Exploiting B Cell Receptor Analyses to Inform on HIV-1 Vaccination Strategies
Abstract
1. Introduction
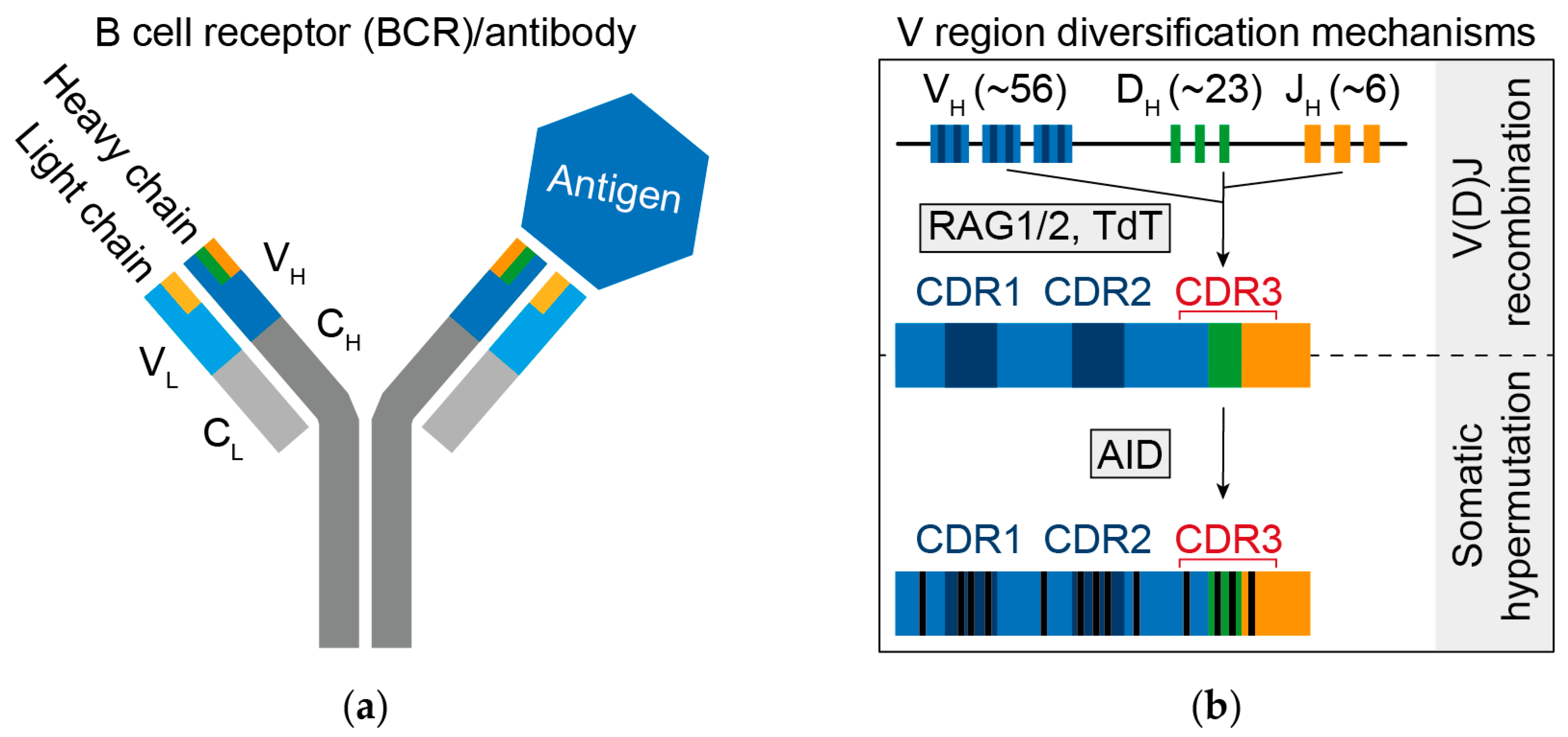
2. B Cell Receptor Diversity
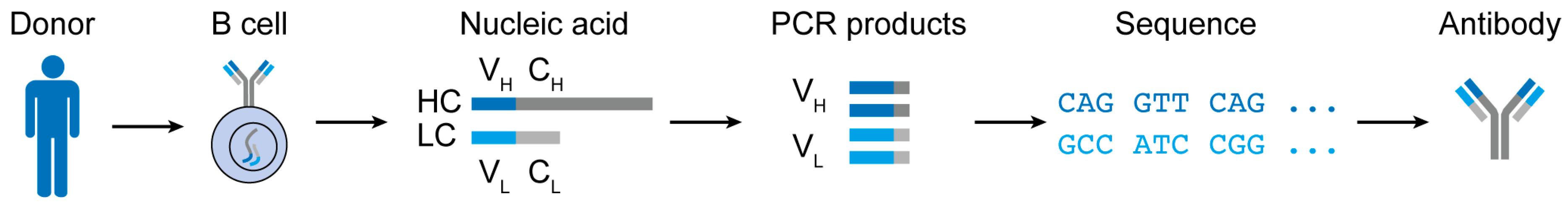
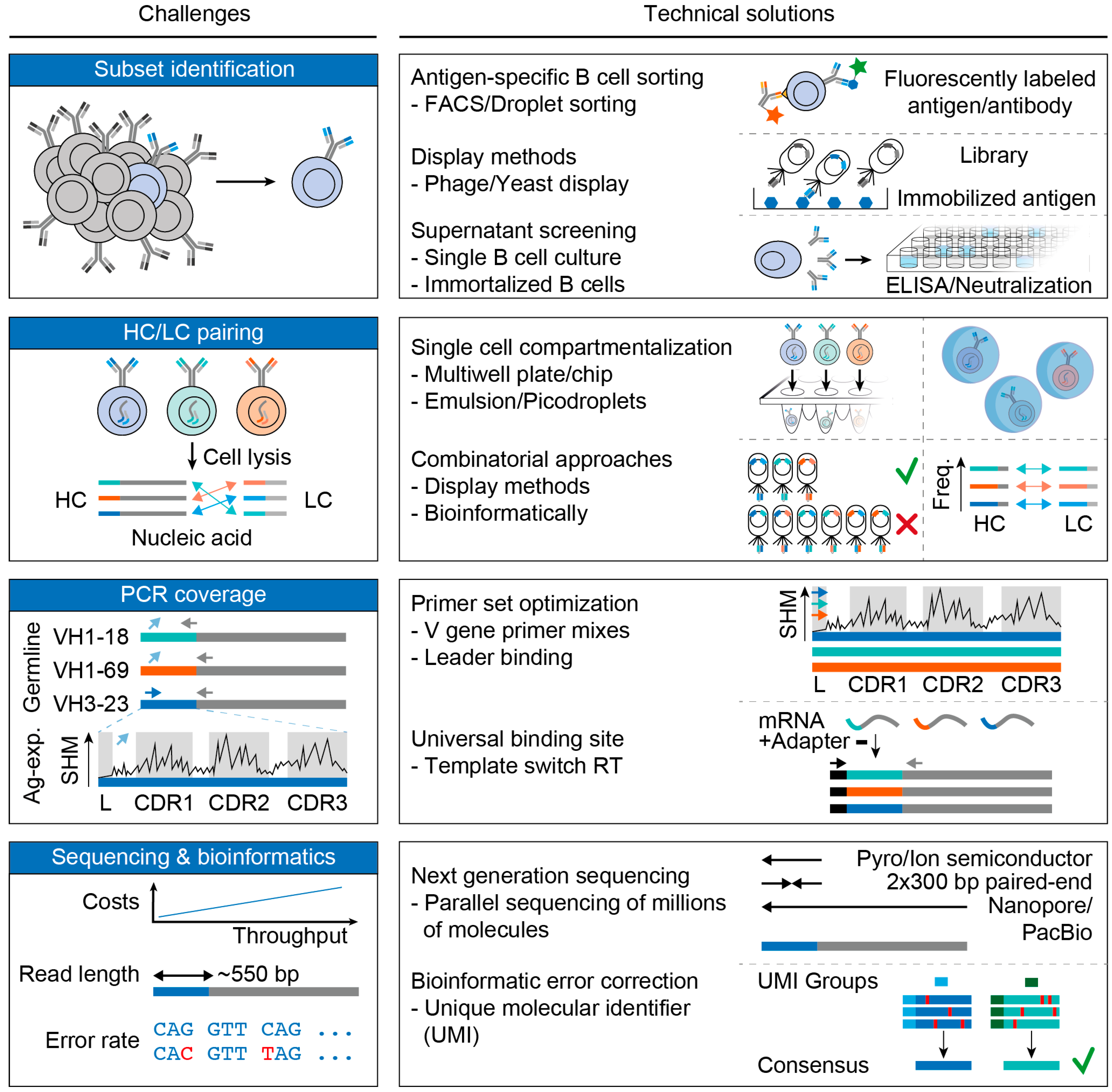
3. Challenges and Advances in B Cell Receptor Analyses
3.1. Subset Identification
3.2. Pairing of Heavy and Light Chains
3.3. PCR Coverage
3.4. Sequencing and Bioinformatics
4. Informing about Vaccination Strategies (I): Molecular Characterization of Broadly HIV-1 Neutralizing Antibodies
5. Informing about Vaccination Strategies (II): B Cell Receptor Repertoire Analyses
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Parvin, J.D.; Moscona, A.; Pan, W.T.; Leider, J.M.; Palese, P. Measurement of the mutation rates of animal viruses: Influenza A virus and poliovirus type 1. J. Virol. 1986, 59, 377–383. [Google Scholar] [CrossRef]
- Starcich, B.R.; Hahn, B.H.; Shaw, G.M.; McNeely, P.D.; Modrow, S.; Wolf, H.; Parks, E.S.; Parks, W.P.; Josephs, S.F.; Gallo, R.C.; et al. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 1986, 45, 637–648. [Google Scholar] [CrossRef]
- Vidarsson, G.; Dekkers, G.; Rispens, T. IgG subclasses and allotypes: From structure to effector functions. Front. Immunol. 2014, 5, 520. [Google Scholar] [CrossRef]
- Alt, F.W.; Zhang, Y.; Meng, F.L.; Guo, C.; Schwer, B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 2013, 152, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Komori, T.; Okada, A.; Stewart, V.; Alt, F.W. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science 1993, 261, 1171–1175. [Google Scholar] [CrossRef] [PubMed]
- Boyer, S.; Biswas, D.; Kumar Soshee, A.; Scaramozzino, N.; Nizak, C.; Rivoire, O. Hierarchy and extremes in selections from pools of randomized proteins. Proc. Natl. Acad. Sci. USA 2016, 113, 3482–3487. [Google Scholar] [CrossRef] [PubMed]
- Klein, F.; Diskin, R.; Scheid, J.F.; Gaebler, C.; Mouquet, H.; Georgiev, I.S.; Pancera, M.; Zhou, T.; Incesu, R.B.; Fu, B.Z.; et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 2013, 153, 126–138. [Google Scholar] [CrossRef]
- Lefranc, M.P.; Giudicelli, V.; Ginestoux, C.; Bodmer, J.; Muller, W.; Bontrop, R.; Lemaitre, M.; Malik, A.; Barbie, V.; Chaume, D. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 1999, 27, 209–212. [Google Scholar] [CrossRef]
- Sethna, Z.; Elhanati, Y.; Callan, C.G.; Walczak, A.M.; Mora, T. OLGA: Fast computation of generation probabilities of B- and T-cell receptor amino acid sequences and motifs. Bioinformatics 2019, 35, 2974–2981. [Google Scholar] [CrossRef]
- Marcou, Q.; Mora, T.; Walczak, A.M. High-throughput immune repertoire analysis with IGoR. Nat. Commun. 2018, 9, 561. [Google Scholar] [CrossRef]
- Elhanati, Y.; Sethna, Z.; Marcou, Q.; Callan, C.G., Jr.; Mora, T.; Walczak, A.M. Inferring processes underlying B-cell repertoire diversity. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140243. [Google Scholar] [CrossRef] [PubMed]
- Vollmers, C.; Sit, R.V.; Weinstein, J.A.; Dekker, C.L.; Quake, S.R. Genetic measurement of memory B-cell recall using antibody repertoire sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 13463–13468. [Google Scholar] [CrossRef] [PubMed]
- Mora, T.; Walczak, A.M. How many different clonotypes do immune repertoires contain? Curr. Opin. Syst. Biol. 2019, 18, 104–110. [Google Scholar] [CrossRef]
- Schroeder, H.W.; Cavacini, L., Jr. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 2010, 125, S41–S52. [Google Scholar] [CrossRef] [PubMed]
- Victora, G.D.; Nussenzweig, M.C. Germinal centers. Annu. Rev. Immunol. 2012, 30, 429–457. [Google Scholar] [CrossRef]
- Teng, G.; Papavasiliou, F.N. Immunoglobulin somatic hypermutation. Annu. Rev. Genet. 2007, 41, 107–120. [Google Scholar] [CrossRef]
- Yaari, G.; Vander Heiden, J.A.; Uduman, M.; Gadala-Maria, D.; Gupta, N.; Stern, J.N.; O’Connor, K.C.; Hafler, D.A.; Laserson, U.; Vigneault, F.; et al. Models of somatic hypermutation targeting and substitution based on synonymous mutations from high-throughput immunoglobulin sequencing data. Front. Immunol. 2013, 4, 358. [Google Scholar] [CrossRef]
- Schramm, C.A.; Douek, D.C. Beyond Hot Spots: Biases in Antibody Somatic Hypermutation and Implications for Vaccine Design. Front. Immunol. 2018, 9, 1876. [Google Scholar] [CrossRef]
- LeBien, T.W.; Tedder, T.F. B lymphocytes: How they develop and function. Blood 2008, 112, 1570–1580. [Google Scholar] [CrossRef]
- De Costa, D.; Broodman, I.; Vanduijn, M.M.; Stingl, C.; Dekker, L.J.; Burgers, P.C.; Hoogsteden, H.C.; Sillevis Smitt, P.A.; van Klaveren, R.J.; Luider, T.M. Sequencing and quantifying IgG fragments and antigen-binding regions by mass spectrometry. J. Proteome Res. 2010, 9, 2937–2945. [Google Scholar] [CrossRef]
- Plouffe, B.D.; Murthy, S.K.; Lewis, L.H. Fundamentals and application of magnetic particles in cell isolation and enrichment: A review. Rep. Prog. Phys. 2015, 78, 016601. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.F.; van den Engh, G. Flow cytometry and cell sorting. Adv. Biochem. Eng. Biotechnol. 2007, 106, 19–39. [Google Scholar] [PubMed]
- Eyer, K.; Doineau, R.C.L.; Castrillon, C.E.; Briseno-Roa, L.; Menrath, V.; Mottet, G.; England, P.; Godina, A.; Brient-Litzler, E.; Nizak, C.; et al. Single-cell deep phenotyping of IgG-secreting cells for high-resolution immune monitoring. Nat. Biotechnol. 2017, 35, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Shembekar, N.; Hu, H.; Eustace, D.; Merten, C.A. Single-Cell Droplet Microfluidic Screening for Antibodies Specifically Binding to Target Cells. Cell Rep. 2018, 22, 2206–2215. [Google Scholar] [CrossRef] [PubMed]
