of the particles in the gradient-separated fractions ranged from 135 nm ± 19.3 to 342 nm ± 113.9 and size of particles in the 15 k pellet vesicle fraction was 274 nm ± 40.7. Gradient-separated fractions having an appropriate purity and density corresponding to 120 k pellet vesicles (1.09–1.22 g/mL) [20,27] (fractions 5–9) were selected for further analysis (Figure 1).
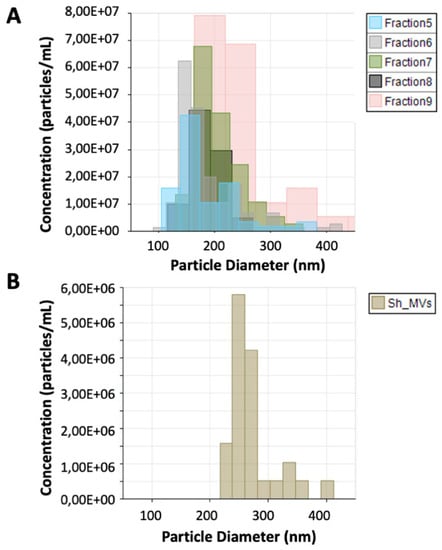
Figure 1.
Tunable resistive pulse sensing analysis of 120 k pellet vesicles and 15 k pellet vesicles from Schistosoma haematobium. The size and number of extracellular vesicles (EVs) secreted by S. haematobium were analyzed by qNano (iZon). (A) Size and concentration of particles in fractions containing 120 k pellet vesicles (5–9). (B) Size and particle concentration of S. haematobium 15 k pellet vesicles.
3.2. Proteomic Analysis of Schistosoma haematobium 120 k and 15 k Pellet Vesicles
The proteome composition of S. haematobium adult worm EVs was characterized by LC-MS/MS. After combining the protein identified in fractions 5–9 (fractions containing the highest purity of 120 k pellet vesicles), a total of 133 proteins matching S. haematobium proteins and common contaminants from the cRAP database were identified. From these, 80 proteins were identified with at least two validated unique peptides and 57 of them matched S. haematobium proteins. From the 57 identified proteins, eight (14%) contained a transmembrane domain and seven (12%) had a signal peptide. In a similar fashion, 506 proteins were identified from analysis of the 15 k pellet vesicles. From these, 344 proteins were identified with at least two validated unique peptides and 330 matched S. haematobium proteins. From these identified proteins, 54 (16.3%) contained a transmembrane domain and 30 (9%) had a signal peptide. Forty proteins were identified in both types of vesicles. The identity of the most abundant proteins in each type of vesicle, as well as proteins having homologues typically found in other helminth EVs, are shown in Table 1. A full list of proteins identified in both 15 k and 120 k pellet vesicles is shown in Supplementary Tables S2 and S3.

Table 1.
Proteins found in 120 k and 15 k pellet vesicles isolated from the ES products of adult S. haematobium *.
3.3. Protein Families Present in Schistosoma haematobium 120 k and 15 k Pellet Vesicles
Identified proteins were subjected to a Pfam analysis using default parameters in HMMER v3.1b1 and proteins containing an identified Pfam domain with an E-value < 1 × 10−5 were selected. A total of 70 and 387 domains were identified from 120 k and 15 k pellet vesicles, respectively. In 120 k pellet vesicles, the three most abundant domains were proteasome subunit domains (PF00227) (14%), TSP family domains (PF00335) (7%), and ferritin-like domains (PF12902) (4%) (Figure 2A). The most abundant protein domains from 15 k pellet vesicles were EF-hand domains (PF00036) (3%), Ras family domains (PF00071) (3%), TCP-1/cpn60 chaperonin family domains (PF00118) (2%), and TSP family domains (PF00335) (2%) (Figure 2B). From these, TSP family domains and 14-3-3 protein domains were common to both types of vesicles.
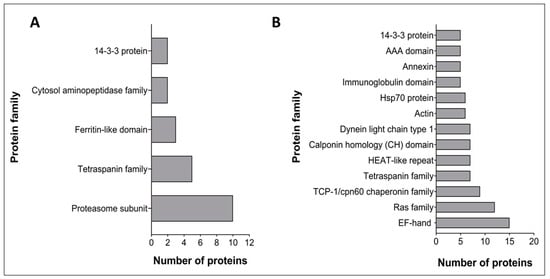
Figure 2.
Pfam analysis of the most abundant Schistosoma haematobium vesicle proteins. The X-axis represents the number of proteins containing at least one of those domains. (A) 120 k pellet vesicles (B) 15 k pellet vesicles.
3.4. Gene Ontology of Proteins Identified from Schistosoma haematobium 120 k and 15 k Pellet Vesicles
The proteins of adult S. haematobium 120 k and 15 k pellet vesicles were annotated using Blast2GO. To avoid redundancy in the analysis and better comprehend the represented GO terms in the vesicles, the parental GO terms were removed and children GO terms were visualized using ReviGO based on semantic similarity-based scatterplots. The GO terms were ranked by the nodescore provided by Blast2GO and plotted using their nodescore and frequency. Semantically similar GO terms plot close together and increasing heatmap score signifies increasing nodescore from Blast2GO. The circle size denotes the frequency of the GO term from the underlying database. In 120 k pellet vesicles, several biological processes were highly represented, such as the ubiquitin-dependent protein catabolic process, oxidation-reduction process, and gluconeogenesis and glycolytic process (Figure 3A). Similarly, in 15 k pellet vesicles, several biological processes were highly represented, such as the carbohydrate metabolic process, transport process, organonitrogen compound metabolic process, and microtubule-based process (Figure 3B). The oxidation–reduction process was common to both types of vesicles; six proteins from 120 k pellet vesicles and 22 proteins from 15 k pellet vesicles were predicted to be involved in this process.
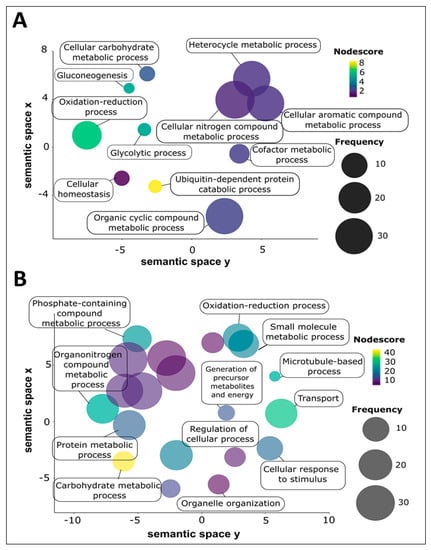
Figure 3.
