Mutations at the Serine Hydroxymethyltransferase Impact Its Interaction with a Soluble NSF Attachment Protein and a Pathogenesis-Related Protein in Soybean
Abstract
1. Introduction
2. Materials and Methods
2.1. Development of the EMS Mutagenesis Forrest Population
2.2. Genotyping of ExF RIL Population
2.3. SCN-Infection Phenotyping
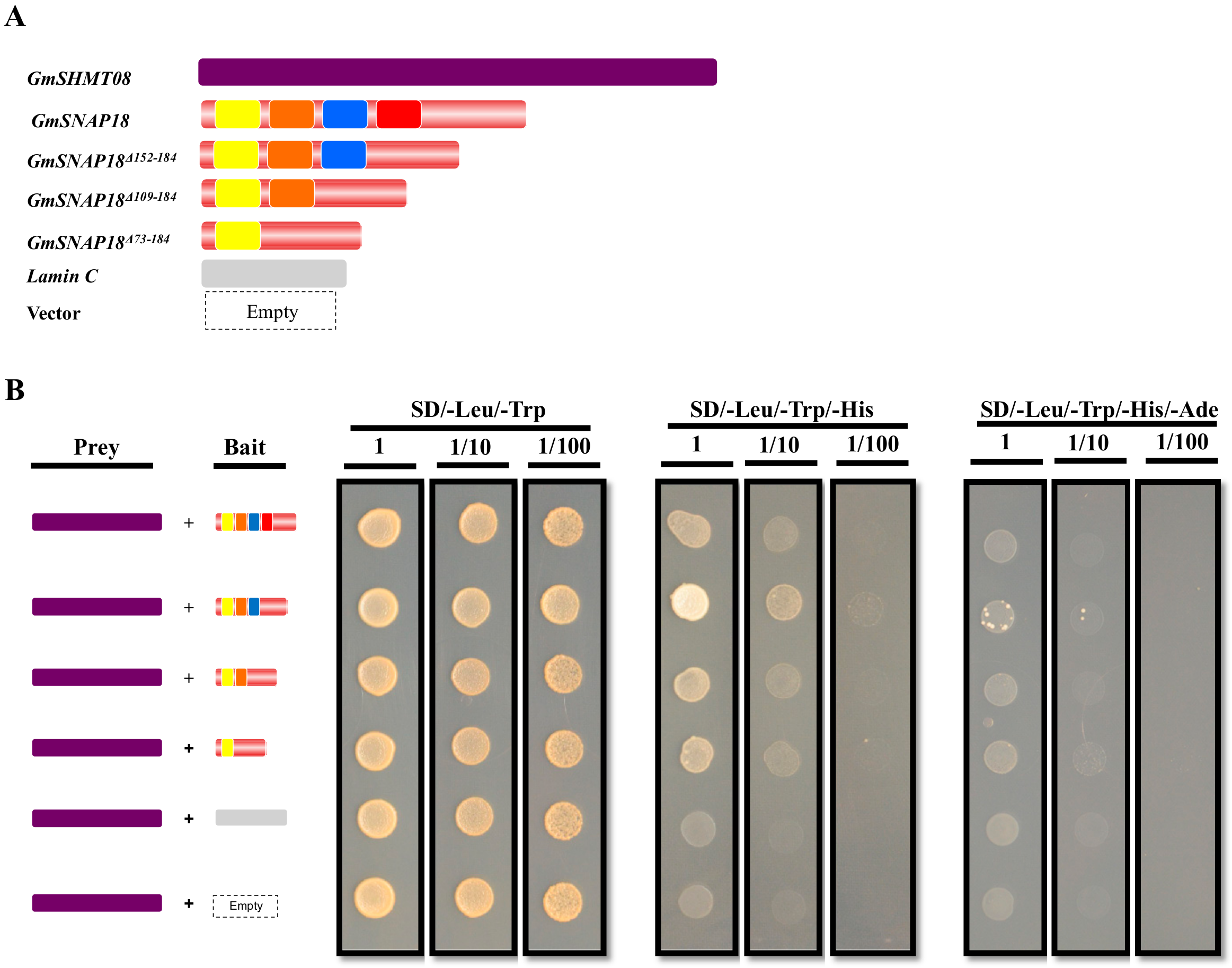
2.4. Plasmid Construction for Y2H Analysis
2.5. Yeast Co-Transformation Assay
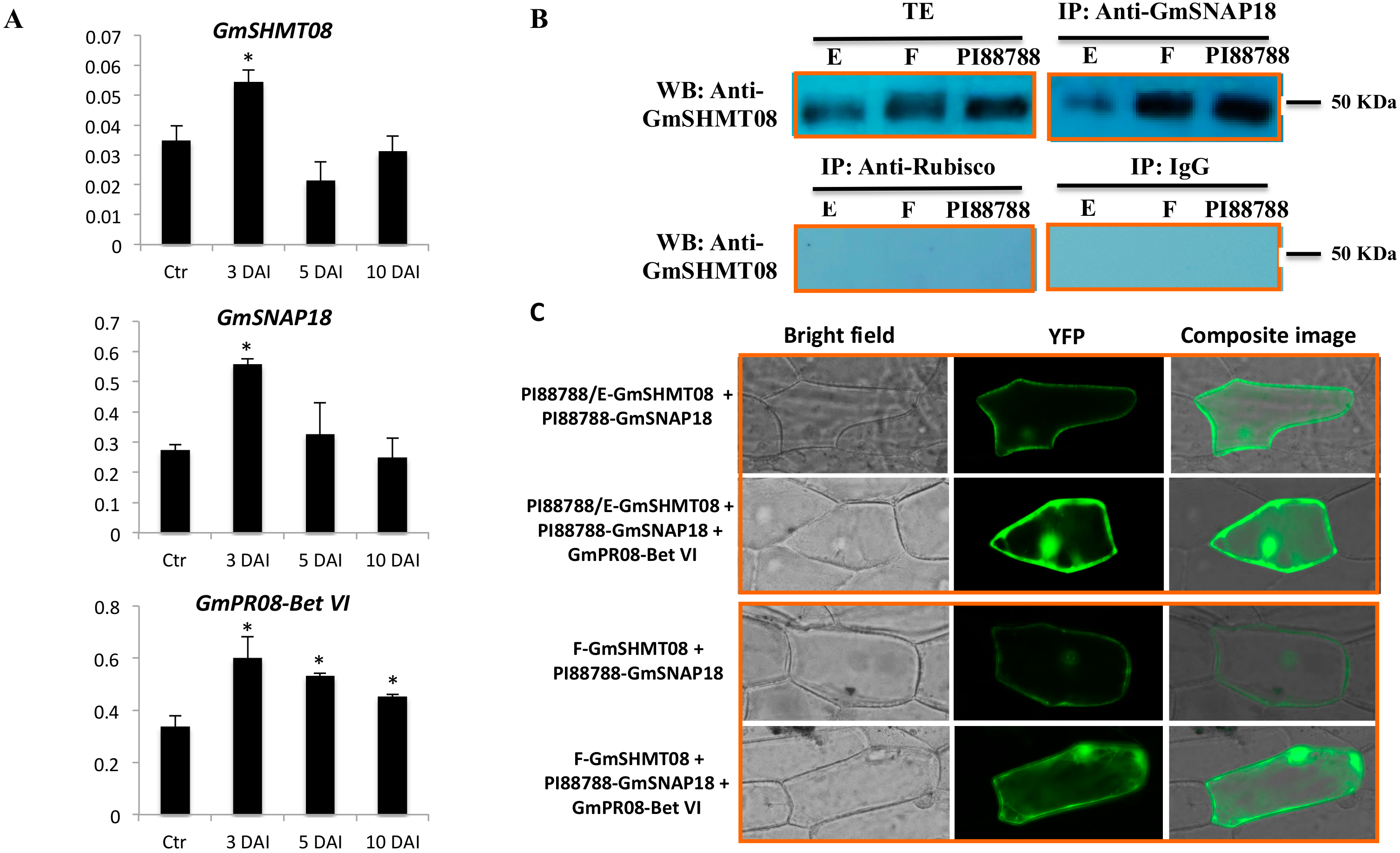
2.6. qRT-PCR Analysis
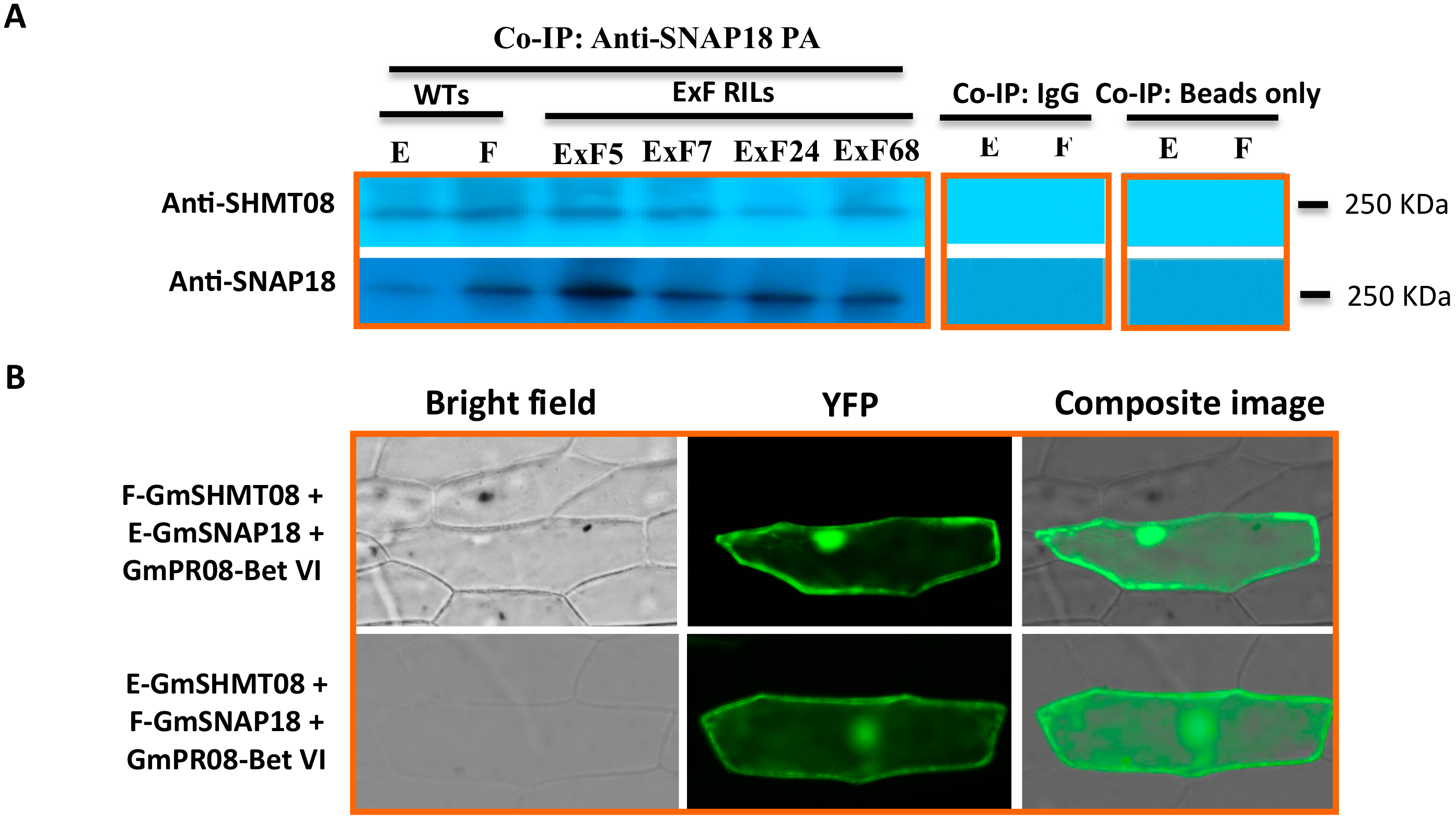
2.7. Protein Extraction and Co-Immunoprecipiation Analysis
2.8. BiFC Assay
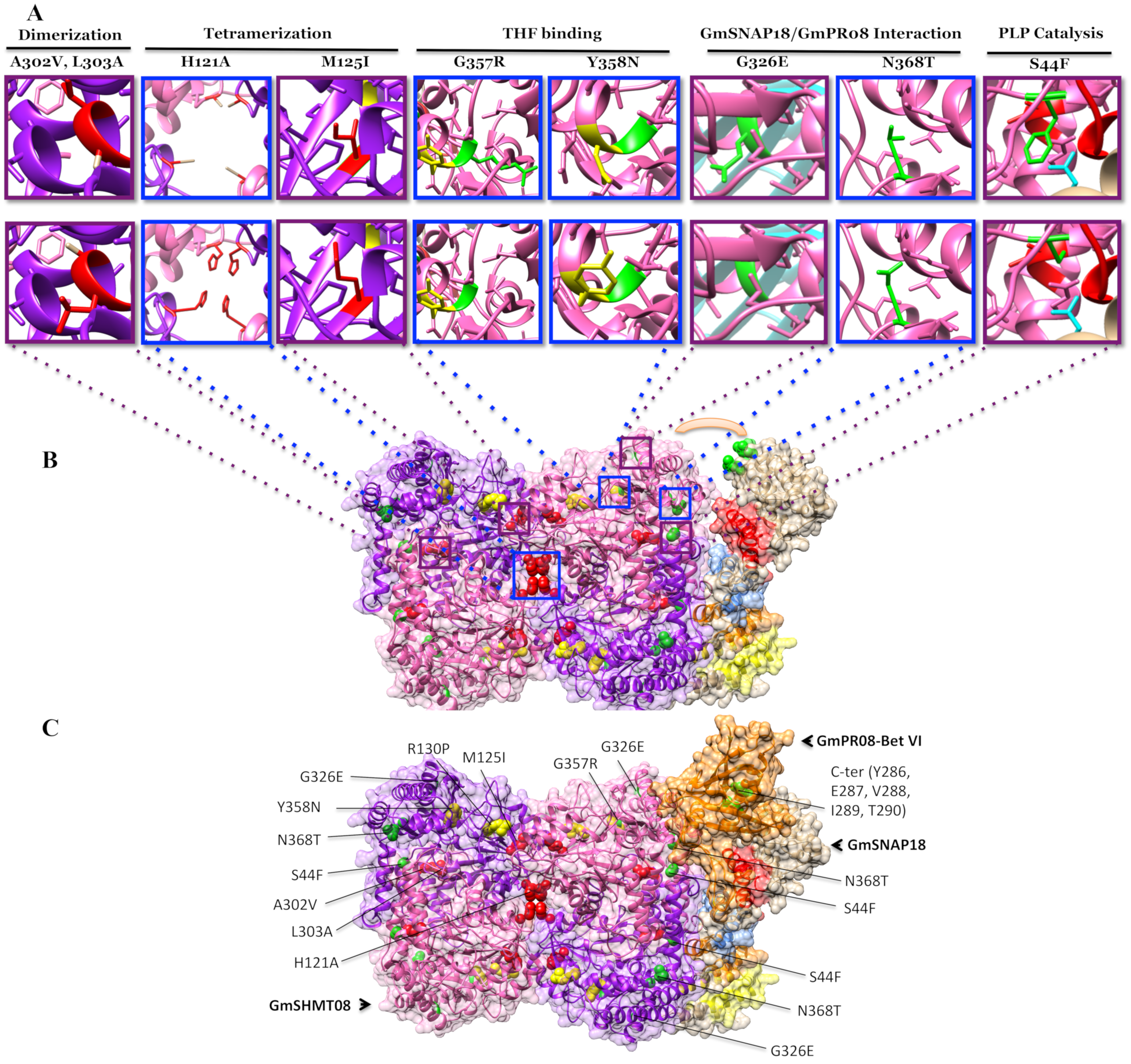
2.9. Modeling of GmSNAP18, GmSNAP18, and GmPR08-Bet VI Proteins and Mutational Analysis
2.10. Interaction Analysis of Homology Models
3. Results
3.1. GmSNAP18 and GmSHMT08 Interaction in Yeast
3.2. Resistant and Susceptible Alleles of GmSNAP18 and GmSHMT08 from Forrest and Essex can Physically Associate with Each Other
3.3. Expression and Interaction Analysis of GmSHMT08, GmSNAP18, and GmPR08-Bet VI in PI88788-Type Resistance
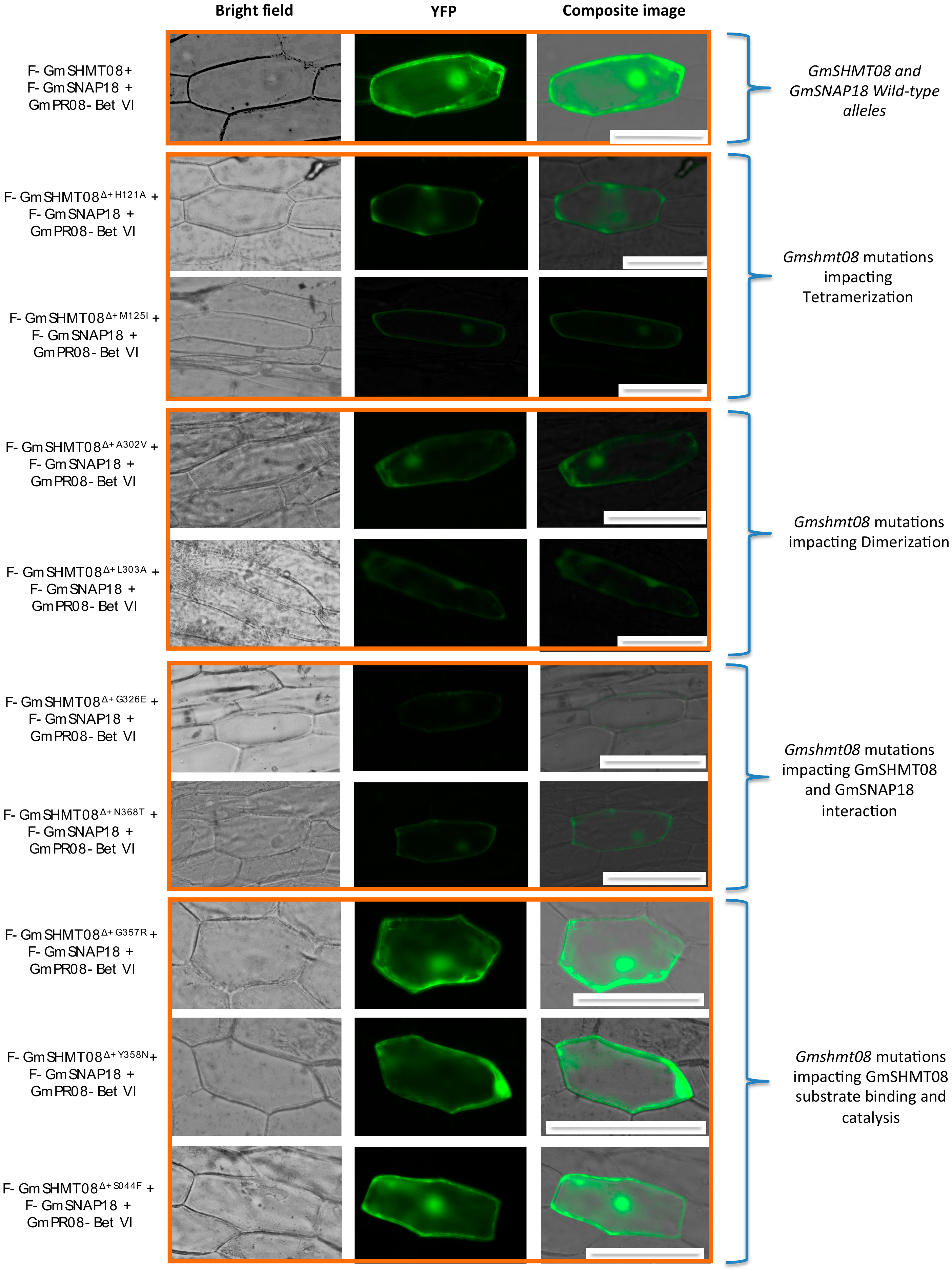
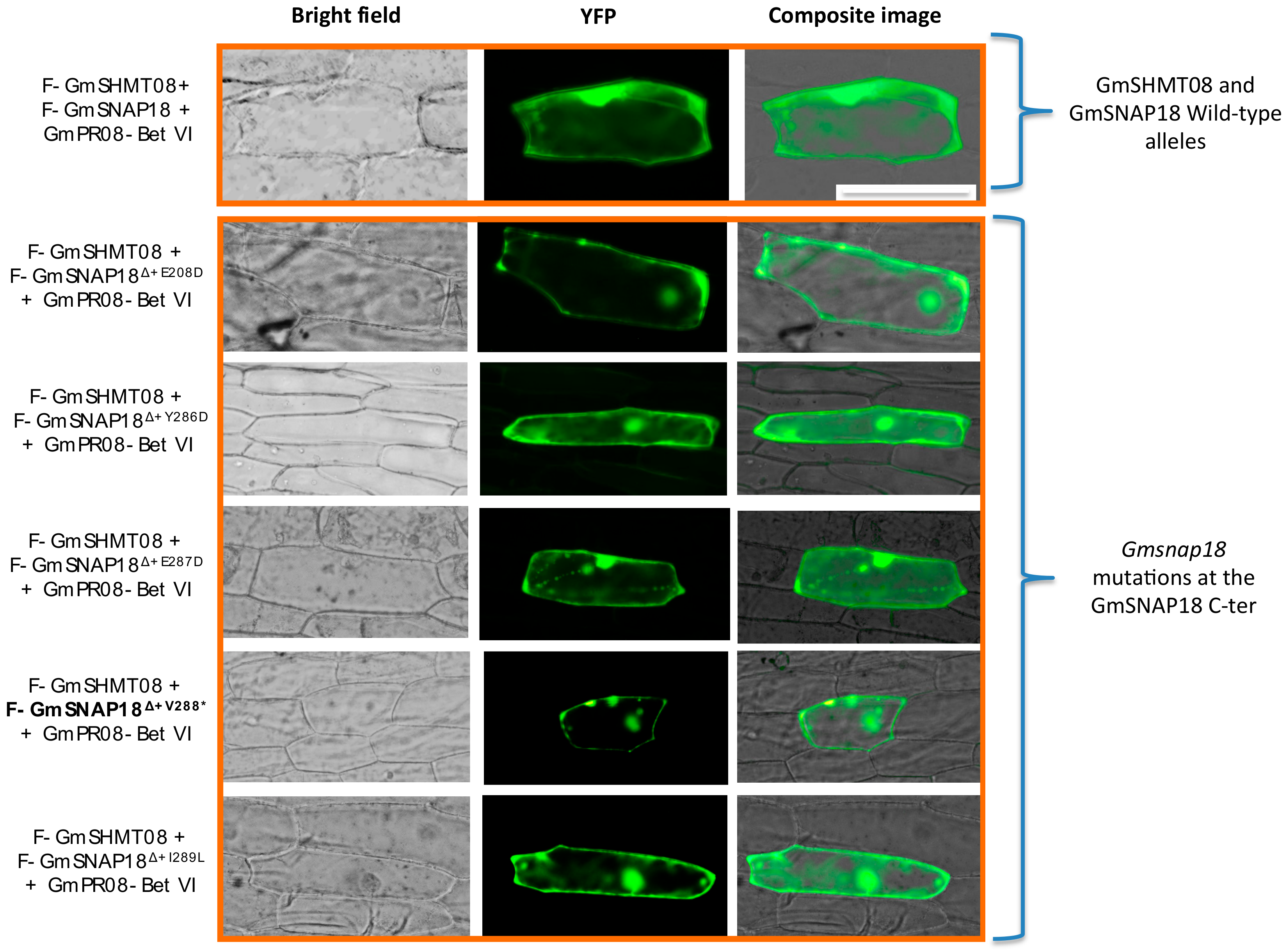
3.4. Mutational Analysis Supported the Predicted Interaction Model
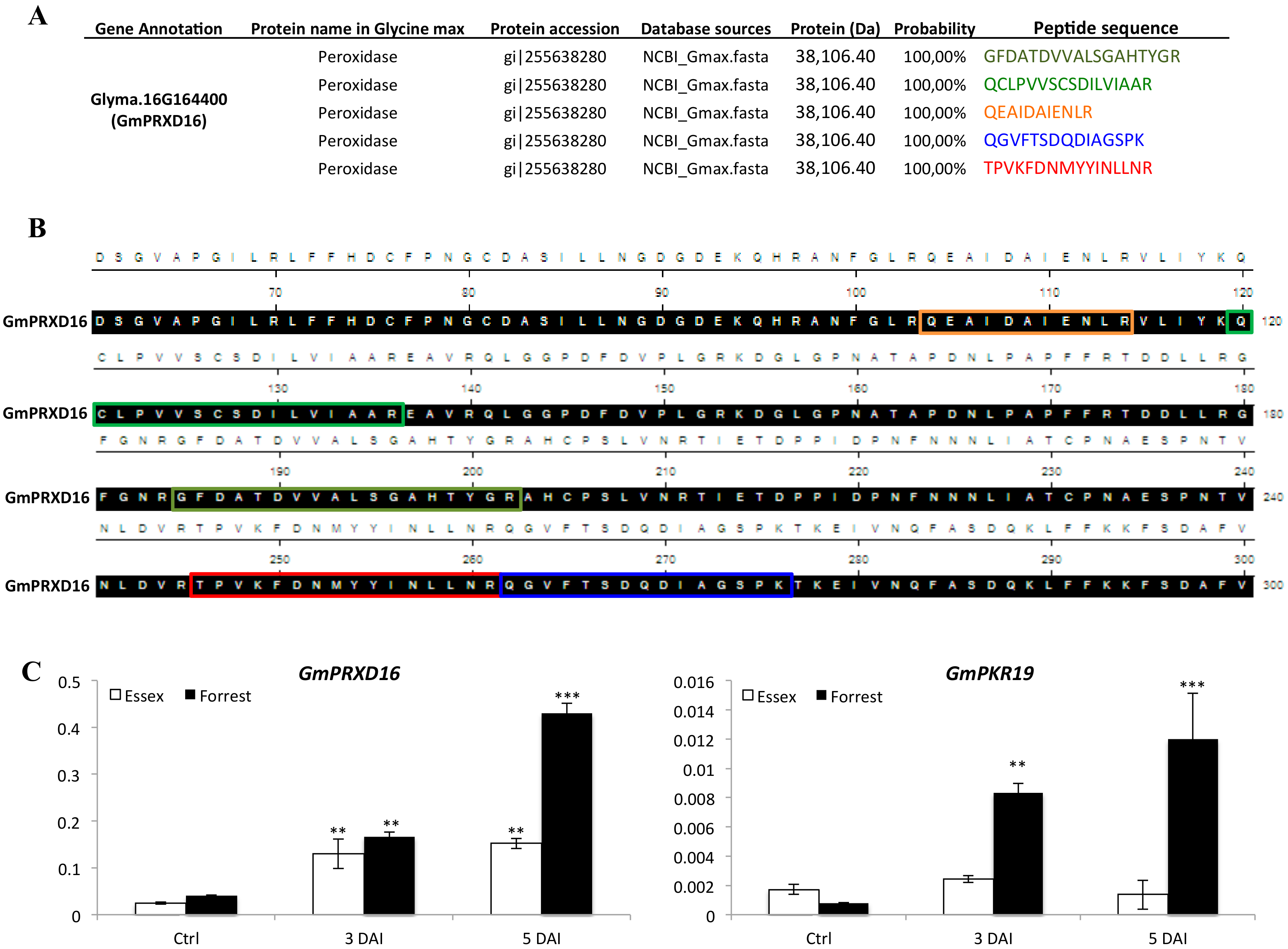
3.5. Genes Encoding Key Components of ROS Signaling Pathway were Induced Under SCN Infection
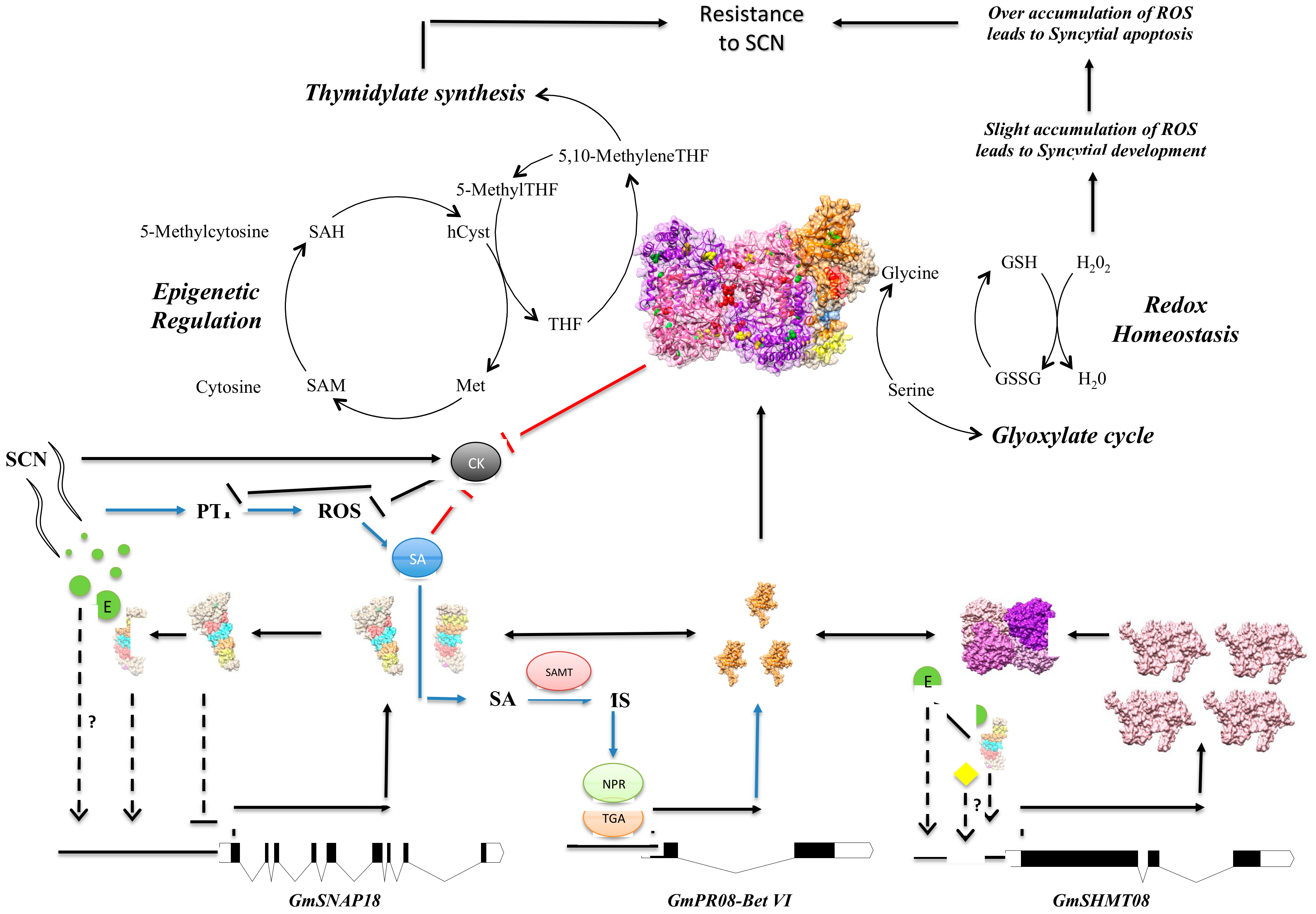
4. Discussion
4.1. The Presence of a Tetrameric GmSHMT08 Protein within the Multi-Protein Complex
4.2. Mutational Analysis Reveals the Importance of the Gmshmt08 Tetrameric Structure in Maintaining the Multi-Protein Complex
4.3. Induced and Natural Gmshmt08 Mutations at the PLP Catalysis and THF Substrate Binding Result in SCN Susceptibility but not Necessary Impacting the Multi-Protein Complex Interactions
4.4. Mutations at GmSHMT08 Residues Mapped at the GmSNAP18/GmPR08-Bet VI Interacting Sites Negatively Impacted the Multi-Protein Complex
4.5. GmSNAP18 C-Terminal Involvement in Driving the Multi-Protein Complex Toward SCN Infected Sites
4.6. GmSHMT08 as Mediator of Peking-Type SCN Resistance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pham, A.-T.; Lee, J.-D.; Shannon, J.G.; Bilyeu, K. A novel FAD2-1 A allele in a soybean plant introduction offers an alternate means to produce soybean seed oil with 85% oleic acid content. Theor. Appl. Genet. 2011, 123, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Lakhssassi, N.; Colantonio, V.; Flowers, N.D.; Zhou, Z.; Henry, J.; Liu, S.; Meksem, K. Stearoyl-Acyl Carrier Protein Desaturase Mutations Uncover an Impact of Stearic Acid in Leaf and Nodule Structure. Plant Physiol. 2017, 174, 1531–1543. [Google Scholar] [CrossRef] [PubMed]
- Lakhssassi, N.; Zhou, Z.; Liu, S.; Colantonio, V.; AbuGhazaleh, A.; Meksem, K. Characterization of the FAD2 Gene Family in Soybean Reveals the Limitations of Gel-Based TILLING in Genes with High Copy Number. Front. Plant Sci. 2017, 8, 324. [Google Scholar] [CrossRef]
- Koenning, S.R.; Wrather, J.A. Suppression of Soybean Yield Potential in the Continental United States by Plant Diseases from 2006 to 2009. Plant Heal. Prog. 2010, 11, 5. [Google Scholar] [CrossRef]