- Xi, H.D.; Zheng, H.; Guo, W.; Ganan-Calvo, A.M.; Ai, Y.; Tsao, C.W.; Zhou, J.; Li, W.; Huang, Y.; Nguyen, N.T.; et al. Active droplet sorting in microfluidics: A review. Lab Chip 2017, 17, 751–771. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.C.; Andrews, S.F. Tools to therapeutically harness the human antibody response. Nat. Rev. Immunol. 2012, 12, 709–719. [Google Scholar] [CrossRef]
- Walker, L.M.; Burton, D.R. Passive immunotherapy of viral infections: ’Super-antibodies’ enter the fray. Nat. Rev. Immunol. 2018, 18, 297–308. [Google Scholar] [CrossRef]
- Scheid, J.F.; Mouquet, H.; Feldhahn, N.; Walker, B.D.; Pereyra, F.; Cutrell, E.; Seaman, M.S.; Mascola, J.R.; Wyatt, R.T.; Wardemann, H.; et al. A method for identification of HIV gp140 binding memory B cells in human blood. J. Immunol. Methods 2009, 343, 65–67. [Google Scholar] [CrossRef]
- Scheid, J.F.; Mouquet, H.; Feldhahn, N.; Seaman, M.S.; Velinzon, K.; Pietzsch, J.; Ott, R.G.; Anthony, R.M.; Zebroski, H.; Hurley, A.; et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 2009, 458, 636–640. [Google Scholar] [CrossRef]
- Wu, X.; Yang, Z.Y.; Li, Y.; Hogerkorp, C.M.; Schief, W.R.; Seaman, M.S.; Zhou, T.; Schmidt, S.D.; Wu, L.; Xu, L.; et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 2010, 329, 856–861. [Google Scholar] [CrossRef]
- Sok, D.; van Gils, M.J.; Pauthner, M.; Julien, J.P.; Saye-Francisco, K.L.; Hsueh, J.; Briney, B.; Lee, J.H.; Le, K.M.; Lee, P.S.; et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc. Natl. Acad. Sci. USA 2014, 111, 17624–17629. [Google Scholar] [CrossRef] [PubMed]
- Joyce, M.G.; Wheatley, A.K.; Thomas, P.V.; Chuang, G.Y.; Soto, C.; Bailer, R.T.; Druz, A.; Georgiev, I.S.; Gillespie, R.A.; Kanekiyo, M.; et al. Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses. Cell 2016, 166, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Robbiani, D.F.; Bozzacco, L.; Keeffe, J.R.; Khouri, R.; Olsen, P.C.; Gazumyan, A.; Schaefer-Babajew, D.; Avila-Rios, S.; Nogueira, L.; Patel, R.; et al. Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico. Cell 2017, 169, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, S.A.; Zehner, M.; Krahling, V.; Cohen-Dvashi, H.; Kreer, C.; Elad, N.; Gruell, H.; Ercanoglu, M.S.; Schommers, P.; Gieselmann, L.; et al. Polyclonal and convergent antibody response to Ebola virus vaccine rVSV-ZEBOV. Nat. Med. 2019, 25, 1589–1600. [Google Scholar] [CrossRef] [PubMed]
- Hicar, M.D.; Chen, X.; Briney, B.; Hammonds, J.; Wang, J.J.; Kalams, S.; Spearman, P.W.; Crowe, J.E., Jr. Pseudovirion particles bearing native HIV envelope trimers facilitate a novel method for generating human neutralizing monoclonal antibodies against HIV. J. Acquir. Immune Defic. Syndr. 2010, 54, 223–235. [Google Scholar] [CrossRef]
- Cale, E.M.; Gorman, J.; Radakovich, N.A.; Crooks, E.T.; Osawa, K.; Tong, T.; Li, J.; Nagarajan, R.; Ozorowski, G.; Ambrozak, D.R.; et al. Virus-like Particles Identify an HIV V1V2 Apex-Binding Neutralizing Antibody that Lacks a Protruding Loop. Immunity 2017, 46, 777–791. [Google Scholar] [CrossRef]
- Klein, F.; Gaebler, C.; Mouquet, H.; Sather, D.N.; Lehmann, C.; Scheid, J.F.; Kraft, Z.; Liu, Y.; Pietzsch, J.; Hurley, A.; et al. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. J. Exp. Med. 2012, 209, 1469–1479. [Google Scholar] [CrossRef]
- Woda, M.; Mathew, A. Fluorescently labeled dengue viruses as probes to identify antigen-specific memory B cells by multiparametric flow cytometry. J. Immunol. Methods 2015, 416, 167–177. [Google Scholar] [CrossRef][Green Version]
- Burton, D.R.; Barbas, C.F.; Persson, M.A., 3rd; Koenig, S.; Chanock, R.M.; Lerner, R.A. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. USA 1991, 88, 10134–10137. [Google Scholar] [CrossRef]
- Walker, L.M.; Bowley, D.R.; Burton, D.R. Efficient recovery of high-affinity antibodies from a single-chain Fab yeast display library. J. Mol. Biol. 2009, 389, 365–375. [Google Scholar] [CrossRef]
- Traggiai, E.; Becker, S.; Subbarao, K.; Kolesnikova, L.; Uematsu, Y.; Gismondo, M.R.; Murphy, B.R.; Rappuoli, R.; Lanzavecchia, A. An efficient method to make human monoclonal antibodies from memory B cells: Potent neutralization of SARS coronavirus. Nat. Med. 2004, 10, 871–875. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.M.; Phogat, S.K.; Chan-Hui, P.Y.; Wagner, D.; Phung, P.; Goss, J.L.; Wrin, T.; Simek, M.D.; Fling, S.; Mitcham, J.L.; et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 2009, 326, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011, 333, 850–856. [Google Scholar] [CrossRef] [PubMed]
- De Benedictis, P.; Minola, A.; Rota Nodari, E.; Aiello, R.; Zecchin, B.; Salomoni, A.; Foglierini, M.; Agatic, G.; Vanzetta, F.; Lavenir, R.; et al. Development of broad-spectrum human monoclonal antibodies for rabies post-exposure prophylaxis. EMBO Mol. Med. 2016, 8, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R.W.; Derking, R.; Cupo, A.; Julien, J.P.; Yasmeen, A.; de Val, N.; Kim, H.J.; Blattner, C.; de la Pena, A.T.; Korzun, J.; et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013, 9, e1003618. [Google Scholar] [CrossRef] [PubMed]