Biological process GO term categories of adult Schistosoma haematobium vesicle proteins. Biological processes were ranked by nodescore (Blast2GO) and plotted using REViGO. Semantically similar GO terms plot close together, increasing heatmap score signifies increasing nodescore from Blast2GO, while circle size denotes the frequency of the GO term from the underlying database. (A) 120 k pellet vesicles, (B) 15 k pellet vesicles.
In S. haematobium 120 k vesicles, several molecular functions were highly represented, such as threonine-type endopeptidase activity, protein binding activity, endopeptidase activity, and transition metal ion binding activity. In 15 k vesicles, molecular functions such as protein binding activity, ATP binding activity, nucleoside-triphosphatase activity, and calcium ion binding activity were highly represented. From these highly represented molecular function terms, protein binding was common to both 120 k and 15 k vesicles.
3.5. Parasite Burdens in Vaccinated and Control Mice
Vaccination of mice with MS3_01370, MS3_09198 showed a trend towards reduced adult S. mansoni burden in trial 1 by 22% and 12%, respectively, and in trial 2 by 14% and 5%, respectively, compared to controls. Sh-TSP2-vaccinated mice showed a reduction in trial 1 only of 2%, compared to controls. None of these differences were statistically significant except for MS3_01370 in trial 1 (p < 0.05) (Figure 4A,D). However, in trial 1, vaccination of mice with MS3_01370, MS3_09198 and Sh-TSP-2 significantly reduced liver egg burdens by 39% (p < 0.05), 49% (p < 0.001), and 32% (p < 0.01), respectively (Figure 4B), and in trial 2, MS3_01370, MS3_09198, and Sh-TSP-2 vaccination significantly reduced liver egg burdens by 54% (p < 0.001), 27% (p < 0.05), and 49% (p < 0.001), respectively (Figure 4E). Similarly, immunization of mice with MS3_01370, MS3_09198 and Sh-TSP-2 reduced the intestinal egg burden by 57% (p < 0.01), 51% (p < 0.01), and 54% (p < 0.001) in trial 1, respectively (Figure 4C), and in trial 2, MS3_01370, MS3_09198, and Sh-TSP-2 vaccination reduced the intestinal egg burden by 36% (p < 0.01), 39% (p < 0.05), and 27% (p < 0.05), respectively (Figure 4F).
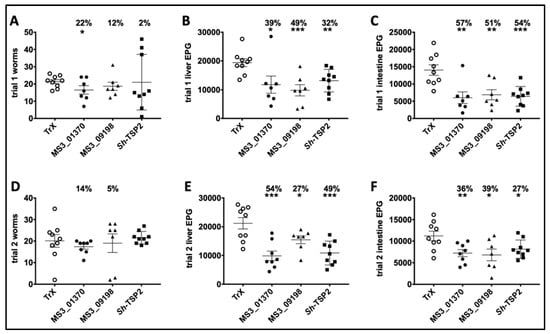
Figure 4.
Schistosoma mansoni worm and egg burden reduction of vaccinated and control mice vaccinated with S. haematobium recombinant tetraspanins. (A) Adult worm reduction trial 1, (B) liver egg reduction trial 1, (C) intestinal egg reduction trial 1, (D) Adult worm reduction trial 2, (E) liver egg reduction trial 2, and (F) intestinal egg reduction trial 2. The percentage of reductions in parasite burden are above each dataset. Differences between each vaccinated group and the control group were analyzed with a student’s t-test. * p < 0.05, ** p < 0.01, *** p < 0.001.
4. Discussion
More than 100 million people are infected with S. haematobium and 150,000 people die every year [4]. Efforts have been made to reduce the prevalence of schistosomiasis, but the parasite is spreading to new areas [3] and is now the second most prevalent of the neglected tropical diseases [63]. Praziquantel is the only drug available for schistosomiasis treatment, although it does not protect against re-infection and the risk of resistance emerging in the field is high [64,65]. Furthermore, there is no licensed and effective vaccine for this devastating disease, and there is an urgent need to develop one to eliminate urogenital schistosomiasis as well as the other species of schistosomes.
We have shown previously that S. haematobium secretes at least two populations of vesicles with distinct differences in size and proteomic composition, similar to other schistosome species such as S. mansoni [20]. In this paper, we now show the first proteomic analysis of two types of extracellular vesicles (exosome-like, 120 k pellet vesicles and microvesicle-like, 15 k pellet vesicles) from adult Schistosoma haematobium worms.
The most represented domains contained within 120 k pellet vesicle proteins were the proteasome subunit domains, TSP domains, and ferritin-like domains, among others, while the most represented domains in the 15 k pellet vesicles were EF-hand, Ras family, TCP-1/cpn60 chaperonin family and TSP family domains. The proteasome is involved in the biogenesis of EVs [66] and also controls protein homeostasis and degradation of damaged proteins [67]. Furthermore, in schistosomes, the proteasome plays an important role in the cellular stress response and survival of the parasite [68]; it has been shown that treating mice with a proteasome inhibitor prior to infection with S. mansoni cercariae significantly impaired parasite development [69] and in vitro treatment of schistosomula with siRNAs targeting a deubiquitinase subunit of the 19S regulatory particle significantly reduced parasite viability [70].
Ferritins are iron-storage proteins, involved in maintaining intracellular iron balance [71], which minimize free-radical reactions and prevent cellular damage caused by iron accumulation in the cell [72]. Iron also plays an important role in the eggshell formation of schistosomes, and fer-1, one of the two ferritin isoforms of the parasite, is highly expressed in female worms in comparison to males [73]. Since female worms produce many eggs per day and eggs are the primary cause of pathology, vaccination using ferritins could disrupt the formation of eggs and reduce egg-induced disease. Indeed, ferritins have been tested as vaccine candidates against schistosomes; immunization of mice with the recombinant Fer-1 of S. japonicum caused 35.5% and 52.1% reduction in adult worm and liver egg burden, respectively [74]. Ferritins have been identified in the proteomic analysis of other blood feeding helminth EVs [20,27], suggesting a role for EVs in iron storage and acquisition.
