Overexpression of VviPGIP1 and NtCAD14 in Tobacco Screened Using Glycan Microarrays Reveals Cell Wall Reorganisation in the Absence of Fungal Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Plant Growth Conditions
2.3. Cell Wall Isolation Protocol
2.4. Pyrolysis Gas Chromatography Mass Spectrometry (Py-GC–MS) for Lignin Analysis
2.5. Gas Chromatography–Mass Spectrometry (GC–MS) for Monosaccharides
2.6. Comprehensive Microarray Polymer Profiling (CoMPP) for Polysaccharides-Proteins
2.7. Univariate Statistical and Multivariate Data Analysis
3. Results
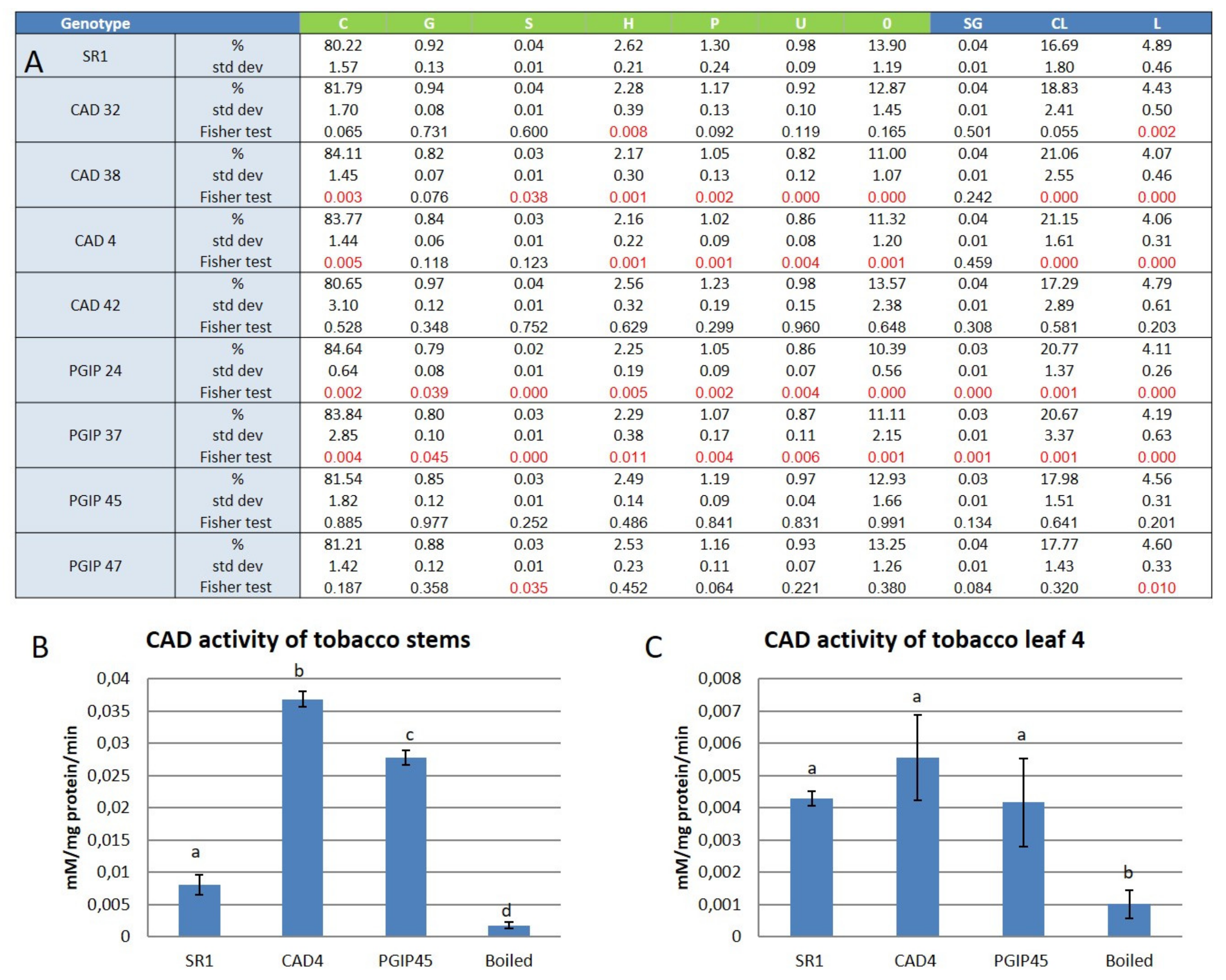
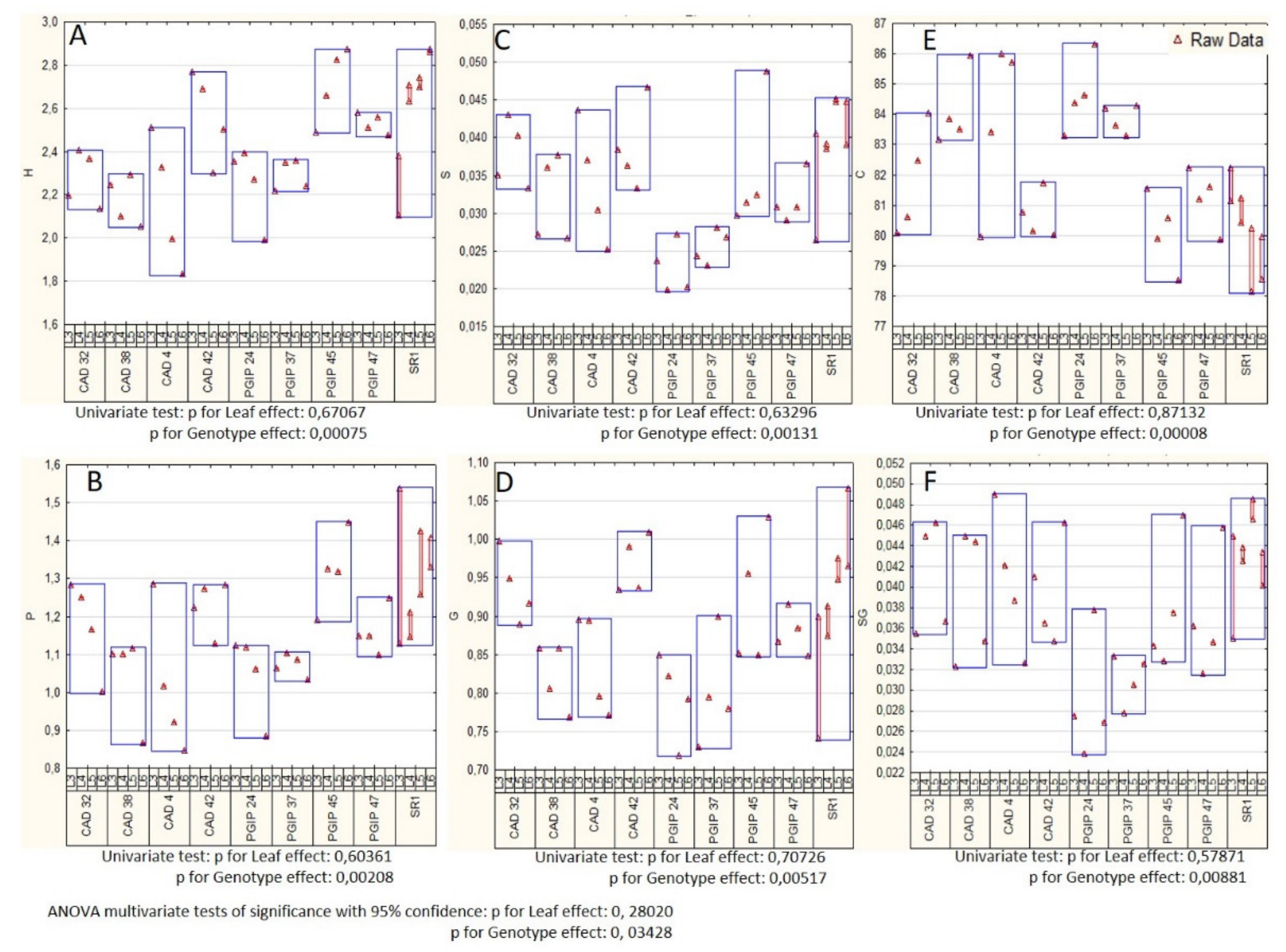
3.1. Lignin Compositional Analysis of Wild Type Versus Transgenic Tobacco Using Py-GC–MS
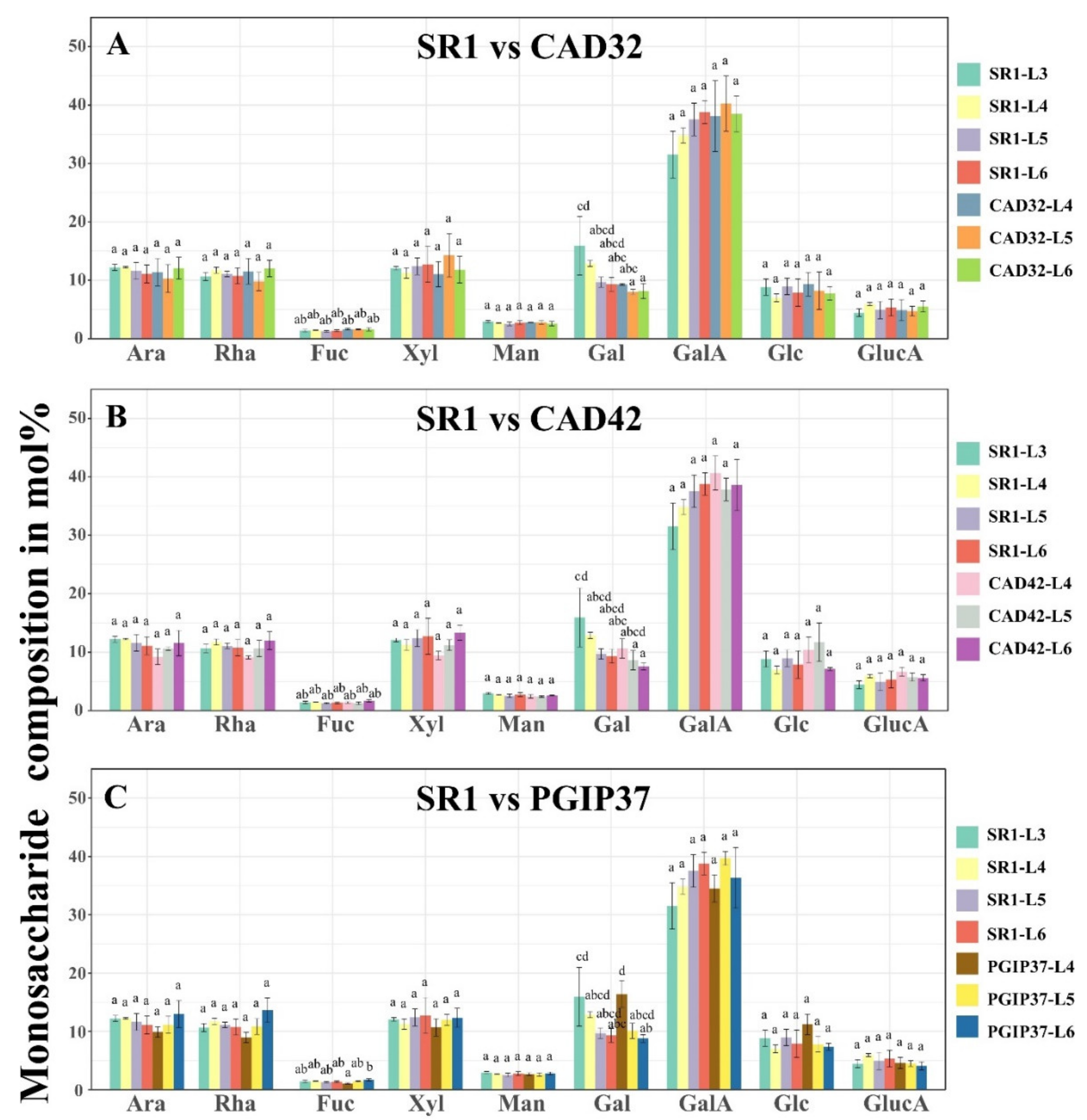
3.2. Monosaccharide Composition Analysis of Wild Type Versus Transgenic Tobacco Using GC–MS
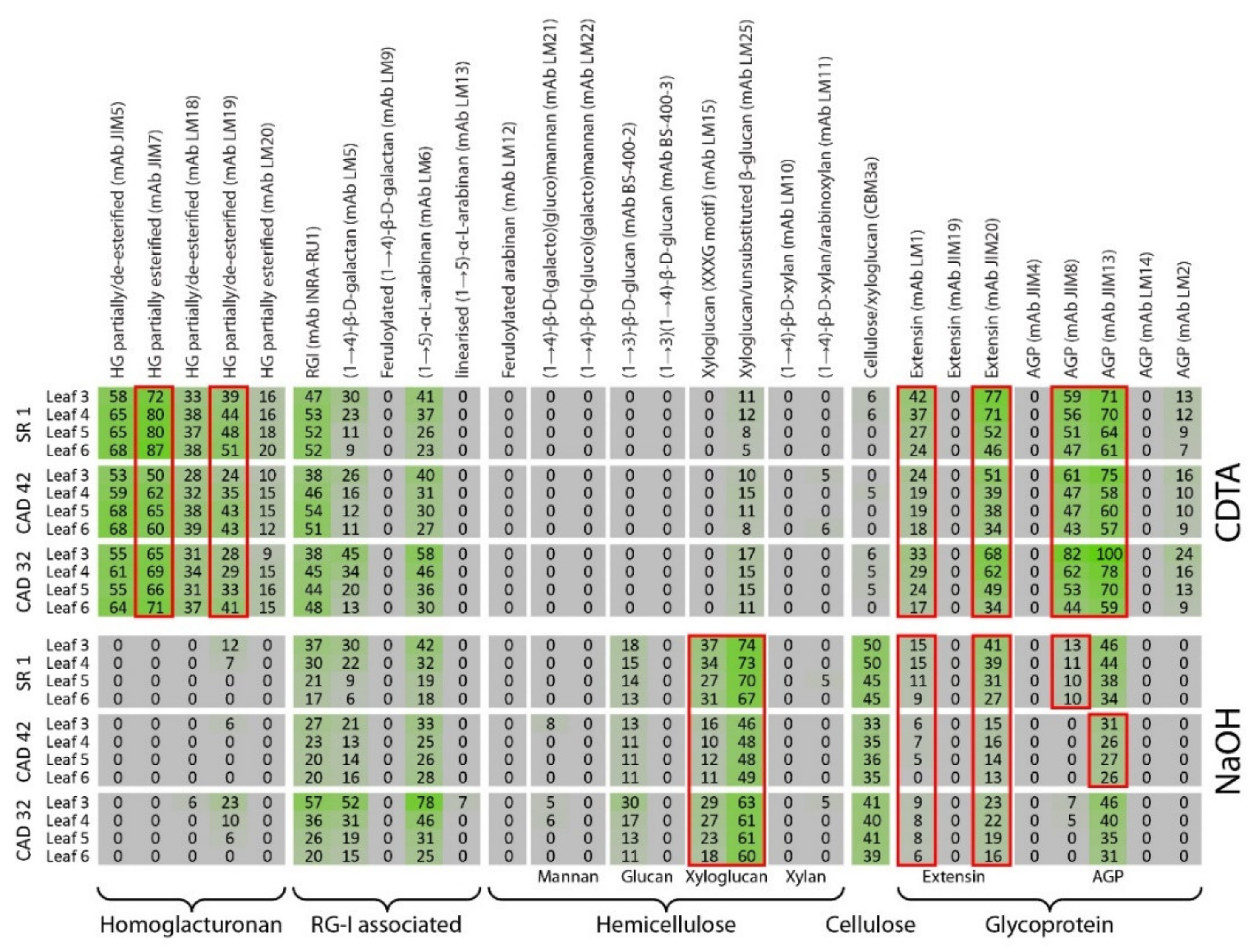
3.3. Comprehensive Microarray Polymer Profiling (CoMPP) of SR1 Versus CAD Transgenic Tobacco Lines
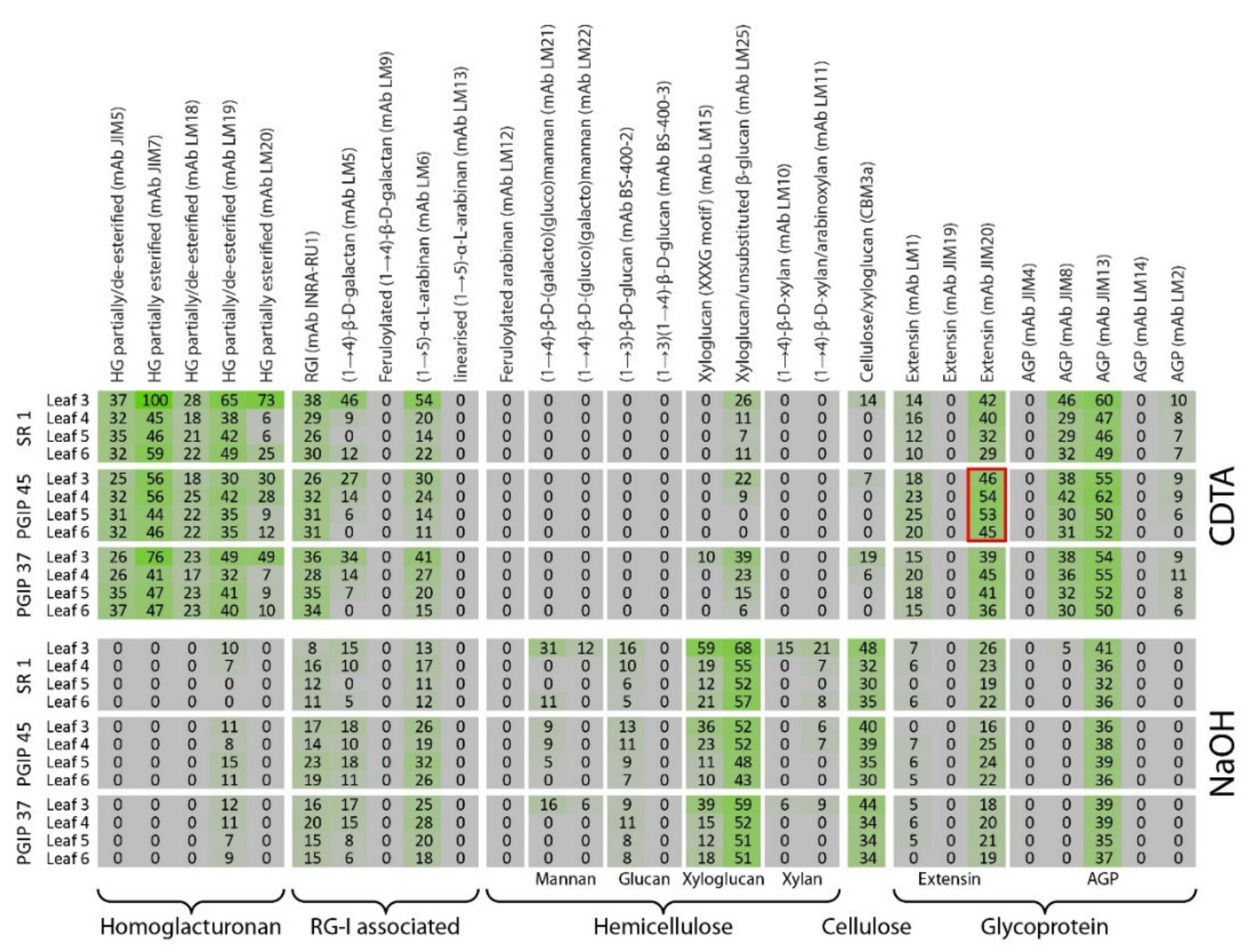
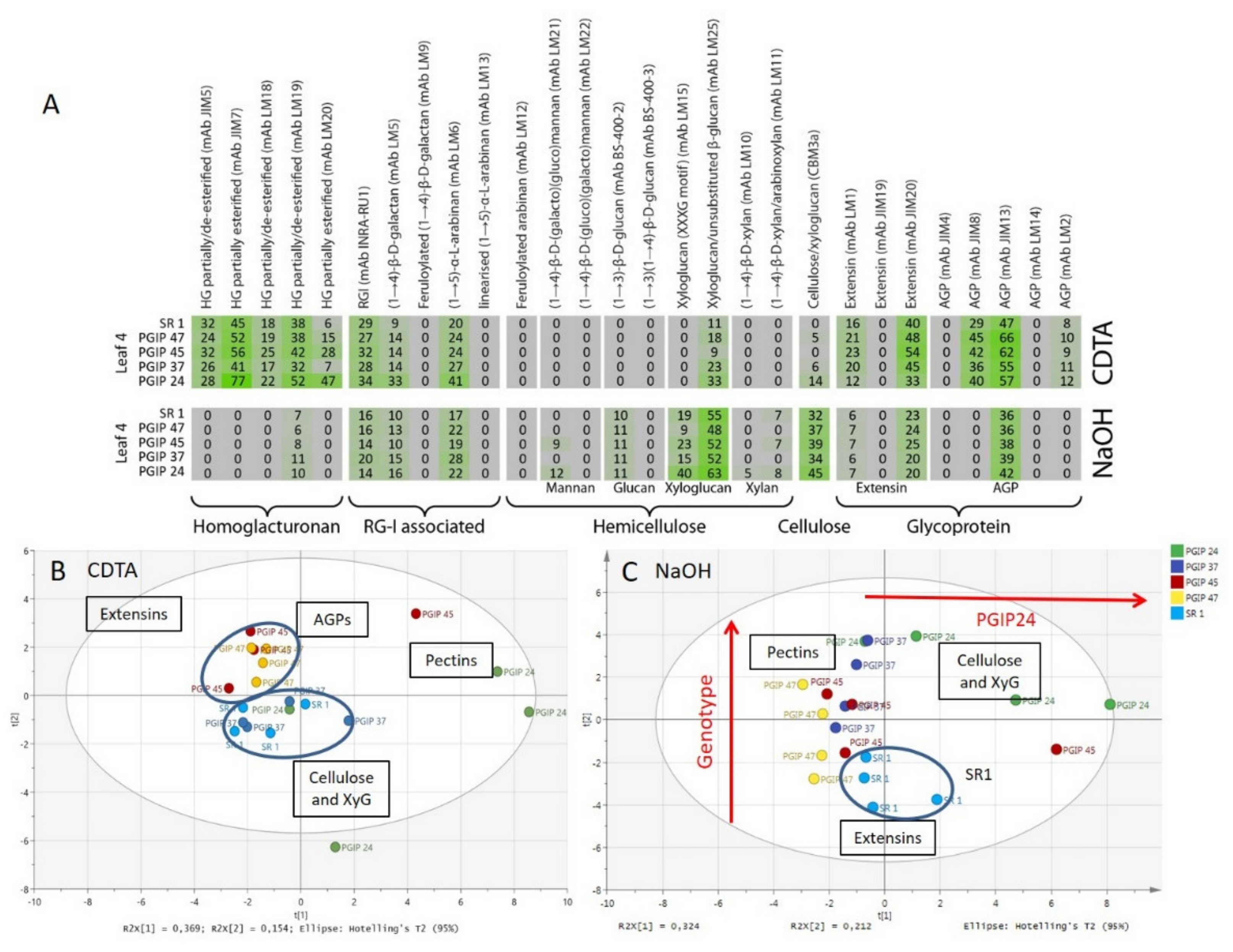
3.4. Comprehensive Microarray Polymer Profiling (CoMPP) of SR1 Versus Grapevine PGIP1 Transgenic Tobacco Lines
4. Discussion
4.1. Lignin Composition was Mostly Unaltered in Leaves of Plants Overexpressing Either CAD or the Grapevine PGIP1 Gene
4.2. Pectin Organisation Varied between the SR1 and the Transgenic CAD and PGIP Lines
4.3. Extensin and AGP Epitope Distribution Varies between Wild Type and Transgenic Lines Suggesting a Role in Cell Wall Resistance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Albersheim, P.; Darvill, A.G.; Roberts, K.; Sederoff, R.; Staehelin, A. Cell Walls and Plant Anatomy. In Plant Cell Walls; Garland Science: New York, NY, USA, 2011; pp. 1–42. ISBN 9780815319962. [Google Scholar]
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef] [PubMed]
- Petrasch, S.; Silva, C.J.; Mesquida-Pesci, S.D.; Gallegos, K.; van den Abeele, C.; Papin, V.; Fernandez-Acero, F.J.; Knapp, S.J.; Blanco-Ulate, B. Infection strategies deployed by Botrytis cinerea, Fusarium acuminatum, and Rhizopus stolonifer as a function of tomato fruit ripening stage. Front. Plant Sci. 2019, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Castilleux, R.; Plancot, B.; Ropitaux, M.; Carreras, A.; Leprince, J.; Boulogne, I.; Follet-Gueye, M.-L.; Popper, Z.A.; Driouich, A.; Vicré-Gibouin, M. Cell wall extensins in root–microbe interactions and root secretions. J. Exp. Bot. 2018, 69, 4235–4247. [Google Scholar] [CrossRef]
- Castilleux, R.; Plancot, B.; Gügi, B.; Attard, A.; Loutelier-Bourhis, C.; Lefranc, B.; Nguema-Ona, E.; Arkoun, M.; Yvin, J.-C.; Driouich, A.; et al. Extensin arabinosylation is involved in root response to elicitors and limits oomycete colonization. Ann. Bot. 2019, 22, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ryder, L.S.; Talbot, N.J. Regulation of appressorium development in pathogenic fungi. Curr. Opin. Plant Biol. 2015, 26, 8–13. [Google Scholar] [CrossRef]
- Prins, T.W.; Tudzynski, P.; von Tiedemann, A.; Tudzynski, B.; Ten Have, A.; Hansen, M.E.; Tenberge, K.; van Kan, J.A.L. Infection strategies of Botrytis cinerea and related necrotrophic pathogens. In Fungal Pathology; Springer: Dordrecht, The Netherlands, 2000; pp. 33–64. [Google Scholar]