- Mitchum, M.G. Soybean Resistance to the Soybean Cyst NematodeHeterodera glycines: An Update. Phytopathology 2016, 106, 1444–1450. [Google Scholar] [CrossRef]
- Lakhssassi, N.; Liu, S.; Bekal, S.; Zhou, Z.; Colantonio, V.; Lambert, K.; Barakat, A.; Meksem, K. Characterization of the Soluble NSF Attachment Protein gene family identifies two members involved in additive resistance to a plant pathogen. Sci. Rep. 2017, 7, 45226. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Kandoth, P.K.; Lakhssassi, N.; Kang, J.; Colantonio, V.; Heinz, R.; Yeckel, G.; Zhou, Z.; Bekal, S.; Dapprich, J.; et al. The soybean GmSNAP18 gene underlies two types of resistance to soybean cyst nematode. Nat. Commun. 2017, 8, 14822. [Google Scholar] [CrossRef]
- Liu, S.; Kandoth, P.K.; Warren, S.D.; Yeckel, G.; Heinz, R.; Alden, J.; Yang, C.; Jamai, A.; El-Mellouki, T.; Juvale, P.S.; et al. A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 2012, 492, 256–260. [Google Scholar] [CrossRef]
- Kandoth, P.K.; Liu, S.; Prenger, E.; Ludwig, A.; Lakhssassi, N.; Heinz, R.; Zhou, Z.; Howland, A.; Gunther, J.; Eidson, S.; et al. Systematic Mutagenesis of Serine Hydroxymethyltransferase Reveals an Essential Role in Nematode Resistance. Plant Physiol. 2017, 175, 1370–1380. (In press) [CrossRef]
- Lakhssassi, N.; Patil, G.; Piya, S.; Zhou, Z.; Baharlouei, A.; Kassem, M.A.; Lightfoot, D.A.; Hewezi, T.; Barakat, A.; Nguyen, H.; et al. Genome reorganization of the GmSHMT gene family in soybean showed a lack of functional redundancy in resistance to soybean cyst nematode. Sci. Rep. 2019, 9, 1506. [Google Scholar] [CrossRef]
- Cook, D.E.; Lee, T.G.; Guo, X.; Melito, S.; Wang, K.; Bayless, A.M.; Wang, J.; Hughes, T.J.; Willis, D.K.; Clemente, T.E.; et al. Copy Number Variation of Multiple Genes at Rhg1 Mediates Nematode Resistance in Soybean. Science 2012, 338, 1206–1209. [Google Scholar] [CrossRef] [PubMed]
- Bayless, A.M.; Smith, J.M.; Song, J.; McMINN, P.H.; Teillet, A.; August, B.K.; Bent, A. Disease resistance through impairment of α-SNAP–NSF interaction and vesicular trafficking by soybean Rhg1. Proc. Natl. Acad. Sci. USA 2016, 113, E7375–E7382. [Google Scholar] [CrossRef] [PubMed]
- Bayless, A.M.; Zapotocny, R.W.; Grunwald, D.J.; Amundson, K.K.; Diers, B.W.; Bent, A. An atypical N-ethylmaleimide sensitive factor enables the viability of nematode-resistant Rhg1 soybeans. Proc. Natl. Acad. Sci. USA 2018, 115, E4512–E4521. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.E.; Bayless, A.M.; Wang, K.; Guo, X.; Song, Q.; Jiang, J.; Bent, A. Distinct Copy Number, Coding Sequence, and Locus Methylation Patterns Underlie Rhg1-Mediated Soybean Resistance to Soybean Cyst Nematode. Plant Physiol. 2014, 165, 630–647. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.G.; Kumar, I.; Diers, B.W.; Hudson, M. Evolution and selection ofRhg1,a copy-number variant nematode-resistance locus. Mol. Ecol. 2015, 24, 1774–1791. [Google Scholar] [CrossRef]
- Yu, N.; Lee, T.G.; Rosa, D.P.; Hudson, M.; Diers, B. Impact of Rhg1 copy number, type, and interaction with Rhg4 on resistance to Heterodera glycines in soybean. Theor. Appl. Genet. 2016, 129, 2403–2412. [Google Scholar] [CrossRef]