Proteins containing EF-hand domains are involved in a number of protein-protein interactions for the uptake and release of calcium [75]. The influx of calcium in the cell induces the redistribution of phospholipids in the cell membrane, resulting in increased release of microvesicles [76]. EF-hand domains are also among the most predominant protein domains found within other helminth EV proteins [77]. This is consistent with the GO analysis herein, in which proteins involved in protein binding and calcium ion binding are the highest represented molecular function terms.
Ras proteins serve as signaling nodes activated in response to diverse extracellular stimuli [78] and are involved in biogenesis and release of microvesicles [79]. In S. mansoni, Ras proteins are involved in the male-directed maturation of the female worms [80], which could suggest a potential role of EVs in parasite-parasite communication.
TCP-1/cpn60 chaperonin family proteins play an important role in the folding of proteins, including actin and tubulin [81], which bind and hydrolyze ATP using magnesium ions [82]. This is consistent with the molecular function GO terms, in which ATP binding and nucleoside-triphosphatase was the most represented.
The second and fourth most abundant protein domains in 120 k and 15 k pellet vesicles, respectively, were the TSPs. TSPs are involved in EV biogenesis [79], are present on the surface membrane of EVs from many different organisms, and are considered a molecular marker of EVs [83]. TSPs are also found from the proteomic analysis of other helminth EVs (reviewed in [77]). In trematodes, TSPs are involved in tegument development [84,85,86]. TSP LELs have been tested as vaccine candidates in trematode models of infection [42,47,87] and antibodies produced against TSP vaccine candidates present in S. mansoni and O. viverrini EVs blocked the internalization of EVs through host cells in both parasites, and decreased pathogenesis in the case of O. viverrini [29,42,88], suggesting a possible mechanism of vaccine efficacy.
Tetraspanins were highly represented in the analysis and found in both vesicle types. Since there is evidence that these molecules orchestrate interactions between parasite EVs and host tissues, we aimed to assess the vaccine efficacy of some of the most abundant EV TSPs in a challenge model of schistosome infection. Difficulties in generating the large amount of cercariae needed for mouse infection (the use of hamsters is prohibited in Australia) precluded the use of S. haematobium as the challenge parasite, and thus, we used an S. mansoni challenge model instead. Although this meant that a heterologous infection model would be employed to assess the efficacy of S. haematobium proteins, we reasoned that the homology of the candidates between the two species could be sufficiently high (Sh-TSP2—69.6% identity, MS3_09198—98.6% identity, and MS3_01370—79.5% identity) (Figure S1) as to afford a level of cross-species protection and provide informative data with regards to the vaccine efficacy of these candidates.
Vaccination with any of the TSPs did not elicit significant reductions in worm burden compared to controls (except for MS3_01370 in trial 1); however, significant reductions in tissue egg burdens (both liver and intestinal) were observed in all groups of vaccinated mice in both trials. Decreases in tissue egg loads are arguably the most important hallmarks of an effective vaccine against schistosomiasis given that (1) pathology due to the disease is egg-induced [89] and (2) disease transmission is dependent on the excretion of eggs from the host into the environment [10], thus, a vaccine that reduces egg burden in the host would ameliorate both disease pathology and transmission.
Other schistosome antigens have been reported to elicit a primarily anti-fecundity protective effect upon vaccination. Glutathione-S-transferases (GSTs) from S. japonicum, S. haematobium and the bovine schistosome S. bovis have all been reported to induce decreases in tissue egg loads, despite no observable reduction in worm burden [90,91,92]. Interestingly, GSTs have been prominently identified from the vesicles in this study and other proteomic analyses of schistosome EVs [20,27,28]. This manifestation of protective immunity is seen in cattle that have had repeated field and laboratory exposure to S. bovis [93,94] and the closely-related S. matheei [95], in Rhesus monkeys experimentally infected with S. mansoni [96], and in baboons following laboratory exposure to S. haematobium [97]. In the cattle/S. bovis and baboon/S. haematobium models, this observation was confirmed by surgically transplanting “suppressed” worms into hosts with no prior exposure and showing that the parasites resumed high levels of egg production [91,97]. It could be that vaccination with the EV molecules described herein is eliciting similar immune mechanisms to engender the observed anti-fecundity effects, which may be manifesting in a reduction of the fitness and fecundity of adult worms and/or a shortening of the lifespan of eggs embolized in the tissues [91]. Furthermore, we have reported on the capacity of helminth EVs to promote pathogenesis via their endocytosis by host cells and subsequent manipulation of host immune effector mechanisms, such as an increase in IL-6 secretion [29] and differential expression of genes involved in immune cell trafficking [88], and the ability of antibodies against helminth EV proteins, such as TSPs, to block vesicle uptake [29,42,88] provides a plausible mechanism for protective efficacy.
In this study we provide the first characterization of EVs secreted by S. haematobium, their expression as vaccine candidates present on the surface of these EVs, and the evaluation of these candidates in a heterologous challenge model of schistosomiasis vaccination of mice with recombinant versions of three of these tetraspanins-induced protection in a heterologous challenge (S. mansoni) model of infection, resulting in significant reductions (averaged across two independent trials) in liver (47%, 38%, and 41%) and intestinal (47%, 45%, and 41%) egg burdens. The significant reduction in tissue egg burden described here indicates that these EV-derived vaccine candidates could be effective in reducing the pathology and transmission of S. mansoni and S. haematobium (due to the cross-protective efficacy observed in the heterologous vaccine/challenge model herein) and they could potentially be incorporated into a pan-schistosome vaccine, due to the geographical overlap between the two species.
Currently, the World Health Organization (WHO) roadmap regarding schistosomiasis is to eliminate disease morbidity and mortality as a public health problem in the coming decade [98]. Vaccine-linked chemotherapy has been deemed one of the most effective control measures for combating the disease, with this strategy combining the mainstay of schistosomiasis intervention (mass administration of PZQ) with the more long-term benefits of vaccination, such as the one based on the molecules described herein. Modeling has predicted that chemotherapy linked with a vaccine that has an effective duration of protection (transmission interruption and pathology reduction) is the best intervention strategy for achievement of the WHO’s goals to significantly decrease the burden of disease caused by schistosomiasis [99].