- Ten Have, A. The Botrytis Cinerea Endopolygalacturonase Gene Family. PhD Thesis, Wageningen University, Wageningen, The Netherlands, 2000. [Google Scholar]
- Esquerré-Tugayé, M.T.; Boudart, G.; Dumas, B. Cell wall degrading enzymes, inhibitory proteins, and oligosaccharides participate in the molecular dialogue between plants and pathogens. Plant Physiol. 2000, 38, 157–163. [Google Scholar] [CrossRef]
- Kars, I.; Krooshof, G.H.; Wagemakers, L.; Joosten, R.; Benen, J.A.E.; Van Kan, J.A.L. Necrotizing activity of five Botrytis cinerea endopolygalacturonases produced in Pichia pastoris. Plant J. 2005, 43, 213–225. [Google Scholar] [CrossRef]
- Kühnel, S. Characterization of Cell Wall Degrading Enzymes from Chrysosporium Lucknowense C1 and Their Use to Degrade Sugar Beet Pulp. PhD Thesis, Wageningen University, Wageningen, The Netherlands, 2011. [Google Scholar]
- Tudzynski, P.; Kokkelink, L. Botrytis cinerea: molecular aspects of a necrotrophic life style. Style DeKalb IL 2009, 5, 29–50. [Google Scholar] [CrossRef]
- Barnes, W.J.; Anderson, C.T. Release, recycle, rebuild: cell-wall remodeling, autodegradation, and sugar salvage for new wall biosynthesis during plant development. Mol. Plant 2018, 11, 31–46. [Google Scholar] [CrossRef]
- De Lorenzo, G.; Ferrari, S. Polygalacturonase-inhibiting proteins in defense against phytopathogenic fungi. Curr. Opin. Plant Biol. 2002, 5, 295–299. [Google Scholar] [CrossRef]
- Gomathi, V.; Gnanamanickam, S.S. Polygalacturonase-inhibiting proteins in plant defence. Current 2004, 87, 1211–1217. [Google Scholar]
- Kalunke, R.M.; Tundo, S.; Benedetti, M.; Cervone, F.; De Lorenzo, G.; D’Ovidio, R. An update on polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein that protects crop plants against pathogens. Front. Plant Sci. 2015, 6, 146. [Google Scholar] [CrossRef]
- Alexandersson, E.; Becker, J.V.W.; Jacobson, D.; Nguema-Ona, E.; Steyn, C.; Denby, K.J.; Vivier, M.A. Constitutive expression of a grapevine polygalacturonase-inhibiting protein affects gene expression and cell wall properties in uninfected tobacco. BMC Res. Notes 2011, 4, 493. [Google Scholar] [CrossRef]
- Joubert, D.A.; Kars, I.; Wagemakers, L.; Bergmann, C.; Kemp, G.; Vivier, M.A.; Van Kan, J.A.L. A polygalacturonase-inhibiting protein from grapevine reduces the symptoms of the endopolygalacturonase BcPG2 from Botrytis cinerea in Nicotiana benthamiana leaves without any evidence for in vitro interaction. Mol. Plant-Microbe Interact. 2007, 20, 392–402. [Google Scholar] [CrossRef] [PubMed]
- Federici, L.; Caprari, C.; Mattei, B.; Savino, C.; Di Matteo, A.; De Lorenzo, G.; Cervone, F.; Tsernoglou, D.; Staskawicz, B.J. Structural requirements of endopolygalacturonase for the interaction with PGIP (polygalacturonase- inhibiting protein). Proc. Natl. Acad. Sci. USA 2001, 98, 13425–13430. [Google Scholar] [CrossRef] [PubMed]