- Van Gils, M.J.; van den Kerkhof, T.L.; Ozorowski, G.; Cottrell, C.A.; Sok, D.; Pauthner, M.; Pallesen, J.; de Val, N.; Yasmeen, A.; de Taeye, S.W.; et al. An HIV-1 antibody from an elite neutralizer implicates the fusion peptide as a site of vulnerability. Nat. Microbiol. 2016, 2, 16199. [Google Scholar] [CrossRef]
- Kong, R.; Xu, K.; Zhou, T.; Acharya, P.; Lemmin, T.; Liu, K.; Ozorowski, G.; Soto, C.; Taft, J.D.; Bailer, R.T.; et al. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 2016, 352, 828–833. [Google Scholar] [CrossRef]
- Doria-Rose, N.A.; Bhiman, J.N.; Roark, R.S.; Schramm, C.A.; Gorman, J.; Chuang, G.Y.; Pancera, M.; Cale, E.M.; Ernandes, M.J.; Louder, M.K.; et al. New Member of the V1V2-Directed CAP256-VRC26 Lineage That Shows Increased Breadth and Exceptional Potency. J. Virol. 2016, 90, 76–91. [Google Scholar] [CrossRef]
- Setliff, I.; Shiakolas, A.R.; Pilewski, K.A.; Murji, A.A.; Mapengo, R.E.; Janowska, K.; Richardson, S.; Oosthuysen, C.; Raju, N.; Ronsard, L.; et al. High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity. Cell 2019, 179, 1636–1646. [Google Scholar] [CrossRef]
- Boyd, S.D.; Marshall, E.L.; Merker, J.D.; Maniar, J.M.; Zhang, L.N.; Sahaf, B.; Jones, C.D.; Simen, B.B.; Hanczaruk, B.; Nguyen, K.D.; et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci. Transl. Med. 2009, 1, 12ra23. [Google Scholar] [CrossRef]
- Wu, Y.C.; Kipling, D.; Leong, H.S.; Martin, V.; Ademokun, A.A.; Dunn-Walters, D.K. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood 2010, 116, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Glanville, J.; Kuo, T.C.; von Budingen, H.C.; Guey, L.; Berka, J.; Sundar, P.D.; Huerta, G.; Mehta, G.R.; Oksenberg, J.R.; Hauser, S.L.; et al. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc. Natl. Acad. Sci. USA 2011, 108, 20066–20071. [Google Scholar] [CrossRef] [PubMed]
- Soto, C.; Bombardi, R.G.; Branchizio, A.; Kose, N.; Matta, P.; Sevy, A.M.; Sinkovits, R.S.; Gilchuk, P.; Finn, J.A.; Crowe, J.E., Jr. High frequency of shared clonotypes in human B cell receptor repertoires. Nature 2019, 566, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.C.A.; Roskin, K.M.; Jackson, K.J.L.; Joshi, S.A.; Nejad, P.; Lee, J.Y.; Wagar, L.E.; Pham, T.D.; Hoh, R.A.; Nguyen, K.D.; et al. Shaping of infant B cell receptor repertoires by environmental factors and infectious disease. Sci. Transl. Med. 2019, 11, eaat2004. [Google Scholar] [CrossRef] [PubMed]
- Briney, B.; Inderbitzin, A.; Joyce, C.; Burton, D.R. Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature 2019, 566, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Ofek, G.; Yang, Y.; Zhang, B.; Louder, M.K.; Lu, G.; McKee, K.; Pancera, M.; Skinner, J.; Zhang, Z.; et al. Mining the antibodyome for HIV-1-neutralizing antibodies with next-generation sequencing and phylogenetic pairing of heavy/light chains. Proc. Natl. Acad. Sci. USA 2013, 110, 6470–6475. [Google Scholar] [CrossRef]
- Wardemann, H.; Yurasov, S.; Schaefer, A.; Young, J.W.; Meffre, E.; Nussenzweig, M.C. Predominant autoantibody production by early human B cell precursors. Science 2003, 301, 1374–1377. [Google Scholar] [CrossRef]
- Tiller, T.; Meffre, E.; Yurasov, S.; Tsuiji, M.; Nussenzweig, M.C.; Wardemann, H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 2008, 329, 112–124. [Google Scholar] [CrossRef]
- Busse, C.E.; Czogiel, I.; Braun, P.; Arndt, P.F.; Wardemann, H. Single-cell based high-throughput sequencing of full-length immunoglobulin heavy and light chain genes. Eur. J. Immunol. 2014, 44, 597–603. [Google Scholar] [CrossRef]
- Tan, Y.C.; Blum, L.K.; Kongpachith, S.; Ju, C.H.; Cai, X.; Lindstrom, T.M.; Sokolove, J.; Robinson, W.H. High-throughput sequencing of natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clin. Immunol. 2014, 151, 55–65. [Google Scholar] [CrossRef]
- Mazutis, L.; Gilbert, J.; Ung, W.L.; Weitz, D.A.; Griffiths, A.D.; Heyman, J.A. Single-cell analysis and sorting using droplet-based microfluidics. Nat. Protoc. 2013, 8, 870–891. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, J.R.; DeKosky, B.J.; Tanno, H.; Ellington, A.D.; Georgiou, G. Ultra-high-throughput sequencing of the immune receptor repertoire from millions of lymphocytes. Nat. Protoc. 2016, 11, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Al-Eryani, G.; Carswell, S.; Ferguson, J.M.; Blackburn, J.; Barton, K.; Roden, D.; Luciani, F.; Giang Phan, T.; Junankar, S.; et al. High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat. Commun. 2019, 10, 3120. [Google Scholar] [CrossRef] [PubMed]
- Briggs, A.; Goldfless, S.; Timberlake, S.; Belmont, B.; Clouser, C.; Koppstein, D.; Sok, D.; Heiden, J.V.; Tamminen, M.; Kleinstein, S.; et al. Tumor-infiltrating immune repertoires captured by single-cell barcoding in emulsion. BioRxiv 2017, 134841. [Google Scholar] [CrossRef]
- Goldstein, L.D.; Chen, Y.J.; Wu, J.; Chaudhuri, S.; Hsiao, Y.C.; Schneider, K.; Hoi, K.H.; Lin, Z.; Guerrero, S.; Jaiswal, B.S.; et al. Massively parallel single-cell B-cell receptor sequencing enables rapid discovery of diverse antigen-reactive antibodies. Commun. Biol. 2019, 2, 304. [Google Scholar] [CrossRef]
- Bradbury, A.R.; Sidhu, S.; Dubel, S.; McCafferty, J. Beyond natural antibodies: The power of in vitro display technologies. Nat. Biotechnol. 2011, 29, 245–254. [Google Scholar] [CrossRef]