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-393X/8/3/416/s1, Figure S1: Protein alignment of the large extracellular loop from each of the Schistosoma haematobium tetraspanins used in the vaccine trials with their homologue from Schistosoma mansoni, Table S1: Density, protein concentration, particle concentration, purity, and size of the 15 k pellet vesicle fraction and gradient-separated 120 k vesicle pellet fractions, Table S2: Proteins identified from Schistosoma haematobium-derived 15 k pellet vesicles, Table S3: Proteins identified from Schistosoma haematobium-derived 120 k pellet vesicles.
Author Contributions
Conceptualization, A.L., J.S., and M.S.P.; Formal analysis, B.A.T., G.G.M., A.L., J.S., M.E.B., and M.S.P.; Funding acquisition, A.L.; Investigation, B.A.T., G.G.M., D.P., L.B., B.Z., L.W., J.S., and M.S.P.; Methodology, J.S., B.A.T., G.G.M., D.P., L.W., B.Z., and M.S.P.; Supervision, A.L., J.S., and M.S.P.; Validation, J.S. and M.S.P.; Writing—original draft, J.S. and M.S.P.; Writing—review and editing, A.L., M.S.P., J.S., B.Z., and M.E.B. Authorship must be limited to those who have contributed substantially to the work reported. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the NHMRC program grant APP1037304. A.L. was funded by an NHMRC Senior Principal Research Fellowship (APP1117504). B.A.T. was funded by a James Cook University Postgraduate Scholarship. G.G.M. was funded by an AITHM Postgraduate Scholarship.
Acknowledgments
The authors thank Atik Susianto for maintenance of the parasites and laboratory animals. They also gratefully acknowledge the NIAID Schistosomiasis Research Center of the Biomedical Research Institute, Rockville, MD, USA for the provision of S. mansoni-infected B. glabrata snails for this work through NIH-NIAID contract HHSN2722017000141 for distribution through BEI resources.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- McManus, D.P.; Dunne, D.W.; Sacko, M.; Utzinger, J.; Vennervald, B.; Zhou, X.N. Schistosomiasis. Nat. Rev. Dis. Prim. 2018, 4, 13. [Google Scholar] [CrossRef]
- Chitsulo, L.; Engels, D.; Montresor, A.; Savioli, L. The global status of schistosomiasis and its control. Acta Trop. 2000, 77, 41–51. [Google Scholar] [CrossRef]
- Boissier, J.; Grech-Angelini, S.; Webster, B.L.; Allienne, J.-F.; Huyse, T.; Mas-Coma, S.; Toulza, E.; Barré-Cardi, H.; Rollinson, D.; Kincaid-Smith, J.; et al. Outbreak of urogenital schistosomiasis in Corsica (France): An epidemiological case study. Lancet Infect. Dis. 2016, 16, 971–979. [Google Scholar] [CrossRef]
- Berger, S. Schistosoma Haematobium, 2017 ed.; GIDEON Informatics Inc.: Los Angeles, CA, USA, 2017. [Google Scholar]
- World Health Organization. Schistosomiasis: Number of People Treated Worldwide in 2014. Weekly Epidemiol. Record 2016, 91, 53–60. [Google Scholar]
- Berry, A.; Moné, H.; Iriart, X.; Mouahid, G.; Aboo, O.; Boissier, J.; Fillaux, J.; Cassaing, S.; Debuisson, C.; Valentin, A.; et al. schistosomiasis haematobium, Corsica, France. Emerg. Infect. Dis. 2014, 20, 1595–1597. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, L.; Liang, Y.S. Susceptibility or resistance of praziquantel in human schistosomiasis: A review. Parasitol. Res. 2012, 111, 1871–1877. [Google Scholar] [CrossRef] [PubMed]
- Vale, N.; Gouveia, M.J.; Rinaldi, G.; Brindley, P.J.; Gartner, F.; da Correia Costa, J.M. Praziquantel for Schistosomiasis: Single-Drug Metabolism Revisited, Mode of Action, and Resistance. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Gönnert, R.; Andrews, P. Praziquantel, a new broad-spectrum antischistosomal agent. Z. Parasitenkunde 1977, 52, 129–150. [Google Scholar] [CrossRef]
- McManus, D.P.; Bergquist, R.; Cai, P.; Ranasinghe, S.; Tebeje, B.M.; You, H. Schistosomiasis—From immunopathology to vaccines. Semin. Immunopathol. 2020, 42, 355–371. [Google Scholar] [CrossRef]
- Wilson, R.; Coulson, P. Why Don’t We Have a Schistosomiasis Vaccine? Parasitol. Today 1998, 14, 97–99. [Google Scholar] [CrossRef]
- Tebeje, B.M.; Harvie, M.; You, H.; Loukas, A.; McManus, D.P. Schistosomiasis vaccines: Where do we stand? Parasites Vectors 2016, 9, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Riveau, G.; Schacht, A.-M.; Dompnier, J.-P.; Deplanque, D.; Seck, M.; Waucquier, N.; Senghor, S.; Delcroix-Genete, D.; Hermann, E.; Idris-Khodja, N.; et al. Safety and efficacy of the rSh28GST urinary schistosomiasis vaccine: A phase 3 randomized, controlled trial in Senegalese children. PLoS Negl. Trop. Dis. 2018, 12, e0006968. [Google Scholar] [CrossRef] [PubMed]
- Crowe, J.; Lumb, F.E.; Harnett, M.M.; Harnett, W. Parasite excretory-secretory products and their effects on metabolic syndrome. Parasite Immunol. 2017, 39, e12410. [Google Scholar] [CrossRef] [PubMed]
- Delcroix, M.; Medzihradsky, K.; Caffrey, C.R.; Fetter, R.D.; McKerrow, J.H. Proteomic analysis of adult S. mansoni gut contents. Mol. Biochem. Parasitol. 2007, 154, 95–97. [Google Scholar] [CrossRef]
- Hall, S.L.; Braschi, S.; Truscott, M.; Mathieson, W.; Cesari, I.M.; Wilson, R.A. Insights into blood feeding by schistosomes from a proteomic analysis of worm vomitus. Mol. Biochem. Parasitol. 2011, 179, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Cui, S.-J.; Hu, W.; Feng, Z.; Wang, Z.-Q.; Han, Z.-G. Excretory/Secretory Proteome of the Adult Developmental Stage of Human Blood Fluke, Schistosoma japonicum. Mol. Cell. Proteom. 2009, 8, 1236–1251. [Google Scholar] [CrossRef]