- Sicilia, F.; Fernandez-Recio, J.; Caprari, C.; De Lorenzo, G.; Tsernoglou, D.; Cervone, F.; Federici, L. The Polygalacturonase-inhibiting protein pgip2 of Phaseolus vulgaris has evolved a mixed mode of inhibition of endopolygalacturonase pg1 of Botrytis cinerea 1. Plant Physiol. 2005, 139, 1380–1388. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhang, X.; Sun, Y.; Wang, P.; Li, X.; Pei, Y.; Li, F.; Hou, Y. Molecular evidence for the involvement of a polygalacturonase-inhibiting protein, GhPGIP1, in enhanced resistance to Verticillium and Fusarium wilts in cotton. Sci. Rep. 2017, 7, 39840. [Google Scholar] [CrossRef]
- Liu, N.; Ma, X.; Zhou, S.; Wang, P.; Sun, Y.; Li, X.; Hou, Y. Molecular and functional characterization of a polygalacturonase-inhibiting protein from Cynanchum komarovii that confers fungal resistance in Arabidopsis. PLoS ONE 2016, 11, e0146959. [Google Scholar] [CrossRef][Green Version]
- Joubert, D.A.; Slaughter, A.R.; Kemp, G.; Becker, J.V.W.; Krooshof, G.H.; Bergmann, C.; Benen, J.; Pretorius, I.S.; Vivier, M.A. The grapevine polygalacturonase-inhibiting protein (VvPGIP1) reduces Botrytis cinerea susceptibility in transgenic tobacco and differentially inhibits fungal polygalacturonases. Transgenic Res. 2006, 15, 687–702. [Google Scholar] [CrossRef]
- Spadoni, S.; Zabotina, O.; Di Matteo, A.; Dalgaard Mikkelsen, J.; Cervone, F.; De Lorenzo, G.; Mattei, B.; Bellincampi, D. Polygalacturonase-inhibiting protein interacts with pectin through a binding site formed by four clustered residues of arginine and lysine 1. Plant Physiol. 2006. [Google Scholar] [CrossRef] [PubMed]
- D’Ovidio, R.; Mattei, B.; Roberti, S.; Bellincampi, D. Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant-pathogen interactions. Biochim. Biophys. Acta Proteins Proteom. 2004, 1696, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Davidsson, P.; Broberg, M.; Kariola, T.; Sipari, N.; Pirhonen, M.; Palva, E.T. Short oligogalacturonides induce pathogen resistance-associated gene expression in Arabidopsis thaliana. BMC Plant Biol. 2017, 17, 19. [Google Scholar] [CrossRef]
- Benedetti, M.; Pontiggia, D.; Raggi, S.; Cheng, Z.; Scaloni, F.; Ferrari, S.; Ausubel, F.M.; Cervone, F.; De Lorenzo, G. Plant immunity triggered by engineered in vivo release of oligogalacturonides, damage-associated molecular patterns. Proc. Natl. Acad. Sci. USA 2015, 112, 5533–5538. [Google Scholar] [CrossRef]
- Ferrari, S.; Savatin, D.V.; Sicilia, F.; Gramegna, G.; Cervone, F.; Lorenzo, G. De Oligogalacturonides: Plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 2013, 4, 49. [Google Scholar] [CrossRef]
- Vorwerk, S.; Somerville, S.; Somerville, C. The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci. 2004, 9, 203–209. [Google Scholar] [CrossRef]
- Voxeur, A.; Habrylo, O.; Guénin, S.; Miart, F.; Soulié, M.-C.; Rihouey, C.; Pau-Roblot, C.; Domon, J.-M.; Gutierrez, L.; Pelloux, J.; et al. Oligogalacturonide production upon Arabidopsis thaliana-Botrytis cinerea interaction. Proc. Natl. Acad. Sci. USA 2019, 116, 19743–19752. [Google Scholar] [CrossRef]
- Joubert, D.A.; de Lorenzo, G.; Vivier, M.A. Regulation of the grapevine polygalacturonase-inhibiting protein encoding gene: Expression pattern, induction profile and promoter analysis. J. Plant Res. 2013, 126, 267–281. [Google Scholar] [CrossRef]
- Mbewana, S. Functional Analysis of a lignin biosynthetic gene in transgenic tobacco. Master’s Thesis, University of Stellenbosch, Stellenbosch, South Africa, 2010. [Google Scholar]