- Larrick, J.W.; Danielsson, L.; Brenner, C.A.; Abrahamson, M.; Fry, K.E.; Borrebaeck, C.A. Rapid cloning of rearranged immunoglobulin genes from human hybridoma cells using mixed primers and the polymerase chain reaction. Biochem. Biophys. Res. Commun. 1989, 160, 1250–1256. [Google Scholar] [CrossRef]
- Kuppers, R.; Zhao, M.; Hansmann, M.L.; Rajewsky, K. Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO J. 1993, 12, 4955–4967. [Google Scholar] [CrossRef]
- Sblattero, D.; Bradbury, A. A definitive set of oligonucleotide primers for amplifying human V regions. Immunotechnology 1998, 3, 271–278. [Google Scholar] [CrossRef]
- Lim, T.S.; Mollova, S.; Rubelt, F.; Sievert, V.; Dubel, S.; Lehrach, H.; Konthur, Z. V-gene amplification revisited—An optimised procedure for amplification of rearranged human antibody genes of different isotypes. N. Biotechnol. 2010, 27, 108–117. [Google Scholar] [CrossRef]
- Ippolito, G.C.; Hoi, K.H.; Reddy, S.T.; Carroll, S.M.; Ge, X.; Rogosch, T.; Zemlin, M.; Shultz, L.D.; Ellington, A.D.; Vandenberg, C.L.; et al. Antibody repertoires in humanized NOD-scid-IL2Rgamma(null) mice and human B cells reveals human-like diversification and tolerance checkpoints in the mouse. PLoS ONE 2012, 7, e35497. [Google Scholar] [CrossRef] [PubMed]
- Scheid, J.F.; Mouquet, H.; Ueberheide, B.; Diskin, R.; Klein, F.; Oliveira, T.Y.; Pietzsch, J.; Fenyo, D.; Abadir, A.; Velinzon, K.; et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 2011, 333, 1633–1637. [Google Scholar] [CrossRef] [PubMed]
- Klein, F.; Mouquet, H.; Dosenovic, P.; Scheid, J.F.; Scharf, L.; Nussenzweig, M.C. Antibodies in HIV-1 vaccine development and therapy. Science 2013, 341, 1199–1204. [Google Scholar] [CrossRef] [PubMed]
- Kreer, C.; Doering, M.; Lehnen, N.; Ercanoglu, M.S.; Gieselmann, L.; Luca, D.; Jain, K.; Schommers, P.; Pfeifer, N.; Klein, F. OpenPrimer for multiplex amplification of highly diverse templates. BioRxiv 2019, 847574. [Google Scholar] [CrossRef]
- He, L.; Sok, D.; Azadnia, P.; Hsueh, J.; Landais, E.; Simek, M.; Koff, W.C.; Poignard, P.; Burton, D.R.; Zhu, J. Toward a more accurate view of human B-cell repertoire by next-generation sequencing, unbiased repertoire capture and single-molecule barcoding. Sci. Rep. 2014, 4, 6778. [Google Scholar] [CrossRef]
- Frohman, M.A.; Dush, M.K.; Martin, G.R. Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sc.i USA 1988, 85, 8998–9002. [Google Scholar] [CrossRef]
- Huang, C.; Stollar, B.D. Construction of representative immunoglobulin variable region cDNA libraries from human peripheral blood lymphocytes without in vitro stimulation. J. Immunol. Methods 1991, 141, 227–236. [Google Scholar] [CrossRef]
- Ozawa, T.; Kishi, H.; Muraguchi, A. Amplification and analysis of cDNA generated from a single cell by 5′-RACE: Application to isolation of antibody heavy and light chain variable gene sequences from single B cells. Biotechniques 2006, 40, 469–470. [Google Scholar] [CrossRef]
- Rollenske, T.; Szijarto, V.; Lukasiewicz, J.; Guachalla, L.M.; Stojkovic, K.; Hartl, K.; Stulik, L.; Kocher, S.; Lasitschka, F.; Al-Saeedi, M.; et al. Cross-specificity of protective human antibodies against Klebsiella pneumoniae LPS O-antigen. Nat. Immunol. 2018, 19, 617–624. [Google Scholar] [CrossRef]
- Olbrich, A.; Wardemann, H.; Bohm, S.; Rother, K.; Colpitts, C.C.; Wrensch, F.; Baumert, T.F.; Berg, T.; Benckert, J. Repertoire and Neutralizing Activity of Antibodies Against Hepatitis C Virus E2 Peptide in Patients with Spontaneous Resolution of Hepatitis, C. J. Infect. Dis. 2019, 220, 1209–1218. [Google Scholar] [CrossRef]
- Greiff, V.; Miho, E.; Menzel, U.; Reddy, S.T. Bioinformatic and Statistical Analysis of Adaptive Immune Repertoires. Trends Immunol. 2015, 36, 738–749. [Google Scholar] [CrossRef] [PubMed]
- DeKosky, B.J.; Kojima, T.; Rodin, A.; Charab, W.; Ippolito, G.C.; Ellington, A.D.; Georgiou, G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat. Med. 2015, 21, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; Wesemann, D.R. Analyzing Immunoglobulin Repertoires. Front. Immunol. 2018, 9, 462. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, J.A.; Marquez, S.; Marthandan, N.; Bukhari, S.A.C.; Busse, C.E.; Corrie, B.; Hershberg, U.; Kleinstein, S.H.; Matsen Iv, F.A.; Ralph, D.K.; et al. AIRR Community Standardized Representations for Annotated Immune Repertoires. Front. Immunol. 2018, 9, 2206. [Google Scholar] [CrossRef]
- Corrie, B.D.; Marthandan, N.; Zimonja, B.; Jaglale, J.; Zhou, Y.; Barr, E.; Knoetze, N.; Breden, F.M.W.; Christley, S.; Scott, J.K.; et al. iReceptor: A platform for querying and analyzing antibody/B-cell and T-cell receptor repertoire data across federated repositories. Immunol. Rev. 2018, 284, 24–41. [Google Scholar] [CrossRef]
- Miho, E.; Yermanos, A.; Weber, C.R.; Berger, C.T.; Reddy, S.T.; Greiff, V. Computational Strategies for Dissecting the High-Dimensional Complexity of Adaptive Immune Repertoires. Front. Immunol. 2018, 9, 224. [Google Scholar] [CrossRef]
- Williams, L.D.; Ofek, G.; Schatzle, S.; McDaniel, J.R.; Lu, X.; Nicely, N.I.; Wu, L.; Lougheed, C.S.; Bradley, T.; Louder, M.K.; et al. Potent and broad HIV-neutralizing antibodies in memory B cells and plasma. Sci. Immunol. 2017, 2, eaal2200. [Google Scholar] [CrossRef]
- Sajadi, M.M.; Dashti, A.; Rikhtegaran Tehrani, Z.; Tolbert, W.D.; Seaman, M.S.; Ouyang, X.; Gohain, N.; Pazgier, M.; Kim, D.; Cavet, G.; et al. Identification of Near-Pan-neutralizing Antibodies against HIV-1 by Deconvolution of Plasma Humoral Responses. Cell 2018, 173, 1783–1795. [Google Scholar] [CrossRef]