- Sotillo, J.; Pearson, M.S.; Becker, L.; Mekonnen, G.G.; Amoah, A.S.; Van Dam, G.J.; Corstjens, P.L.A.M.; Murray, J.; Mduluza, T.; Mutapi, F.; et al. In-depth proteomic characterization of Schistosoma haematobium: Towards the development of new tools for elimination. PLoS Negl. Trop. Dis. 2019, 13, e0007362. [Google Scholar] [CrossRef]
- Eichenberger, R.M.; Sotillo, J.; Loukas, A. Immunobiology of parasitic worm extracellular vesicles. Immunol. Cell Biol. 2018, 96, 704–713. [Google Scholar] [CrossRef]
- Kifle, D.W.; Pearson, M.S.; Becker, L.; Pickering, D.; Loukas, A.; Sotillo, J. Proteomic analysis of two populations of Schistosoma mansoni-derived extracellular vesicles: 15 k pellet and 120 k pellet vesicles. Mol. Biochem. Parasitol. 2020, 236, 111264. [Google Scholar] [CrossRef]
- Mekonnen, G.G.; Pearson, M.S.; Loukas, A.; Sotillo, J. Extracellular vesicles from parasitic helminths and their potential utility as vaccines. Expert Rev. Vaccines 2018, 17, 1–9. [Google Scholar] [CrossRef]
- Kifle, D.W.; Sotillo, J.; Pearson, M.S.; Loukas, A. Extracellular vesicles as a target for the development of anti-helminth vaccines. Emerg. Top. Life Sci. 2017, 1, 659–665. [Google Scholar] [CrossRef]
- Kim, K.M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M. RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA 2017, 8, e1413. [Google Scholar] [CrossRef] [PubMed]
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019, 47, D516–D519. [Google Scholar] [CrossRef] [PubMed]
- Coakley, G.; McCaskill, J.L.; Borger, J.G.; Simbari, F.; Robertson, E.; Millar, M.; Harcus, Y.; McSorley, H.J.; Maizels, R.M.; Buck, A.H. Extracellular vesicles from a helminth parasite suppress macrophage activation and constitute an effective vaccine for protective immunity. Cell Rep. 2017, 19, 1545–1557. [Google Scholar] [CrossRef]
- Tritten, L.; Geary, T.G. Helminth extracellular vesicles in host-parasite interactions. Curr. Opin. Microbiol. 2018, 46, 73–79. [Google Scholar] [CrossRef]
- Sotillo, J.; Pearson, M.S.; Potriquet, J.; Becker, L.; Pickering, D.; Mulvenna, J.; Loukas, A. Extracellular vesicles secreted by Schistosoma mansoni contain protein vaccine candidates. Int. J. Parasitol. 2016, 46, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Nowacki, F.C.; Swain, M.T.; Klychnikov, O.I.; Niazi, U.; Ivens, A.; Quintana, J.F.; Hensbergen, P.J.; Hokke, C.H.; Buck, A.H.; Hoffmann, K.F. Protein and small non-coding RNA-enriched extracellular vesicles are released by the pathogenic blood fluke Schistosoma mansoni. J. Extracell. Vesicles 2015, 4, 28665. [Google Scholar] [CrossRef]
- Chaiyadet, S.; Sotillo, J.; Smout, M.J.; Cantacessi, C.; Jones, M.K.; Johnson, M.S.; Turnbull, L.; Whitchurch, C.B.; Potriquet, J.; Laohaviroj, M.; et al. Carcinogenic Liver Fluke Secretes Extracellular Vesicles That Promote Cholangiocytes to Adopt a Tumorigenic Phenotype. J. Infect. Dis. 2015, 212, 1636–1645. [Google Scholar] [CrossRef]
- Cwiklinski, K.; De La Torre-Escudero, E.; Trelis, M.; Bernal, D.; Dufresne, P.J.; Brennan, G.P.; O’Neill, S.; Tort, J.F.; Paterson, S.; Marcilla, A.; et al. The Extracellular Vesicles of the Helminth Pathogen, Fasciola hepatica: Biogenesis Pathways and Cargo Molecules Involved in Parasite Pathogenesis. Mol. Cell. Proteom. 2015, 14, 3258–3273. [Google Scholar] [CrossRef]
- Zhu, L.; Liu, J.; Dao, J.; Lu, K.; Li, H.; Gu, H.; Liu, J.; Feng, X.; Cheng, G. Molecular characterization of S. japonicum exosome-like vesicles reveals their regulatory roles in parasite-host interactions. Sci. Rep. 2016, 6, 25885. [Google Scholar] [CrossRef]
- Zheng, Y.; Guo, X.; Su, M.; Guo, A.; Ding, J.; Yang, J.; Xiang, H.; Cao, X.; Zhang, S.; Ayaz, M.; et al. Regulatory effects of Echinococcus multilocularis extracellular vesicles on RAW264.7 macrophages. Veter. Parasitol. 2017, 235, 29–36. [Google Scholar] [CrossRef]
- Nicolao, M.C.; Rodrigues, C.R.; Cumino, A.C. Extracellular vesicles from Echinococcus granulosus larval stage: Isolation, characterization and uptake by dendritic cells. PLoS Negl. Trop. Dis. 2019, 13, e0007032. [Google Scholar] [CrossRef] [PubMed]
- Eichenberger, R.M.; Ryan, S.; Jones, L.; Buitrago, G.; Polster, R.; De Oca, M.M.; Zuvelek, J.; Giacomin, P.R.; Dent, L.A.; Engwerda, C.R.; et al. Hookworm Secreted Extracellular Vesicles Interact With Host Cells and Prevent Inducible Colitis in Mice. Front. Immunol. 2018, 9, 850. [Google Scholar] [CrossRef]
- Eichenberger, R.M.; Talukder, H.; Field, M.A.; Wangchuk, P.; Giacomin, P.R.; Loukas, A.; Sotillo, J. Characterization of Trichuris muris secreted proteins and extracellular vesicles provides new insights into host–parasite communication. J. Extracell. Vesicles 2018, 7, 1428004–1428016. [Google Scholar] [CrossRef] [PubMed]
- Bernal, L.; Trelis, M.; Montaner, S.; Cantalapiedra, F.; Galiano, A.; Hackenberg, M.; Gomis, A.M.; Tarbes, S.M. Surface analysis of Dicrocoelium dendriticum. The molecular characterization of exosomes reveals the presence of miRNAs. J. Proteom. 2014, 105, 232. [Google Scholar] [CrossRef] [PubMed]
- Buck, A.H.; Coakley, G.; Simbari, F.; McSorley, H.J.; Quintana, J.F.; Le Bihan, T.; Kumar, S.; Abreu-Goodger, C.; Lear, M.; Harcus, Y.; et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 2014, 5, 5488. [Google Scholar] [CrossRef]
- Hansen, E.P.; Fromm, B.; Andersen, S.D.; Marcilla, A.; Andersen, K.L.; Borup, A.; Williams, A.R.; Jex, A.R.; Gasser, R.B.; Young, N.D.; et al. Exploration of extracellular vesicles from Ascaris suum provides evidence of parasite–host cross talk. J. Extracell. Vesicles 2019, 8, 1578116. [Google Scholar] [CrossRef]