- Nguema-Ona, E.; Moore, J.P.; Fagerström, A.D.; Fangel, J.U.; Willats, W.G.T.; Hugo, A.; Vivier, M.A. Overexpression of the grapevine PGIP1 in tobacco results in compositional changes in the leaf arabinoxyloglucan network in the absence of fungal infection. BMC Plant Biol. 2013, 13, 46. [Google Scholar] [CrossRef]
- Claverie, J.; Balacey, S.; Lemaître-Guillier, C.; Brulé, D.; Chiltz, A.; Granet, L.; Noirot, E.; Daire, X.; Darblade, B.; Héloir, M.-C.; et al. The cell wall-derived xyloglucan is a new damp triggering plant immunity in Vitis vinifera and Arabidopsis thaliana. Front. Plant Sci. 2018, 9, 1725. [Google Scholar] [CrossRef]
- Basson, C.E. Transcriptomic analysis of disease resistance responses using a tobacco-Botrytis cinerea pathosystem. Ph.D. Thesis, Stellenbosch University, Stellenbosch, South Africa, 2017. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Nguema-Ona, E.; Moore, J.P.; Fagerström, A.; Fangel, J.U.; Willats, W.G.T.; Hugo, A.; Vivier, M.A. Profiling the main cell wall polysaccharides of tobacco leaves using high-throughput and fractionation techniques. Carbohydr. Polym. 2012, 88, 939–949. [Google Scholar] [CrossRef]
- Gerber, L.; Eliasson, M.; Trygg, J.; Moritz, T.; Sundberg, B. Multivariate curve resolution provides a high-throughput data processing pipeline for pyrolysis-gas chromatography/mass spectrometry. J. Anal. Appl. Pyrolysis 2012, 95, 95–100. [Google Scholar] [CrossRef]
- York, W.S.; Darvill, A.G.; McNeil, M.; Stevenson, T.T.; Albersheim, P. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 1986, 118, 3–40. [Google Scholar]
- Kračun, S.K.; Fangel, J.U.; Rydahl, M.G.; Pedersen, H.L.; Vidal-Melgosa, S.; Willats, W.G.T. Carbohydrate microarray technology applied to high-throughput mapping of plant cell wall glycans using comprehensive microarray polymer profiling (CoMPP). Methods Mol. Biol. (Clifton NJ) 2017, 1503, 147–165. [Google Scholar]
- Knox, J.P.; Linstead, P.; King, J.; Cooper, C.; Roberts, K. Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 1990, 181, 512–521. [Google Scholar] [CrossRef]
- Clausen, M.H.; Willats, W.G.T.; Knox, J.P. Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr. Res. 2003, 338, 1797–1800. [Google Scholar] [CrossRef]
- Verhertbruggen, Y.; Marcus, S.E.; Haeger, A.; Ordaz-Ortiz, J.J.; Knox, J.P. An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr. Res. 2009, 344, 1858–1862. [Google Scholar] [CrossRef]
- Ralet, M.-C.; Tranquet, O.; Poulain, D.; Moïse, A.; Guillon, F. Monoclonal antibodies to rhamnogalacturonan I backbone. Planta 2010, 231, 1373–1383. [Google Scholar] [CrossRef]
- Jones, L.; Seymour, G.B.; Knox, J.P. Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1→4)-[beta]-D-galactan. Plant Physiol. 1997, 113, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Clausen, M.H.; Ralet, M.-C.; Willats, W.G.T.; McCartney, L.; Marcus, S.E.; Thibault, J.-F.; Knox, J.P. A monoclonal antibody to feruloylated-(1→4)-β-d-galactan. Planta 2004, 219, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Willats, W.G.T.; Marcus, S.E.; Knox, J.P. Generation of a monoclonal antibody specific to (1→5)-α-l-arabinan. Carbohydr. Res. 1998, 308, 149–152. [Google Scholar] [CrossRef]