- Lee, T.G.; Diers, B.W.; Hudson, M. An efficient method for measuring copy number variation applied to improvement of nematode resistance in soybean. Plant J. 2016, 88, 143–153. [Google Scholar] [CrossRef]
- Patil, G.; Lakhssassi, N.; Wan, J.; Song, L.; Zhou, Z.; Klepadlo, M.; Vuong, T.D.; Stec, A.O.; Kahil, S.S.; Colantonio, V.; et al. Whole-genome re-sequencing reveals the impact of the interaction of copy number variants of the rhg1 and Rhg4 genes on broad-based resistance to soybean cyst nematode. Plant Biotechnol. J. 2019, 17, 1595–1611. [Google Scholar] [CrossRef]
- Schirch, L. Serine hydroxymethyltransferase. Adv. Enzymol. Relat. Areas Mol. Biol. 1982, 53, 83–112. [Google Scholar]
- Hanson, A.D.; Gage, D.A.; Shachar-Hill, Y. Plant one-carbon metabolism and its engineering. Trends Plant Sci. 2000, 5, 206–213. [Google Scholar] [CrossRef]
- Appaji Rao, N.A.; Ambili, M.; Jala, V.R.; Subramanya, H.; Savithri, H. Structure–function relationship in serine hydroxymethyltransferase. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2003, 1647, 24–29. [Google Scholar] [CrossRef]
- Stover, P.; Schirch, V. Serine hydroxymethyltransferase catalyzes the hydrolysis of 5,10-methenyltetrahydrofolate to 5-formyltetrahydrofolate. J. Biol. Chem. 1990, 265, 14227–14233. [Google Scholar]
- Narkewicz, M.R.; Sauls, S.D.; Tjoa, S.S.; Teng, C.; Fennessey, P.V. Evidence for intracellular partitioning of serine and glycine metabolism in Chinese hamster ovary cells. Biochem. J. 1996, 313, 991–996. [Google Scholar] [CrossRef] [PubMed]
- Voll, L.M.; Jamai, A.; Renné, P.; Voll, H.; McClung, C.; Weber, A. The Photorespiratory Arabidopsis shm1 Mutant Is Deficient in SHM11. Plant Physiol. 2005, 140, 59–66. [Google Scholar] [CrossRef]
- Skibola, C.F.; Smith, M.T.; Hubbard, A.; Shane, B.; Roberts, A.C.; Law, G.; Rollinson, S.; Roman, E.; Cartwright, R.A.; Morgan, G. Polymorphisms in the thymidylate synthase and serine hydroxymethyltransferase genes and risk of adult acute lymphocytic leukemia. Blood 2002, 99, 3786–3791. [Google Scholar] [CrossRef]
- Lim, U.; Peng, K.; Shane, B.; Stover, P.J.; A Litonjua, A.; Weiss, S.T.; Gaziano, J.M.; Strawderman, R.L.; Raiszadeh, F.; Selhub, J.; et al. Polymorphisms in cytoplasmic serine hydroxymethyltransferase and methylenetetrahydrofolate reductase affect the risk of cardiovascular disease in men. J. Nutr. 2005, 135, 1989–1994. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Amelio, I.; Cutruzzolà, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014, 39, 191–198. [Google Scholar] [CrossRef]
- Schapire, A.L.; Valpuesta, V.; Botella, J.R. TPR Proteins in Plant Hormone Signaling. Plant Signal. Behav. 2006, 1, 229–230. [Google Scholar] [CrossRef] [PubMed]
- Lakhssassi, N.; Piya, S.; Bekal, S.; Liu, S.; Zhou, Z.; Bergounioux, C.; Miao, L.; Meksem, J.; Lakhssassi, A.; Jones, K.; et al. A pathogenesis-related protein GmPR08-Bet VI promotes a molecular interaction between the GmSHMT08 and GmSNAP18 in resistance to Heterodera glycines. Plant Biotechnol. J. 2020. [Google Scholar] [CrossRef]
- Blatch, G.L.; Lassle, M. The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. Bioessays 1999, 21, 932–939. [Google Scholar] [CrossRef]