- Wang, L.; Li, Z.; Shen, J.; Liu, Z.; Liang, J.; Wu, X.; Sun, X.; Wu, Z. Exosome-like vesicles derived by Schistosoma japonicum adult worms mediates M1 type immune-activity of macrophage. Parasitol. Res. 2015, 114, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Zamanian, M.; Fraser, L.M.; Agbedanu, P.N.; Harischandra, H.; Moorhead, A.R.; Day, T.; Bartholomay, L.C.; Kimber, M.J. Release of Small RNA-containing Exosome-like Vesicles from the Human Filarial Parasite Brugia malayi. PLoS Negl. Trop. Dis. 2015, 9, e0004069. [Google Scholar] [CrossRef] [PubMed]
- Shears, R.K.; Bancroft, A.J.; Hughes, G.W.; Grencis, R.K.; Thornton, D.J. Extracellular vesicles induce protective immunity against Trichuris muris. Parasite Immunol. 2018, 40, e12536. [Google Scholar] [CrossRef]
- Chaiyadet, S.; Sotillo, J.; Krueajampa, W.; Thongsen, S.; Brindley, P.J.; Sripa, B.; Loukas, A.; Laha, T. Vaccination of hamsters with Opisthorchis viverrini extracellular vesicles and vesicle-derived recombinant tetraspanins induces antibodies that block vesicle uptake by cholangiocytes and reduce parasite burden after challenge infection. PLoS Negl. Trop. Dis. 2019, 13, e0007450. [Google Scholar] [CrossRef] [PubMed]
- Trelis, M.; Galiano, A.; Bolado, A.; Toledo, R.; Gomis, A.M.; Bernal, D. Subcutaneous injection of exosomes reduces symptom severity and mortality induced by Echinostoma caproni infection in BALB/c mice. Int. J. Parasitol. 2016, 46, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Coakley, G.; Maizels, R.M.; Buck, A.H. Exosomes and Other Extracellular Vesicles: The New Communicators in Parasite Infections. Trends Parasitol. 2015, 31, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Da’Dara, A.; Skelly, P.J.; Wang, M.; Harn, D.A. Immunization with plasmid DNA encoding the integral membrane protein, Sm23, elicits a protective immune response against schistosome infection in mice. Vaccine 2001, 20, 359–369. [Google Scholar] [CrossRef]
- Pearson, M.S.; Pickering, D.A.; McSorley, H.J.; Bethony, J.M.; Tribolet, L.; Dougall, A.M.; Hotez, P.J.; Loukas, A. Enhanced Protective Efficacy of a Chimeric Form of the Schistosomiasis Vaccine Antigen Sm-TSP-2. PLoS Negl. Trop. Dis. 2012, 6, e1564. [Google Scholar] [CrossRef]
- Tran, M.H.; Pearson, M.S.; Bethony, J.; Smyth, D.J.; Jones, M.K.; Duke, M.; Don, A.T.; McManus, D.P.; Correa-Oliveira, R.; Loukas, A. Tetraspanins on the surface of Schistosoma mansoni are protective antigens against schistosomiasis. Nat. Med. 2006, 12, 835–840. [Google Scholar] [CrossRef]
- Keitel, W.; Potter, G.; Diemert, D.; Bethony, J.; El Sahly, H.; Kennedy, J.; Patel, S.; Plieskatt, J.; Jones, W.; Deye, G.; et al. A phase 1 study of the safety, reactogenicity, and immunogenicity of a Schistosoma mansoni vaccine with or without glucopyranosyl lipid A aqueous formulation (GLA-AF) in healthy adults from a non-endemic area. Vaccine 2019, 37, 6500–6509. [Google Scholar] [CrossRef]
- Yuan, C.; Fu, Y.-J.; Li, J.; Yue, Y.-F.; Cai, L.-L.; Xiao, W.-J.; Chen, J.-P.; Yang, L. Schistosoma japonicum: Efficient and rapid purification of the tetraspanin extracellular loop 2, a potential protective antigen against schistosomiasis in mammalian. Exp. Parasitol. 2010, 126, 456–461. [Google Scholar] [CrossRef]
- Tucker, M.S.; Karunaratne, L.B.; Lewis, F.A.; Frietas, T.C.; Liang, Y.-S. Schistosomiasis. In Current Protocols in Immunology; Coico, R., Ed.; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2013; pp. 19.11.11–19.11.57. [Google Scholar] [CrossRef]
- Stroehlein, A.J.; Korhonen, P.K.; Chong, T.M.; Lim, Y.L.; Chan, K.-G.; Webster, L.B.; Rollinson, D.; Brindley, P.J.; Gasser, R.; Young, N.D. High-quality Schistosoma haematobium genome achieved by single-molecule and long-range sequencing. GigaScience 2019, 8. [Google Scholar] [CrossRef]
- Young, N.D.; Jex, A.R.; Li, B.; Liu, S.; Yang, L.; Xiong, Z.; Li, Y.; Cantacessi, C.; Hall, R.S.; Xu, X.; et al. Whole-genome sequence of Schistosoma haematobium. Nat. Genet. 2012, 44, 221–225. [Google Scholar] [CrossRef]
- Craig, R.; Cortens, A.J.P.; Beavis, R.C. Open Source System for Analyzing, Validating, and Storing Protein Identification Data. J. Proteome Res. 2004, 3, 1234–1242. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Pevzner, P.A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014, 5, 5277. [Google Scholar] [CrossRef] [PubMed]
- Diament, B.J.; Noble, W.S. Faster SEQUEST Searching for Peptide Identification from Tandem Mass Spectra. J. Proteome Res. 2011, 10, 3871–3879. [Google Scholar] [CrossRef] [PubMed]
- Vaudel, M.; Barsnes, H.; Berven, F.S.; Sickmann, A.; Martens, L. SearchGUI: An open? Source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics 2011, 11, 996–999. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39 (Suppl. 2), W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Goetz, S.; García-Gómez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformation 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Supek, F.; Bosnjak, M.; Skunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Sonnhammer, E.L.; von Heijne, G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998, 6, 175–182. [Google Scholar]
- Petersen, T.N.; Brunak, S.; Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Tedla, B.A.; Pickering, D.; Becker, L.; Loukas, A.; Pearson, M.S. Vaccination with Schistosoma mansoni Cholinesterases Reduces the Parasite Burden and Egg Viability in a Mouse Model of Schistosomiasis. Vaccines 2020, 8, 162. [Google Scholar] [CrossRef]
- WHO. Fact Sheet on Neglected Tropical Diseases. Available online: https://www.who.int/en/news-room/fact-sheets/detail/schistosomiasis (accessed on 20 January 2019).