- Verhertbruggen, Y.; Marcus, S.E.; Haeger, A.; Verhoef, R.; Schols, H.A.; McCleary, B.V.; McKee, L.; Gilbert, H.J.; Knox, J.P. Developmental complexity of arabinan polysaccharides and their processing in plant cell walls. Plant J. 2009, 59, 413–425. [Google Scholar] [CrossRef]
- Pedersen, H.L.; Fangel, J.U.; McCleary, B.; Ruzanski, C.; Rydahl, M.G.; Ralet, M.-C.; Farkas, V.; von Schantz, L.; Marcus, S.E.; Andersen, M.C.F.; et al. Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. J. Biol. Chem. 2012, 287, 39429–39438. [Google Scholar] [CrossRef]
- Marcus, S.E.; Blake, A.W.; Benians, T.A.S.; Lee, K.J.D.; Poyser, C.; Donaldson, L.; Leroux, O.; Rogowski, A.; Petersen, H.L.; Boraston, A.; et al. Restricted access of proteins to mannan polysaccharides in intact plant cell walls. Plant J. 2010, 64, 191–203. [Google Scholar] [CrossRef]
- Meikle, P.J.; Bonig, I.; Hoogenraad, N.J.; Clarke, A.E.; Stone, B.A. The location of (1→3)-β-glucans in the walls of pollen tubes of Nicotiana alata using a (1→3)-β-glucan-specific monoclonal antibody. Planta 1991, 185, 1–8. [Google Scholar] [CrossRef]
- Marcus, S.E.; Verhertbruggen, Y.; Hervé, C.; Ordaz-Ortiz, J.J.; Farkas, V.; Pedersen, H.L.; Willats, W.G.T.; Knox, J.P. Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 2008, 8, 60. [Google Scholar] [CrossRef]
- McCartney, L.; Marcus, S.E.; Knox, J.P. Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J. Histochem. Cytochem. 2005, 53, 543–546. [Google Scholar] [CrossRef]
- Tormo, J.; Lamed, R.; Chirino, A.J.; Morag, E.; Bayer, E.A.; Shoham, Y.; Steitz, T.A. Crystal structure of a bacterial family-III cellulose-binding domain: A general mechanism for attachment to cellulose. EMBO J. 1996, 15, 5739–5751. [Google Scholar] [CrossRef]
- Smallwood, M.; Martin, H.; Knox, J.P. An epitope of rice threonine- and hydroxyproline-rich glycoprotein is common to cell wall and hydrophobic plasma-membrane glycoproteins. Planta 1995, 196, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, M.; Beven, A.; Donovan, N.; Neill, S.J.; Peart, J.; Roberts, K.; Knox, J.P. Localization of cell wall proteins in relation to the developmental anatomy of the carrot root apex. Plant J. 1994, 5, 237–246. [Google Scholar] [CrossRef]
- Knox, J.P.; Peart, J.; Neill, S. Identification of novel cell surface epitopes using a leaf epidermal-strip assay system. Planta 1995, 196. [Google Scholar] [CrossRef]
- Knox, J.P.; Day, S.; Roberts, K. A set of cell surface glycoproteins forms an early position, but not cell type, in the root Apical Carota L. Development 1989, 106. [Google Scholar]
- Yates, E.A.; Valdor, J.-F.; Haslam, S.M.; Morris, H.R.; Dell, A.; Mackie, W.; Knox, J.P. Characterization of carbohydrate structural features recognized by anti-arabinogalactan-protein monoclonal antibodies. Glycobiology 1996, 6, 131–139. [Google Scholar] [CrossRef] [PubMed]
- McCabe, P.F.; Valentine, T.A.; Forsberg, L.S.; Pennell, R.I. Soluble signals from cells identified at the cell wall establish a developmental pathway in carrot. Plant Cell 1997, 9, 2225–2241. [Google Scholar] [CrossRef]