- D’Andrea, L.D. TPR proteins: The versatile helix. Trends Biochem. Sci. 2003, 28, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Goebl, M.; Yanagida, M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem. Sci. 1991, 16, 173–177. [Google Scholar] [CrossRef]
- Prasad, B.D.; Goel, S.; Krishna, P. In silico identification of carboxylate clamp type tetratricopeptide repeat proteins in arabidopsis and rice as putative co-chaperones of hsp90/hsp70. PLoS ONE 2010, 5, e12761. [Google Scholar] [CrossRef] [PubMed]
- Sohoki, M.; Browne, S.; Sullivan, L.S.; Blackshaw, S.; Cepko, C.L.; Payne, A.M.; Bhattacharya, S.S.; Khaliq, S.; Mehdi, S.Q.; Birch, D.G.; et al. Mutations in a new photoreceptor-pineal gene on 17p cause Leber congenital amaurosis. Am. J. Ophthalmol. 2000, 129, 834–835. [Google Scholar] [CrossRef]
- Grizot, S.; Fieschi, F.; Dagher, M.C.; Pebay-Peyroula, E. The active n-terminal region of p67phox. Structure at 1.8 a resolution and biochemical characterizations of the a128v mutant implicated in chronic granulomatous disease. J. Biol. Chem. 2001, 276, 21627–21631. [Google Scholar] [CrossRef] [PubMed]
- Tsukahara, F.; Hattori, M.; Muraki, T.; Sakaki, Y. Identification and Cloning of a Novel cDNA Belonging to Tetratricopeptide Repeat Gene Family from Down Syndrome-Critical Region 21q22.2. J. Biochem. 1996, 120, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Rambani, A.; Pantalone, V.; Yang, S.; Rice, J.H.; Song, Q.; Mazarei, M.; Arelli, P.R.; Meksem, K.; Stewart, C.N.; Hewezi, T. Identification of introduced and stably inherited DNA methylation variants in soybean associated with soybean cyst nematode parasitism. N. Phytol. 2020, 227, 168–184. [Google Scholar] [CrossRef]
- Hewezi, T. Epigenetic Mechanisms in Nematode–Plant Interactions. Annu. Rev. Phytopathol. 2020, 58. [Google Scholar] [CrossRef]
- Thompson, T.C. Glioma Pathogenesis-Related Protein 1: Tumor-Suppressor Activities and Therapeutic Potential. Yonsei Med. J. 2010, 51, 479–483. [Google Scholar] [CrossRef]
- Li, L.; Fattah, E.A.; Cao, G.; Ren, C.; Yang, G.; Goltsov, A.A.; Chinault, A.C.; Cai, W.-W.; Timme, T.L.; Thompson, T.C. Glioma Pathogenesis-Related Protein 1 Exerts Tumor Suppressor Activities through Proapoptotic Reactive Oxygen Species c-Jun NH2 Kinase Signaling. Cancer Res. 2008, 68, 434–443. [Google Scholar] [CrossRef]
- Karnik, R.; Grefen, C.; Bayne, R.; Honsbein, A.; Kohler, T.; Kioumourtzoglou, D.; Williams, M.; Bryant, N.J.; Blatt, M.R. Arabidopsis sec1/munc18 protein sec11 is a competitive and dynamic modulator of snare binding and syp121-dependent vesicle traffic. Plant Cell 2013, 25, 1368–1382. [Google Scholar] [CrossRef] [PubMed]
- Kalde, M.; Nühse, T.S.; Findlay, K.; Peck, S.C. The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc. Natl. Acad. Sci. USA 2007, 104, 11850–11855. [Google Scholar] [CrossRef]