- Sok, D.; Burton, D.R. Recent progress in broadly neutralizing antibodies to HIV. Nat. Immunol. 2018, 19, 1179–1188. [Google Scholar] [CrossRef]
- Lynch, R.M.; Boritz, E.; Coates, E.E.; DeZure, A.; Madden, P.; Costner, P.; Enama, M.E.; Plummer, S.; Holman, L.; Hendel, C.S.; et al. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci. Transl. Med. 2015, 7, 319ra206. [Google Scholar] [CrossRef]
- Caskey, M.; Klein, F.; Lorenzi, J.C.; Seaman, M.S.; West, A.P., Jr.; Buckley, N.; Kremer, G.; Nogueira, L.; Braunschweig, M.; Scheid, J.F.; et al. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 2015, 522, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Bar, K.J.; Sneller, M.C.; Harrison, L.J.; Justement, J.S.; Overton, E.T.; Petrone, M.E.; Salantes, D.B.; Seamon, C.A.; Scheinfeld, B.; Kwan, R.W.; et al. Effect of HIV Antibody VRC01 on Viral Rebound after Treatment Interruption. N. Engl. J. Med. 2016, 375, 2037–2050. [Google Scholar] [CrossRef]
- Scheid, J.F.; Horwitz, J.A.; Bar-On, Y.; Kreider, E.F.; Lu, C.L.; Lorenzi, J.C.; Feldmann, A.; Braunschweig, M.; Nogueira, L.; Oliveira, T.; et al. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 2016, 535, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Caskey, M.; Schoofs, T.; Gruell, H.; Settler, A.; Karagounis, T.; Kreider, E.F.; Murrell, B.; Pfeifer, N.; Nogueira, L.; Oliveira, T.Y.; et al. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 2017, 23, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Bar-On, Y.; Gruell, H.; Schoofs, T.; Pai, J.A.; Nogueira, L.; Butler, A.L.; Millard, K.; Lehmann, C.; Suarez, I.; Oliveira, T.Y.; et al. Safety and antiviral activity of combination HIV-1 broadly neutralizing antibodies in viremic individuals. Nat. Med. 2018, 24, 1701–1707. [Google Scholar] [CrossRef]
- Mendoza, P.; Gruell, H.; Nogueira, L.; Pai, J.A.; Butler, A.L.; Millard, K.; Lehmann, C.; Suarez, I.; Oliveira, T.Y.; Lorenzi, J.C.C.; et al. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 2018, 561, 479–484. [Google Scholar] [CrossRef]
- Moldt, B.; Rakasz, E.G.; Schultz, N.; Chan-Hui, P.Y.; Swiderek, K.; Weisgrau, K.L.; Piaskowski, S.M.; Bergman, Z.; Watkins, D.I.; Poignard, P.; et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc. Natl. Acad. Sci. USA 2012, 109, 18921–18925. [Google Scholar] [CrossRef]
- Shingai, M.; Donau, O.K.; Plishka, R.J.; Buckler-White, A.; Mascola, J.R.; Nabel, G.J.; Nason, M.C.; Montefiori, D.; Moldt, B.; Poignard, P.; et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J. Exp. Med. 2014, 211, 2061–2074. [Google Scholar] [CrossRef]
- Gautam, R.; Nishimura, Y.; Pegu, A.; Nason, M.C.; Klein, F.; Gazumyan, A.; Golijanin, J.; Buckler-White, A.; Sadjadpour, R.; Wang, K.; et al. A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature 2016, 533, 105–109. [Google Scholar] [CrossRef]
- Julg, B.; Tartaglia, L.J.; Keele, B.F.; Wagh, K.; Pegu, A.; Sok, D.; Abbink, P.; Schmidt, S.D.; Wang, K.; Chen, X.; et al. Broadly neutralizing antibodies targeting the HIV-1 envelope V2 apex confer protection against a clade C SHIV challenge. Sci. Transl. Med. 2017, 9, eaal1321. [Google Scholar] [CrossRef]
- Zhou, T.; Lynch, R.M.; Chen, L.; Acharya, P.; Wu, X.; Doria-Rose, N.A.; Joyce, M.G.; Lingwood, D.; Soto, C.; Bailer, R.T.; et al. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell 2015, 161, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Zhu, J.; Wu, X.; Moquin, S.; Zhang, B.; Acharya, P.; Georgiev, I.S.; Altae-Tran, H.R.; Chuang, G.Y.; Joyce, M.G.; et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 2013, 39, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhou, T.; Zhu, J.; Zhang, B.; Georgiev, I.; Wang, C.; Chen, X.; Longo, N.S.; Louder, M.; McKee, K.; et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 2011, 333, 1593–1602. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Kang, B.H.; Ishida, E.; Zhou, T.; Griesman, T.; Sheng, Z.; Wu, F.; Doria-Rose, N.A.; Zhang, B.; McKee, K.; et al. Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth. Immunity 2016, 45, 1108–1121. [Google Scholar] [CrossRef] [PubMed]
- Umotoy, J.; Bagaya, B.S.; Joyce, C.; Schiffner, T.; Menis, S.; Saye-Francisco, K.L.; Biddle, T.; Mohan, S.; Vollbrecht, T.; Kalyuzhniy, O.; et al. Rapid and Focused Maturation of a VRC01-Class HIV Broadly Neutralizing Antibody Lineage Involves Both Binding and Accommodation of the N276-Glycan. Immunity 2019, 51, 141–154. [Google Scholar] [CrossRef]
- Georgiev, I.S.; Doria-Rose, N.A.; Zhou, T.; Kwon, Y.D.; Staupe, R.P.; Moquin, S.; Chuang, G.Y.; Louder, M.K.; Schmidt, S.D.; Altae-Tran, H.R.; et al. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 2013, 340, 751–756. [Google Scholar] [CrossRef]
- Godoy-Lozano, E.E.; Tellez-Sosa, J.; Sanchez-Gonzalez, G.; Samano-Sanchez, H.; Aguilar-Salgado, A.; Salinas-Rodriguez, A.; Cortina-Ceballos, B.; Vivanco-Cid, H.; Hernandez-Flores, K.; Pfaff, J.M.; et al. Lower IgG somatic hypermutation rates during acute dengue virus infection is compatible with a germinal center-independent B cell response. Genome Med. 2016, 8, 23. [Google Scholar] [CrossRef]
- Farci, P.; Diaz, G.; Chen, Z.; Govindarajan, S.; Tice, A.; Agulto, L.; Pittaluga, S.; Boon, D.; Yu, C.; Engle, R.E.; et al. B cell gene signature with massive intrahepatic production of antibodies to hepatitis B core antigen in hepatitis B virus-associated acute liver failure. Proc. Natl. Acad. Sci. USA 2010, 107, 8766–8771. [Google Scholar] [CrossRef]