- Knox, J.P.; Linstead, P.; Cooper, J.P.C.; Roberts, K. Developmentally regulated epitopes of cell surface arabinogalactan proteins and their relation to root tissue pattern formation. Plant J. 1991, 1, 317–326. [Google Scholar] [CrossRef]
- Moller, I.; Marcus, S.E.; Haeger, A.; Verhertbruggen, Y.; Verhoef, R.; Schols, H.; Ulvskov, P.; Mikkelsen, J.D.; Knox, J.P.; Willats, W. High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. Glycoconj. J. 2008, 25, 37–48. [Google Scholar] [CrossRef]
- Smallwood, M.; Yates, E.A.; Willats, W.G.T.; Martin, H.; Knox, J.P. Immunochemical comparison of membrane-associated and secreted arabinogalactan-proteins in rice and carrot. Planta 1996, 198, 452–459. [Google Scholar] [CrossRef]
- Yoon, J.; Choi, H.; An, G. Roles of lignin biosynthesis and regulatory genes in plant development. J. Integr. Plant Biol. 2015, 57, 902–912. [Google Scholar] [CrossRef]
- Ma, Q.-H. Functional analysis of a cinnamyl alcohol dehydrogenase involved in lignin biosynthesis in wheat. J. Exp. Bot. 2010, 61, 2735–2744. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.-H.; Zhu, H.-H.; Qiao, M.-Y. Contribution of both lignin content and sinapyl monomer to disease resistance in tobacco. Plant Pathol. 2018, 67, 642–650. [Google Scholar] [CrossRef]
- Menden, B.; Kohlhoff, M.; Moerschbacher, B.M. Wheat cells accumulate a syringyl-rich lignin during the hypersensitive resistance response. Phytochemistry 2007, 68, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Cesarino, I. Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 2019, 56, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Brummell, D.A. Cell wall disassembly in ripening fruit. Funct. Plant Biol. 2006, 33, 103. [Google Scholar] [CrossRef]
- Willats, W.G.T.; Steele-King, C.G.; Marcus, S.E.; Knox, J.P. Side chains of pectic polysaccharides are regulated in relation to cell proliferation and cell differentiation. Plant J. 1999, 20, 619–628. [Google Scholar] [CrossRef]
- Zykwinska, A.W.; Ralet, M.-C.J.; Garnier, C.D.; Thibault, J.-F. Evidence for in vitro binding of pectin side chains to cellulose. Plant Physiol. 2005, 139, 397–407. [Google Scholar] [CrossRef]
- Zykwinska, A.; Thibault, J.-F.; Ralet, M.-C. Organization of pectic arabinan and galactan side chains in association with cellulose microfibrils in primary cell walls and related models envisaged. J. Exp. Bot. 2007, 58, 1795–1802. [Google Scholar] [CrossRef]
- Park, Y.B.; Cosgrove, D.J. Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol. 2015, 56, 180–194. [Google Scholar] [CrossRef]
- Cosgrove, D.J.; Jarvis, M.C. Comparative structure and biomechanics of plant primary and secondary cell walls. Front. Plant Sci. 2012, 3, 204. [Google Scholar] [CrossRef]
- Nishitani, K.; Demura, T. Editorial: An emerging view of plant cell walls as an apoplastic intelligent system. Plant Cell Physiol. 2015, 56, 177–179. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.B.; Cosgrove, D.J. A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol. 2012, 158, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Volpi, C.; Janni, M.; Lionetti, V.; Bellincampi, D.; Favaron, F.; D’Ovidio, R. The ectopic expression of a pectin methyl esterase inhibitor increases pectin methyl esterification and limits fungal diseases in wheat. Mol. Plant. Microbe. Interact. 2011, 24, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, V.; Cervone, F.; Bellincampi, D. Methyl esterification of pectin plays a role during plant-pathogen interactions and affects plant resistance to diseases. J. Plant Physiol. 2012, 169, 1623–1630. [Google Scholar] [CrossRef] [PubMed]