- Matthews, B.F.; Beard, H.; Brewer, E.; Kabir, S.; Macdonald, M.H.; Youssef, R.M. Arabidopsis genes, AtNPR1, AtTGA2 and AtPR-5, confer partial resistance to soybean cyst nematode (Heterodera glycines) when overexpressed in transgenic soybean roots. BMC Plant Biol. 2014, 14, 96. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Jiang, L.; Wu, J.; Li, W.; Fan, S.; Zhang, S. Isolation and characterization of a pathogenesis-related protein 10 gene (GmPR10) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. Mol. Biol. Rep. 2014, 41, 4899–4909. [Google Scholar] [CrossRef] [PubMed]
- Lightfoot, D.A.; Njiti, V.N.; Gibson, P.T.; Kassem, M.A.; Iqbal, J.M.; Meksem, K. Registration of the essex × forrest recombinant inbred line mapping population. Crop Sci. 2005, 45, 1678. [Google Scholar] [CrossRef]
- Hewezi, T.; Howe, P.J.; Maier, T.R.; Hussey, R.S.; Mitchum, M.G.; Davis, E.L.; Baum, T.J. Arabidopsis spermidine synthase is targeted by an effector protein of the cyst nematode Heterodera schachtii. Plant Physiol. 2009, 152, 968–984. [Google Scholar] [CrossRef] [PubMed]
- Schwede, T. SWISS-MODEL: An automated protein homology-modeling server. Nucl. Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef]
- Renwick, S.B.; Snell, K.; Baumann, U. The crystal structure of human cytosolic serine hydroxymethyltransferase: A target for cancer chemotherapy. Structure 1998, 6, 1105–1116. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Yang, Z.; Lasker, K.; Schneidman-Duhovny, D.; Webb, B.; Huang, C.C.; Pettersen, E.F.; Goddard, T.D.; Meng, E.C.; Sali, A.; Ferrin, T. UCSF Chimera, MODELLER, and IMP: An integrated modeling system. J. Struct. Biol. 2011, 179, 269–278. [Google Scholar] [CrossRef]
- Sensoy, O.; Almeida, J.G.; Shabbir, J.; Moreira, I.S.; Morra, G. Computational studies of g protein-coupled receptor complexes: Structure and dynamics. In Methods in Cell Biology; Shukla, A.K., Ed.; Academic Press: Cambridge, MA, USA, 2017; Chapter 16; Volume 142, pp. 205–245. [Google Scholar]
- Korasick, D.A.; Kandoth, P.K.; Tanner, J.J.; Mitchum, M.G.; Beamer, L.J. Impaired folate binding of serine hydroxymethyltransferase 8 from soybean underlies resistance to the soybean cyst nematode. J. Biol. Chem. 2020, 295, 3708–3718. [Google Scholar] [CrossRef] [PubMed]
- Inoguchi, T.; Sonta, T.; Tsubouchi, H.; Etoh, T.; Kakimoto, M.; Sonoda, N.; Sato, N.; Sekiguchi, N.; Kobayashi, K.; Sumimoto, H.; et al. Protein Kinase C-Dependent Increase in Reactive Oxygen Species (ROS) Production in Vascular Tissues of Diabetes: Role of Vascular NAD(P)H Oxidase. J. Am. Soc. Nephrol. 2003, 14, S227–S232. [Google Scholar] [CrossRef] [PubMed]
- Florio, R.; Chiaraluce, R.; Consalvi, V.; Paiardini, A.; Catacchio, B.; Bossa, F.; Contestabile, R. Structural stability of the cofactor binding site in Escherichia coli serine hydroxymethyltransferase—the role of evolutionarily conserved hydrophobic contacts. FEBS J. 2009, 276, 7319–7328. [Google Scholar] [CrossRef] [PubMed]
- Jagath, J.R.; Sharma, B.; Rao, N.A.; Savithri, H.S. The Role of His-134, -147, and -150 Residues in Subunit Assembly, Cofactor Binding, and Catalysis of Sheep Liver Cytosolic Serine Hydroxymethyltransferase. J. Biol. Chem. 1997, 272, 24355–24362. [Google Scholar] [CrossRef]
- Jala, V.R.; Ambili, M.; Prakash, V.; Rao, N.A.; Savithri, H.S. Disruption of distal interactions of arg 262 and of substrate binding to ser 52 affect catalysis of sheep liver cytosolic serine hydroxymethyltransferase. Indian J. Biochem. Biophys. 2003, 40, 226–237. [Google Scholar]