- Bergquist, R.; Utzinger, J.; Keiser, J. Controlling schistosomiasis with praziquantel: How much longer without a viable alternative? Infect. Dis. Poverty 2017, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Cioli, D. Praziquantel: Is there real resistance and are there alternatives? Curr. Opin. Infect. Dis. 2000, 13, 659–663. [Google Scholar] [CrossRef] [PubMed]
- Tucher, C.; Bode, K.; Schiller, P.; Classen, L.; Birr, C.; Souto-Carneiro, M.M.; Blank, N.; Lorenz, H.-M.; Schiller, M. Extracellular Vesicle Subtypes Released From Activated or Apoptotic T-Lymphocytes Carry a Specific and Stimulus-Dependent Protein Cargo. Front. Immunol. 2018, 9, 534. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Yu, H.; Mim, C.; Matouschek, A. Regulated protein turnover: Snapshots of the proteasome in action. Nat. Rev. Mol. Cell Biol. 2014, 15, 122–133. [Google Scholar] [CrossRef]
- De Paula, R.G.; Ornelas, A.M.D.M.; Morais, E.R.; Gomes, M.D.S.; Aguiar, D.D.P.; Magalhães, L.G.; Rodrigues, V. Proteasome stress responses in Schistosoma mansoni. Parasitol. Res. 2015, 114, 1747–1760. [Google Scholar] [CrossRef]
- Guerra-Sá, R.; Castro-Borges, W.; Evangelista, E.; Kettelhut, I.C.; Rodrigues, V. Schistosoma mansoni: Functional proteasomes are required for development in the vertebrate host. Exp. Parasitol. 2005, 109, 228–236. [Google Scholar] [CrossRef]
- Nabhan, J.F.; El-Shehabi, F.; Patocka, N.; Ribeiro, P. The 26S proteasome in Schistosoma mansoni: Bioinformatics analysis, developmental expression, and RNA interference (RNAi) studies. Exp. Parasitol. 2007, 117, 337–347. [Google Scholar] [CrossRef]
- Arosio, P.; Elia, L.; Poli, M. Ferritin, cellular iron storage and regulation. IUBMB Life 2017, 69, 414–422. [Google Scholar] [CrossRef]
- Orino, K.; Lehman, L.; Tsuji, Y.; Ayaki, H.; Torti, S.V.; Torti, F.M. Ferritin and the response to oxidative stress. Biochem. J. 2001, 357, 241–247. [Google Scholar] [CrossRef]
- Jones, M.K.; McManus, D.P.; Sivadorai, P.; Glanfield, A.; Moertel, L.; Belli, S.I.; Gobert, G.N. Tracking the fate of iron in early development of human blood flukes. Int. J. Biochem. Cell Biol. 2007, 39, 1646–1658. [Google Scholar] [CrossRef][Green Version]
- Chen, L.-Y.; Yi, X.-Y.; Zeng, X.-F.; Zhang, S.-K.; McReynolds, L. Mucosal immunization of recombinant Schistosoma japonicum ferritin. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi 2004, 22, 129–132. [Google Scholar] [PubMed]
- Nagamune, K.; Moreno, S.N.; Chini, E.N.; Sibley, L.D. Calcium regulation and signaling in apicomplexan parasites. Subcell. Biochem. 2008, 47, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Bucki, R.; Bachelot-Loza, C.; Zachowski, A.; Giraud, F.; Sulpice, J.-C. Calcium Induces Phospholipid Redistribution and Microvesicle Release in Human Erythrocyte Membranes by Independent Pathways†. Biochemistry 1998, 37, 15383–15391. [Google Scholar] [CrossRef] [PubMed]
- Sotillo, J.; Robinson, M.W.; Kimber, M.J.; Cucher, M.; Ancarola, M.E.; Nejsum, P.; Marcilla, A.; Eichenberger, R.M.; Tritten, L. The protein and microRNA cargo of extracellular vesicles from parasitic helminths—Current status and research priorities. Int. J. Parasitol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Repasky, G.; Chenette, E.J.; Der, C.J. Renewing the conspiracy theory debate: Does Raf function alone to mediate Ras oncogenesis? Trends Cell Biol. 2004, 14, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Schüßler, P.; Grevelding, C.G.; Kunz, W. Identification of Ras, MAP kinases, and a GAP protein in Schistosoma mansoni by immunoblotting and their putative involvement in male–female interaction. Parasitology 1997, 115 Pt 6, 629–634. [Google Scholar] [CrossRef]
- Gómez-Puertas, P.; Martín-Benito, J.; Carrascosa, J.L.; Willison, K.R.; Valpuesta, J.M. The substrate recognition mechanisms in chaperonins. J. Mol. Recognit. 2004, 17, 85–94. [Google Scholar] [CrossRef]
- Frydman, J.; Nimmesgern, E.; Erdjument-Bromage, H.; Wall, J.; Tempst, P.; Hartl, F. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 1992, 11, 4767–4778. [Google Scholar] [CrossRef]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in Extracellular Vesicle Formation and Function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef]