- Micheli, F. Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 2001, 6, 414–419. [Google Scholar] [CrossRef]
- Pelloux, J.; Rustérucci, C.; Mellerowicz, E.J. New insights into pectin methylesterase structure and function. Trends Plant Sci. 2007, 12, 267–277. [Google Scholar] [CrossRef]
- Cabrera, J.C.; Boland, A.; Messiaen, J.; Cambier, P.; Van Cutsem, P. Egg box conformation of oligogalacturonides: The time-dependent stabilization of the elicitor-active conformation increases its biological activity. Glycobiology 2008, 18, 473–482. [Google Scholar] [CrossRef]
- Hocq, L.; Pelloux, J.; Lefebvre, V. Connecting homogalacturonan-type pectin remodeling to acid growth. Trends Plant Sci. 2017, 22, 20–29. [Google Scholar] [CrossRef]
- Deepak, S.; Shailasree, S.; Kini, R.K.; Muck, A.; Mithöfer, A.; Shetty, S.H. Hydroxyproline-rich glycoproteins and plant defence. J. Phytopathol. 2010, 158, 585–593. [Google Scholar] [CrossRef]
- Showalter, A.M. Structure and function of plant cell wall proteins. Plant Cell 1993, 5, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Boudart, G.; Dechamp-Guillaume, G.; Lafitte, C.; Ricart, G.; Barthe, J.-P.; Mazau, D.; Esquerré-Tugayé, M.T. Elicitors and suppressors of hydroxyproline-rich glycoprotein accumulation are solubilized from plant cell walls by endopolygalacturonase. Eur. J. Biochem. 1995, 232, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Cassab, G.I. Plant cell wall proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 281–309. [Google Scholar] [CrossRef]
- Showalter, A.M. Arabinogalactan-proteins: structure, expression and function. Cell. Mol. Life Sci. 2001, 58, 1399–1417. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.; Egelund, J.; Schultz, C.J.; Bacic, A. Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol. 2010, 153, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Mareri, L.; Romi, M.; Cai, G. Arabinogalactan proteins: actors or spectators during abiotic and biotic stress in plants? Plant Biosyst. 2018, 1–13. [Google Scholar] [CrossRef]
- Nguema-Ona, E.; Vicré-Gibouin, M.; Cannesan, M.A.; Driouich, A. Arabinogalactan proteins in root-microbe interactions. Trends Plant Sci. 2013, 18, 1360–1385. [Google Scholar] [CrossRef]
- Cannesan, M.A.; Durand, C.; Burel, C.; Gangneux, C.; Lerouge, P.; Ishii, T.; Laval, K.; Follet-Gueye, M.-L.; Driouich, A.; Vicré-Gibouin, M. Effect of arabinogalactan proteins from the root caps of pea and Brassica napus on Aphanomyces euteiches zoospore chemotaxis and germination. Plant Physiol. 2012, 159, 1658–1670. [Google Scholar] [CrossRef]


| Plant Line | Promotor | Percentage of Successful Infections per Plant Line | Average Lesion Diameter (mm) * | Percentage Decrease in Disease Susceptibility (Compared to SR1) *, ** | Experiments Conducted on Those Lines | Reference |
|---|---|---|---|---|---|---|
| SR1 | - | 97% | 24.0 ± 7.3 (11 dpi) | 0 | ||