- Rao, N.A.; Talwar, R.; Savithri, H. Molecular organization, catalytic mechanism and function of serine hydroxymethyltransferase—A potential target for cancer chemotherapy. Int. J. Biochem. Cell Biol. 2000, 32, 405–416. [Google Scholar] [CrossRef]
- Vivoli, M.; Angelucci, F.; Ilari, A.; Morea, V.; Angelaccio, S.; Di Salvo, M.L.; Contestabile, R. Role of a Conserved Active Site Cation−π Interaction inEscherichia coliSerine Hydroxymethyltransferase. Biochemistry 2009, 48, 12034–12046. [Google Scholar] [CrossRef]
- Jagath, J.R.; Sharma, B.; Bhaskar, B.; Datta, A.; Rao, N.A.; Savithri, H.S. Importance of the amino terminus in maintenance of oligomeric structure of sheep liver cytosolic serine hydroxymethyltransferase. JBIC J. Biol. Inorg. Chem. 1997, 247, 372–379. [Google Scholar] [CrossRef]
- Pogorelko, G.; Juvale, P.S.; Rutter, W.B.; Hewezi, T.; Hussey, R.; Davis, E.L.; Mitchum, M.G.; Baum, T.J. A cyst nematode effector binds to diverse plant proteins, increases nematode susceptibility and affects root morphology. Mol. Plant Pathol. 2016, 17, 832–844. [Google Scholar] [CrossRef]
- Chen, Y.A.; Scales, S.J.; Jagath, J.R.; Scheller, R.H. A Discontinuous SNAP-25 C-terminal Coil Supports Exocytosis. J. Biol. Chem. 2001, 276, 28503–28508. [Google Scholar] [CrossRef]
- Bayless, A.M.; Zapotocny, R.W.; Han, S.; Grunwald, D.J.; Amundson, K.K.; Bent, A. The rhg1-a (Rhg1 low-copy) nematode resistance source harbors a copia-family retrotransposon within the Rhg1-encoded α-SNAP gene. Plant Direct 2019, 3, e00164. [Google Scholar] [CrossRef] [PubMed]
- Hewezi, T.; Baum, T.J. Manipulation of Plant Cells by Cyst and Root-Knot Nematode Effectors. Mol. Plant-Microbe Interact. 2013, 26, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Whitham, S.A.; Qi, M.; Innes, R.W.; Ma, W.; Lopes-Caitar, V.; Hewezi, T. Molecular soybean-pathogen interactions. Ann. Rev. Phytopathol. 2016, 54, 443–468. [Google Scholar] [CrossRef] [PubMed]
- Bekal, S.; Domier, L.L.; Gonfa, B.; Lakhssassi, N.; Meksem, K.; Lambert, K.N. A snare-like protein and biotin are implicated in soybean cyst nematode virulence. PLoS ONE 2015, 10, e0145601. [Google Scholar] [CrossRef] [PubMed]
- Mouillon, J.-M.; Aubert, S.; Bourguignon, J.; Gout, E.; Douce, R.; Rébeillé, F. Glycine and serine catabolism in non-photosynthetic higher plant cells: their role in C1 metabolism. Plant J. 1999, 20, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, D.; Meade, G.; Foley, V.M.; Dowd, C.A. Structure, function and evolution of glutathione transferases: Implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001, 360, 1–16. [Google Scholar] [CrossRef]
- Udomsinprasert, R.; Pongjaroenkit, S.; Wongsantichon, J.; Oakley, A.J.; Prapanthadara, L.-A.; Wilce, M.C.J.; Ketterman, A.J. Identification, characterization and structure of a new Delta class glutathione transferase isoenzyme. Biochem. J. 2005, 388, 763–771. [Google Scholar] [CrossRef]
- Allocati, N.; Federici, L.; Masulli, M.; Di Ilio, C. Glutathione transferases in bacteria. FEBS J. 2008, 276, 58–75. [Google Scholar] [CrossRef]