- Weitkamp, J.H.; Kallewaard, N.; Kusuhara, K.; Bures, E.; Williams, J.V.; LaFleur, B.; Greenberg, H.B.; Crowe, J.E., Jr. Infant and adult human B cell responses to rotavirus share common immunodominant variable gene repertoires. J. Immunol. 2003, 171, 4680–4688. [Google Scholar] [CrossRef]
- Weitkamp, J.H.; Kallewaard, N.L.; Bowen, A.L.; Lafleur, B.J.; Greenberg, H.B.; Crowe, J.E., Jr. VH1-46 is the dominant immunoglobulin heavy chain gene segment in rotavirus-specific memory B cells expressing the intestinal homing receptor alpha4beta7. J. Immunol. 2005, 174, 3454–3460. [Google Scholar] [CrossRef]
- Tian, C.; Luskin, G.K.; Dischert, K.M.; Higginbotham, J.N.; Shepherd, B.E.; Crowe, J.E., Jr. Immunodominance of the VH1-46 antibody gene segment in the primary repertoire of human rotavirus-specific B cells is reduced in the memory compartment through somatic mutation of nondominant clones. J. Immunol. 2008, 180, 3279–3288. [Google Scholar] [CrossRef] [PubMed]
- Scheepers, C.; Shrestha, R.K.; Lambson, B.E.; Jackson, K.J.; Wright, I.A.; Naicker, D.; Goosen, M.; Berrie, L.; Ismail, A.; Garrett, N.; et al. Ability to develop broadly neutralizing HIV-1 antibodies is not restricted by the germline Ig gene repertoire. J. Immunol. 2015, 194, 4371–4378. [Google Scholar] [CrossRef] [PubMed]
- Sui, J.; Hwang, W.C.; Perez, S.; Wei, G.; Aird, D.; Chen, L.M.; Santelli, E.; Stec, B.; Cadwell, G.; Ali, M.; et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009, 16, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Corti, D.; Suguitan, A.L., Jr.; Pinna, D.; Silacci, C.; Fernandez-Rodriguez, B.M.; Vanzetta, F.; Santos, C.; Luke, C.J.; Torres-Velez, F.J.; Temperton, N.J.; et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Invest. 2010, 120, 1663–1673. [Google Scholar] [CrossRef]
- Pappas, L.; Foglierini, M.; Piccoli, L.; Kallewaard, N.L.; Turrini, F.; Silacci, C.; Fernandez-Rodriguez, B.; Agatic, G.; Giacchetto-Sasselli, I.; Pellicciotta, G.; et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature 2014, 516, 418–422. [Google Scholar] [CrossRef]
- Buchacher, A.; Predl, R.; Strutzenberger, K.; Steinfellner, W.; Trkola, A.; Purtscher, M.; Gruber, G.; Tauer, C.; Steindl, F.; Jungbauer, A.; et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 1994, 10, 359–369. [Google Scholar] [CrossRef]
- Galson, J.D.; Truck, J.; Clutterbuck, E.A.; Fowler, A.; Cerundolo, V.; Pollard, A.J.; Lunter, G.; Kelly, D.F. B-cell repertoire dynamics after sequential hepatitis B vaccination and evidence for cross-reactive B-cell activation. Genome Med. 2016, 8, 68. [Google Scholar] [CrossRef]
- Jackson, K.J.; Liu, Y.; Roskin, K.M.; Glanville, J.; Hoh, R.A.; Seo, K.; Marshall, E.L.; Gurley, T.C.; Moody, M.A.; Haynes, B.F.; et al. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe 2014, 16, 105–114. [Google Scholar] [CrossRef]
- Rijal, P.; Elias, S.C.; Machado, S.R.; Xiao, J.; Schimanski, L.; O’Dowd, V.; Baker, T.; Barry, E.; Mendelsohn, S.C.; Cherry, C.J.; et al. Therapeutic Monoclonal Antibodies for Ebola Virus Infection Derived from Vaccinated Humans. Cell Rep. 2019, 27, 172–186. [Google Scholar] [CrossRef]
- Bornholdt, Z.A.; Turner, H.L.; Murin, C.D.; Li, W.; Sok, D.; Souders, C.A.; Piper, A.E.; Goff, A.; Shamblin, J.D.; Wollen, S.E.; et al. Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science 2016, 351, 1078–1083. [Google Scholar] [CrossRef]
- Lucas, A.H.; McLean, G.R.; Reason, D.C.; O’Connor, A.P.; Felton, M.C.; Moulton, K.D. Molecular ontogeny of the human antibody repertoire to the Haemophilus influenzae type B polysaccharide: Expression of canonical variable regions and their variants in vaccinated infants. Clin. Immunol. 2003, 108, 119–127. [Google Scholar] [CrossRef]
- Thomson, C.A.; Bryson, S.; McLean, G.R.; Creagh, A.L.; Pai, E.F.; Schrader, J.W. Germline V-genes sculpt the binding site of a family of antibodies neutralizing human cytomegalovirus. EMBO J. 2008, 27, 2592–2602. [Google Scholar] [CrossRef]
- Bryson, S.; Thomson, C.A.; Risnes, L.F.; Dasgupta, S.; Smith, K.; Schrader, J.W.; Pai, E.F. Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation. J. Immunol. 2016, 196, 4723–4730. [Google Scholar] [CrossRef]
- Stettler, K.; Beltramello, M.; Espinosa, D.A.; Graham, V.; Cassotta, A.; Bianchi, S.; Vanzetta, F.; Minola, A.; Jaconi, S.; Mele, F.; et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 2016, 353, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Sapparapu, G.; Fernandez, E.; Kose, N.; Bin, C.; Fox, J.M.; Bombardi, R.G.; Zhao, H.; Nelson, C.A.; Bryan, A.L.; Barnes, T.; et al. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature 2016, 540, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, S.; Du, L.; Wang, C.; Zou, P.; Hong, B.; Yuan, M.; Ren, X.; Tai, W.; Kong, Y.; et al. Neutralization of Zika virus by germline-like human monoclonal antibodies targeting cryptic epitopes on envelope domain III. Emerg. Microbes Infect. 2017, 6, e89. [Google Scholar] [CrossRef] [PubMed]
- Gorny, M.K.; Wang, X.H.; Williams, C.; Volsky, B.; Revesz, K.; Witover, B.; Burda, S.; Urbanski, M.; Nyambi, P.; Krachmarov, C.; et al. Preferential use of the VH5-51 gene segment by the human immune response to code for antibodies against the V3 domain of HIV-1. Mol. Immunol. 2009, 46, 917–926. [Google Scholar] [CrossRef]