| CAD 4 | CaMV 35Sp | 93% | 17.7 ± 5.2 | 27% | CAD enzyme activity assay | |
| CAD 32 | 93% | 14.5 ± 3.4 | 40% | Whole plant infection assay | [33] | |
| CAD 38 | - | - | - | |||
| CAD 42 | 97% | 16.4 ± 3.6 | 32% | |||
| SR1 | - | 75% | 40.94 ± 3.5 (15 dpi) | 0 | Detached leaves and whole plant infection assay | [24] |
| Gene expression analysis on infected and uninfected leaf tissue On uninfected leaves: | [18] | |||||
| XTH activity | ||||||
| Phytohormones analysis | ||||||
| Lignin staining | [36] | |||||
| PGIP 24 | CaMV 35Sp | 83% | 15.43 ± 0.9 | 62% | Monosaccharide composition | |
| PGIP 37 | 83% | 12.60 ± 1.1 | 69% | Volatile organic compound analysis | ||
| PGIP 45 | 92% | 21.84 ± 3.1 | 47% | CoMPP | [34] | |
| PGIP 47 | 92% | 36.77 ± 3.2 | 10% | Arabinoxyloglucan analysis |
| Homogalacturonan | low DE | JIM5 | [43] [44] | |
| high DE | JIM7 | |||
| Partially ME | LM18 | [45] | ||
| Partially ME | LM19 | |||
| Partially ME | LM20 | |||
| RG-I associated | Backbone of RG-I | INRA-RU1 | [46] | |
| D-galactan | LM5 | [47] | ||
| Feruloylated galactan | LM9 | [48] | ||
| L-arabinan | LM6 | [49] | ||
| Linearised L-arabinan | LM13 | [50] | ||
| Hemicellulose | Feruloylate polymer | LM12 | [51] | |
| Mannan | (galacto)(gluco)mannan | LM21 | [52] | |
| D-(gluco)mannan | LM22 | |||
| Glucan | β-D-glucan | BS-400-2 | [53] | |
| Mixed link β-D-glucan | BS-400-3 | |||
| Xyloglucan | Xyloglucan (XXXG motif) | LM15 | [54] | |
| Xyloglucan | LM25 | [51] | ||
| Xylan | β-D-xylan | LM10 | [55] | |
| β-D-xylan/arabinoxylan | LM11 | |||
| Cellulose | Cellulose (crystalline) | CBM3a | [56] | |
| Proteins | Extensins | LM1 | [57] | |
| JIM19 | [58] [59] | |||
| JIM20 | ||||
| AGP | JIM4 | [60] [61] | ||
| JIM8 | [62] | |||
| JIM13 | [63] [61] | |||
| LM14 | [64] | |||
| β-linked GlcA | LM2 | [65] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weiller, F.; Gerber, L.; Trygg, J.; Fangel, J.U.; Willats, W.G.T.; Driouich, A.; Vivier, M.A.; Moore, J.P. Overexpression of VviPGIP1 and NtCAD14 in Tobacco Screened Using Glycan Microarrays Reveals Cell Wall Reorganisation in the Absence of Fungal Infection. Vaccines 2020, 8, 388. https://doi.org/10.3390/vaccines8030388
Weiller F, Gerber L, Trygg J, Fangel JU, Willats WGT, Driouich A, Vivier MA, Moore JP. Overexpression of VviPGIP1 and NtCAD14 in Tobacco Screened Using Glycan Microarrays Reveals Cell Wall Reorganisation in the Absence of Fungal Infection. Vaccines. 2020; 8(3):388. https://doi.org/10.3390/vaccines8030388
Chicago/Turabian StyleWeiller, Florent, Lorenz Gerber, Johan Trygg, Jonatan U. Fangel, William G.T. Willats, Azeddine Driouich, Melané A. Vivier, and John P. Moore. 2020. "Overexpression of VviPGIP1 and NtCAD14 in Tobacco Screened Using Glycan Microarrays Reveals Cell Wall Reorganisation in the Absence of Fungal Infection" Vaccines 8, no. 3: 388. https://doi.org/10.3390/vaccines8030388
APA StyleWeiller, F., Gerber, L., Trygg, J., Fangel, J. U., Willats, W. G. T., Driouich, A., Vivier, M. A., & Moore, J. P. (2020). Overexpression of VviPGIP1 and NtCAD14 in Tobacco Screened Using Glycan Microarrays Reveals Cell Wall Reorganisation in the Absence of Fungal Infection. Vaccines, 8(3), 388. https://doi.org/10.3390/vaccines8030388