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. Glutathione transferases. Ann. Rev. Pharmacol. Toxicol. 2005, 45, 51–88. [Google Scholar] [CrossRef]
- Josephy, P.D. Genetic Variations in Human Glutathione Transferase Enzymes: Significance for Pharmacology and Toxicology. Hum. Genom. Proteom. 2010, 2010, 1–14. [Google Scholar] [CrossRef]
- Kandoth, P.K.; Ithal, N.; Recknor, J.; Maier, T.; Nettleton, D.S.; Baum, T.J.; Mitchum, M.G. The Soybean Rhg1 locus for resistance to the soybean cyst nematode Heterodera glycines regulates the expression of a large number of stress- and defense-related genes in degenerating feeding cells. Plant Physiol. 2011, 155, 1960–1975. [Google Scholar] [CrossRef]
- Melillo, M.T.; Leonetti, P.; Bongiovanni, M.; Castagnone-Sereno, P.; Bleve-Zacheo, T. Modulation of reactive oxygen species activities and H2O2 accumulation during compatible and incompatible tomato-root-knot nematode interactions. N. Phytol. 2006, 170, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Siddique, S.; Matera, C.; Radakovic, Z.S.; Hasan, M.S.; Gutbrod, P.; Różańska, E.; Sobczak, M.; Torres, M.A.; Grundler, F.M.W. Parasitic Worms Stimulate Host NADPH Oxidases to Produce Reactive Oxygen Species That Limit Plant Cell Death and Promote Infection. Sci. Signal. 2014, 7, ra33. [Google Scholar] [CrossRef] [PubMed]
- Hewezi, T. Cellular Signaling Pathways and Posttranslational Modifications Mediated by Nematode Effector Proteins. Plant Physiol. 2015, 169, 1018–1026. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Dong, L.; Wang, Z.; Hu, H.; Han, Y.; Teng, W.; Zhang, H.; Guo, M.; Li, W. Qtl underlying resistance to two hg types of heterodera glycines found in soybean cultivar ‘l-10’. BMC Gen. 2011, 12, 233. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; A Sleper, D.; Nguyen, H.T.; Arelli, P.R.; Shannon, J.G. Quantitative Trait Loci underlying Resistance to Three Soybean Cyst Nematode Populations in Soybean PI 404198A. Crop. Sci. 2006, 46, 224–233. [Google Scholar] [CrossRef]
- Webb, D.M.; Baltazar, B.M.; Rao-Arelli, A.P.; Schupp, J.; Clayton, K.; Keim, P.; Beavis, W.D. Genetic mapping of soybean cyst nematode race-3 resistance loci in the soybean PI 437.654. Theor. Appl. Genet. 1995, 91, 574–581. [Google Scholar] [CrossRef]
- Yue, P.; A Sleper, D.; Arelli, P.R. Mapping Resistance to Multiple Races of Heterodera glycines in Soybean PI 89772. Crop. Sci. 2001, 41, 1589–1595. [Google Scholar] [CrossRef]
- Jin, J.; Hewezi, T.; Baum, T.J. Arabidopsis peroxidase AtPRX53 influences cell elongation and susceptibility to Heterodera schachtii. Plant Signal. Behav. 2011, 6, 1778–1786. [Google Scholar] [CrossRef]
- León, J.; Lawton, M.A.; Raskin, I. Hydrogen Peroxide Stimulates Salicylic Acid Biosynthesis in Tobacco. Plant Physiol. 1995, 108, 1673–1678. [Google Scholar] [CrossRef]
- Moreno, J.I.; Martín, R.; Castresana, C. Arabidopsis SHMT1, a serine hydroxymethyltransferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. Plant J. 2004, 41, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Paone, A.; Marani, M.; Fiascarelli, A.; Rinaldo, S.; Giardina, G.; Contestabile, R.; Paiardini, A.; Cutruzzolà, F. SHMT1 knockdown induces apoptosis in lung cancer cells by causing uracil misincorporation. Cell Death Dis. 2014, 5, e1525. [Google Scholar] [CrossRef] [PubMed]