- West, A.P., Jr.; Diskin, R.; Nussenzweig, M.C.; Bjorkman, P.J. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc. Natl. Acad. Sci. USA 2012, 109, E2083–E2090. [Google Scholar] [CrossRef]
- McLellan, J.S.; Pancera, M.; Carrico, C.; Gorman, J.; Julien, J.P.; Khayat, R.; Louder, R.; Pejchal, R.; Sastry, M.; Dai, K.; et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 2011, 480, 336–343. [Google Scholar] [CrossRef]
- Lee, J.H.; Andrabi, R.; Su, C.Y.; Yasmeen, A.; Julien, J.P.; Kong, L.; Wu, N.C.; McBride, R.; Sok, D.; Pauthner, M.; et al. A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic beta-Hairpin Structure. Immunity 2017, 46, 690–702. [Google Scholar] [CrossRef]
- DeKosky, B.J.; Lungu, O.I.; Park, D.; Johnson, E.L.; Charab, W.; Chrysostomou, C.; Kuroda, D.; Ellington, A.D.; Ippolito, G.C.; Gray, J.J.; et al. Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc. Natl. Acad. Sci. USA 2016, 113, E2636–E2645. [Google Scholar] [CrossRef]
- Haynes, B.F.; Fleming, J.; St Clair, E.W.; Katinger, H.; Stiegler, G.; Kunert, R.; Robinson, J.; Scearce, R.M.; Plonk, K.; Staats, H.F.; et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 2005, 308, 1906–1908. [Google Scholar] [CrossRef] [PubMed]
- Mascola, J.R.; Haynes, B.F. HIV-1 neutralizing antibodies: Understanding nature’s pathways. Immunol. Rev. 2013, 254, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Simonich, C.A.; Williams, K.L.; Verkerke, H.P.; Williams, J.A.; Nduati, R.; Lee, K.K.; Overbaugh, J. HIV-1 Neutralizing Antibodies with Limited Hypermutation from an Infant. Cell 2016, 166, 77–87. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, D.T.; Choi, N.M.; Briney, B.; Garces, F.; Ver, L.S.; Landais, E.; Murrell, B.; Wrin, T.; Kilembe, W.; Liang, C.H.; et al. Early Antibody Lineage Diversification and Independent Limb Maturation Lead to Broad HIV-1 Neutralization Targeting the Env High-Mannose Patch. Immunity 2016, 44, 1215–1226. [Google Scholar] [CrossRef]
- Kwong, P.D.; Mascola, J.R. HIV-1 Vaccines Based on Antibody Identification, B Cell Ontogeny, and Epitope Structure. Immunity 2018, 48, 855–871. [Google Scholar] [CrossRef]
- Hoot, S.; McGuire, A.T.; Cohen, K.W.; Strong, R.K.; Hangartner, L.; Klein, F.; Diskin, R.; Scheid, J.F.; Sather, D.N.; Burton, D.R.; et al. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS Pathog. 2013, 9, e1003106. [Google Scholar] [CrossRef]
- Zhou, T.; Georgiev, I.; Wu, X.; Yang, Z.Y.; Dai, K.; Finzi, A.; Kwon, Y.D.; Scheid, J.F.; Shi, W.; Xu, L.; et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 2010, 329, 811–817. [Google Scholar] [CrossRef]
- Sok, D.; Laserson, U.; Laserson, J.; Liu, Y.; Vigneault, F.; Julien, J.P.; Briney, B.; Ramos, A.; Saye, K.F.; Le, K.; et al. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS Pathog. 2013, 9, e1003754. [Google Scholar] [CrossRef]
- Haynes, B.F.; Kelsoe, G.; Harrison, S.C.; Kepler, T.B. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotechnol. 2012, 30, 423–433. [Google Scholar] [CrossRef]
- Havenar-Daughton, C.; Abbott, R.K.; Schief, W.R.; Crotty, S. When designing vaccines, consider the starting material: The human B cell repertoire. Curr. Opin. Immunol. 2018, 53, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Briney, B.S.; Willis, J.R.; Crowe, J.E., Jr. Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes. PLoS ONE 2012, 7, e36750. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.R.; Finn, J.A.; Briney, B.; Sapparapu, G.; Singh, V.; King, H.; LaBranche, C.C.; Montefiori, D.C.; Meiler, J.; Crowe, J.E., Jr. Long antibody HCDR3s from HIV-naive donors presented on a PG9 neutralizing antibody background mediate HIV neutralization. Proc. Natl. Acad. Sci. USA 2016, 113, 4446–4451. [Google Scholar] [CrossRef]
- Freund, N.T.; Wang, H.; Scharf, L.; Nogueira, L.; Horwitz, J.A.; Bar-On, Y.; Golijanin, J.; Sievers, S.A.; Sok, D.; Cai, H.; et al. Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller. Sci. Transl. Med. 2017, 9, eaal2144. [Google Scholar] [CrossRef]
- Barnes, C.O.; Gristick, H.B.; Freund, N.T.; Escolano, A.; Lyubimov, A.Y.; Hartweger, H.; West, A.P., Jr.; Cohen, A.E.; Nussenzweig, M.C.; Bjorkman, P.J. Structural characterization of a highly-potent V3-glycan broadly neutralizing antibody bound to natively-glycosylated HIV-1 envelope. Nat. Commun. 2018, 9, 1251. [Google Scholar] [CrossRef]
- Steichen, J.M.; Lin, Y.C.; Havenar-Daughton, C.; Pecetta, S.; Ozorowski, G.; Willis, J.R.; Toy, L.; Sok, D.; Liguori, A.; Kratochvil, S.; et al. A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses. Science 2019, 366, eaax4380. [Google Scholar] [CrossRef]
- Jardine, J.G.; Kulp, D.W.; Havenar-Daughton, C.; Sarkar, A.; Briney, B.; Sok, D.; Sesterhenn, F.; Ereno-Orbea, J.; Kalyuzhniy, O.; Deresa, I.; et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 2016, 351, 1458–1463. [Google Scholar] [CrossRef] [PubMed]
- Havenar-Daughton, C.; Sarkar, A.; Kulp, D.W.; Toy, L.; Hu, X.; Deresa, I.; Kalyuzhniy, O.; Kaushik, K.; Upadhyay, A.A.; Menis, S.; et al. The human naive B cell repertoire contains distinct subclasses for a germline-targeting HIV-1 vaccine immunogen. Sci. Transl. Med. 2018, 10, eaat0381. [Google Scholar] [CrossRef]
- Yacoob, C.; Pancera, M.; Vigdorovich, V.; Oliver, B.G.; Glenn, J.A.; Feng, J.; Sather, D.N.; McGuire, A.T.; Stamatatos, L. Differences in Allelic Frequency and CDRH3 Region Limit the Engagement of HIV Env Immunogens by Putative VRC01 Neutralizing Antibody Precursors. Cell Rep. 2016, 17, 1560–1570. [Google Scholar] [CrossRef]