- Chaiyadet, S.; Krueajampa, W.; Hipkaeo, W.; Plosan, Y.; Piratae, S.; Sotillo, J.; Smout, M.J.; Sripa, B.; Brindley, P.J.; Loukas, A.; et al. Suppression of mRNAs encoding CD63 family tetraspanins from the carcinogenic liver fluke Opisthorchis viverrini results in distinct tegument phenotypes. Sci. Rep. 2017, 7, 14342. [Google Scholar] [CrossRef] [PubMed]
- Piratae, S.; Tesana, S.; Jones, M.K.; Brindley, P.J.; Loukas, A.; Lovas, E.; Eursitthichai, V.; Sripa, B.; Thanasuwan, S.; Laha, T. Molecular Characterization of a Tetraspanin from the Human Liver Fluke, Opisthorchis viverrini. PLoS Negl. Trop. Dis. 2012, 6, e1939. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.H.; Freitas, T.C.; Cooper, L.; Gaze, S.; Gatton, M.; Jones, M.K.; Lovas, E.; Pearce, E.J.; Loukas, A. Suppression of mRNAs Encoding Tegument Tetraspanins from Schistosoma mansoni Results in Impaired Tegument Turnover. PLoS Pathog. 2010, 6, e1000840. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, J.; Duke, M.; Jones, M.K.; Kuang, L.; Zhang, J.; Blair, D.; Li, Y.; McManus, D.P. Inconsistent Protective Efficacy and Marked Polymorphism Limits the Value of Schistosoma japonicum Tetraspanin-2 as a Vaccine Target. PLoS Negl. Trop. Dis. 2011, 5, e1166. [Google Scholar] [CrossRef] [PubMed]
- Kifle, D.W.; Chaiyadet, S.; Waardenberg, A.J.; Wise, I.; Cooper, M.; Becker, L.; Doolan, D.L.; Laha, T.; Sotillo, J.; Pearson, M.S.; et al. Uptake of Schistosoma mansoni extracellular vesicles by human endothelial and monocytic cell lines and impact on vascular endothelial cell gene expression. Int. J. Parasitol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Burke, M.L.; Jones, M.K.; Gobert, G.N.; Li, Y.S.; Ellis, M.K.; McManus, D.P. Immunopathogenesis of human schistosomiasis. Parasite Immunol. 2009, 31, 163–176. [Google Scholar] [CrossRef]
- Beaumier, C.M.; Gillespie, P.M.; Hotez, P.J.; Bottazzi, M.E. New vaccines for neglected parasitic diseases and dengue. Transl. Res. 2013, 162, 144–155. [Google Scholar] [CrossRef]
- Bushara, H.O.; Bashir, M.E.N.; Malik, K.H.E.; Mukhtar, M.M.; Trottein, F.; Capron, A.; Taylor, M.G. Suppression of Schistosoma bovis egg production in cattle by vaccination with either glutathione S-transferase or keyhole limpet haemocyanin. Parasite Immunol. 1993, 15, 383–390. [Google Scholar] [CrossRef]
- Mbanefo, E.; Kumagai, T.; Kodama, Y.; Kurosaki, T.; Furushima-Shimogawara, R.; Cherif, M.S.; Mizukami, S.; Kikuchi, M.; Huy, N.T.; Ohta, N.; et al. Immunogenicity and anti-fecundity effect of nanoparticle coated glutathione S-transferase (SjGST) DNA vaccine against murine Schistosoma japonicum infection. Parasitol. Int. 2015, 64, 24–31. [Google Scholar] [CrossRef]
- Bushara, H.O.; Majid, A.A.; Saad, A.M.; Hussein, M.F.; Taylor, M.G.; Dargie, J.D.; Marshall, T.F.; Nelson, G.S. Observations on cattle schistosomiasis in the Sudan, a study in comparative medicine. II. Experimental demonstration of naturally acquired resistance to Schistosoma bovis. Am. J. Trop. Med. Hyg. 1980, 29, 442–451. [Google Scholar] [CrossRef]
- Saad, A.; Hussein, M.; Dargie, J.; Taylor, M.; Nelson, G. Schistosoma bovis in calves: The development and clinical pathology of primary infections. Res. Veter. Sci. 1980, 28, 105–111. [Google Scholar] [CrossRef]
- Massoud, J. Parasitological and Pathological Observations on Schistosoma bovis Sonsion, 1876, in Calves, Sheep and Goats in Iran. J. Helminthol. 1973, 47, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Cheever, A.W.; Powers, K.G. Schistosoma mansoni infection in rhesus monkeys: Changes in egg production and egg distribution in prolonged infections in intact and splenectomized monkeys. Ann. Trop. Med. Parasitol. 1969, 63, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Webbe, G.; James, C.; Nelson, G.S.; Smithers, S.R.; Terry, R.J. Acquired resistance to Schistosoma haematobium in the baboon (Papio anubis) after cercarial exposure and adult worm transplantation. Ann. Trop. Med. Parasitol. 1976, 70, 411–424. [Google Scholar] [CrossRef] [PubMed]
- WHO. Ending the Neglect to Attain the Sustainable Development Goals: A Roadmap for Neglected Tropical Diseases 2021–2030. 2020. Available online: https://www.who.int/neglected_diseases/Ending-the-neglect-to-attain-the-SDGs--NTD-Roadmap.pdf?ua=1 (accessed on 2 July 2020).
- Kura, K.; Truscott, J.E.; Toor, J.; Anderson, R.M. Modelling the impact of a Schistosoma mansoni vaccine and mass drug administration to achieve morbidity control and transmission elimination. PLoS Negl. Trop. Dis. 2019, 13, e0007349. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).