- Maddocks, O.D.K.; Labuschagne, C.F.; Adams, P.D.; Vousden, K.H. Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Mol. Cell 2016, 61, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Rambani, A.; Rice, J.H.; Liu, J.; Lane, T.; Ranjan, P.; Mazarei, M.; Pantalone, V.; Stewart, C.N.; Staton, M.E.; Hewezi, T. The Methylome of Soybean Roots during the Compatible Interaction with the Soybean Cyst Nematode. Plant Physiol. 2015, 168, 1364–1377. [Google Scholar] [CrossRef]
- Rahalingam, R.; Skorupska, H.T. Cytological expression of early response to infection by Heterodera glycines Ichinohe in resistant PI 437654 soybean. Genome 1996, 39, 986–998. [Google Scholar] [CrossRef]
- Andrews, N.W.; Chakrabarti, S. There’s more to life than neurotransmission: the regulation of exocytosis by synaptotagmin VII. Trends Cell Biol. 2005, 15, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Schapire, A.L.; Voigt, B.; Jasik, J.; Rosado, A.; Lopez-Cobollo, R.; Menzel, D.; Salinas, J.; Mancuso, S.; Valpuesta, V.; Baluska, F.; et al. Arabidopsis Synaptotagmin 1 Is Required for the Maintenance of Plasma Membrane Integrity and Cell Viability. Plant Cell 2008, 20, 3374–3388. [Google Scholar] [CrossRef]
- El Kasmi, F.; Krause, C.; Hiller, U.; Stierhof, Y.-D.; Mayer, U.; Conner, L.; Kong, L.; Reichardt, I.; Sanderfoot, A.; Jürgens, G. SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis. Mol. Biol. Cell 2013, 24, 1593–1601. [Google Scholar] [CrossRef]
- Südhof, T.C. A molecular machine for neurotransmitter release: synaptotagmin and beyond. Nat. Med. 2013, 19, 1227–1231. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lakhssassi, N.; Piya, S.; Knizia, D.; El Baze, A.; Cullen, M.A.; Meksem, J.; Lakhssassi, A.; Hewezi, T.; Meksem, K. Mutations at the Serine Hydroxymethyltransferase Impact Its Interaction with a Soluble NSF Attachment Protein and a Pathogenesis-Related Protein in Soybean. Vaccines 2020, 8, 349. https://doi.org/10.3390/vaccines8030349
Lakhssassi N, Piya S, Knizia D, El Baze A, Cullen MA, Meksem J, Lakhssassi A, Hewezi T, Meksem K. Mutations at the Serine Hydroxymethyltransferase Impact Its Interaction with a Soluble NSF Attachment Protein and a Pathogenesis-Related Protein in Soybean. Vaccines. 2020; 8(3):349. https://doi.org/10.3390/vaccines8030349
Chicago/Turabian StyleLakhssassi, Naoufal, Sarbottam Piya, Dounya Knizia, Abdelhalim El Baze, Mallory A. Cullen, Jonas Meksem, Aicha Lakhssassi, Tarek Hewezi, and Khalid Meksem. 2020. "Mutations at the Serine Hydroxymethyltransferase Impact Its Interaction with a Soluble NSF Attachment Protein and a Pathogenesis-Related Protein in Soybean" Vaccines 8, no. 3: 349. https://doi.org/10.3390/vaccines8030349
APA StyleLakhssassi, N., Piya, S., Knizia, D., El Baze, A., Cullen, M. A., Meksem, J., Lakhssassi, A., Hewezi, T., & Meksem, K. (2020). Mutations at the Serine Hydroxymethyltransferase Impact Its Interaction with a Soluble NSF Attachment Protein and a Pathogenesis-Related Protein in Soybean. Vaccines, 8(3), 349. https://doi.org/10.3390/vaccines8030349