- Zhu, J.; O’Dell, S.; Ofek, G.; Pancera, M.; Wu, X.; Zhang, B.; Zhang, Z.; Program, N.C.S.; Mullikin, J.C.; Simek, M.; et al. Somatic Populations of PGT135-137 HIV-1-Neutralizing Antibodies Identified by 454 Pyrosequencing and Bioinformatics. Front. Microbiol. 2012, 3, 315. [Google Scholar] [CrossRef]
- Zhu, J.; Wu, X.; Zhang, B.; McKee, K.; O’Dell, S.; Soto, C.; Zhou, T.; Casazza, J.P.; Program, N.C.S.; Mullikin, J.C.; et al. De novo identification of VRC01 class HIV-1-neutralizing antibodies by next-generation sequencing of B-cell transcripts. Proc. Natl. Acad. Sci. USA 2013, 110, E4088–E4097. [Google Scholar] [CrossRef]
- Doria-Rose, N.A.; Schramm, C.A.; Gorman, J.; Moore, P.L.; Bhiman, J.N.; DeKosky, B.J.; Ernandes, M.J.; Georgiev, I.S.; Kim, H.J.; Pancera, M.; et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 2014, 509, 55–62. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Z.; Schramm, C.A.; Joyce, M.G.; Kwon, Y.D.; Zhou, T.; Sheng, Z.; Zhang, B.; O’Dell, S.; McKee, K.; et al. Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection. Cell 2015, 161, 470–485. [Google Scholar] [CrossRef] [PubMed]
- Bonsignori, M.; Zhou, T.; Sheng, Z.; Chen, L.; Gao, F.; Joyce, M.G.; Ozorowski, G.; Chuang, G.Y.; Schramm, C.A.; Wiehe, K.; et al. Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody. Cell 2016, 165, 449–463. [Google Scholar] [CrossRef] [PubMed]
- Soto, C.; Ofek, G.; Joyce, M.G.; Zhang, B.; McKee, K.; Longo, N.S.; Yang, Y.; Huang, J.; Parks, R.; Eudailey, J.; et al. Developmental Pathway of the MPER-Directed HIV-1-Neutralizing Antibody 10E8. PLoS ONE 2016, 11, e0157409. [Google Scholar] [CrossRef] [PubMed]
- Bhiman, J.N.; Anthony, C.; Doria-Rose, N.A.; Karimanzira, O.; Schramm, C.A.; Khoza, T.; Kitchin, D.; Botha, G.; Gorman, J.; Garrett, N.J.; et al. Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nat. Med. 2015, 21, 1332–1336. [Google Scholar] [CrossRef]
- Landais, E.; Murrell, B.; Briney, B.; Murrell, S.; Rantalainen, K.; Berndsen, Z.T.; Ramos, A.; Wickramasinghe, L.; Smith, M.L.; Eren, K.; et al. HIV Envelope Glycoform Heterogeneity and Localized Diversity Govern the Initiation and Maturation of a V2 Apex Broadly Neutralizing Antibody Lineage. Immunity 2017, 47, 990–1003. [Google Scholar] [CrossRef]
- Bonsignori, M.; Kreider, E.F.; Fera, D.; Meyerhoff, R.R.; Bradley, T.; Wiehe, K.; Alam, S.M.; Aussedat, B.; Walkowicz, W.E.; Hwang, K.K.; et al. Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Sci. Transl. Med. 2017, 9, eaai7514. [Google Scholar] [CrossRef]
- Rudicell, R.S.; Kwon, Y.D.; Ko, S.Y.; Pegu, A.; Louder, M.K.; Georgiev, I.S.; Wu, X.; Zhu, J.; Boyington, J.C.; Chen, X.; et al. Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J. Virol. 2014, 88, 12669–12682. [Google Scholar] [CrossRef]
- Liao, H.X.; Lynch, R.; Zhou, T.; Gao, F.; Alam, S.M.; Boyd, S.D.; Fire, A.Z.; Roskin, K.M.; Schramm, C.A.; Zhang, Z.; et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 2013, 496, 469–476. [Google Scholar] [CrossRef]
- Gao, F.; Bonsignori, M.; Liao, H.X.; Kumar, A.; Xia, S.M.; Lu, X.; Cai, F.; Hwang, K.K.; Song, H.; Zhou, T.; et al. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell 2014, 158, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Prabakaran, P.; Zhu, Z.; Feng, Y.; Streaker, E.D.; Dimitrov, D.S. Characterization of human IgG repertoires in an acute HIV-1 infection. Exp. Mol. Pathol. 2012, 93, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Prabakaran, P.; Chen, W.; Kessing, B.; Dimitrov, D.S. Deep sequencing and Circos analyses of antibody libraries reveal antigen-driven selection of Ig VH genes during HIV-1 infection. Exp. Mol. Pathol. 2013, 95, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yuan, T.; Li, J.; Zhang, Y.; Xu, J.; Shao, Y.; Chen, Z.; Zhang, M.Y. The potential of the human immune system to develop broadly neutralizing HIV-1 antibodies: Implications for vaccine development. AIDS 2013, 27, 2529–2539. [Google Scholar] [CrossRef]
- Yin, L.; Hou, W.; Liu, L.; Cai, Y.; Wallet, M.A.; Gardner, B.P.; Chang, K.; Lowe, A.C.; Rodriguez, C.A.; Sriaroon, P.; et al. IgM Repertoire Biodiversity is Reduced in HIV-1 Infection and Systemic Lupus Erythematosus. Front. Immunol. 2013, 4, 373. [Google Scholar] [CrossRef]
- Hoehn, K.B.; Gall, A.; Bashford-Rogers, R.; Fidler, S.J.; Kaye, S.; Weber, J.N.; McClure, M.O.; Investigators, S.T.; Kellam, P.; Pybus, O.G. Dynamics of immunoglobulin sequence diversity in HIV-1 infected individuals. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140241. [Google Scholar] [CrossRef][Green Version]
- Waltari, E.; Jia, M.; Jiang, C.S.; Lu, H.; Huang, J.; Fernandez, C.; Finzi, A.; Kaufmann, D.E.; Markowitz, M.; Tsuji, M.; et al. 5′ rapid Amplification of cDNA Ends and Illumina MiSeq Reveals B Cell Receptor Features in Healthy Adults, Adults With Chronic HIV-1 Infection, Cord Blood, and Humanized Mice. Front. Immunol. 2018, 9, 628. [Google Scholar] [CrossRef]
- Setliff, I.; McDonnell, W.J.; Raju, N.; Bombardi, R.G.; Murji, A.A.; Scheepers, C.; Ziki, R.; Mynhardt, C.; Shepherd, B.E.; Mamchak, A.A.; et al. Multi-Donor Longitudinal Antibody Repertoire Sequencing Reveals the Existence of Public Antibody Clonotypes in HIV-1 Infection. Cell Host Microbe 2018, 23, 845–854. [Google Scholar] [CrossRef]
| VH | VL | Pathogen | Donor | Reference |
|---|---|---|---|---|
| IGHV1-2 | - | DENV1 | Acute Dengue Infection | [107] |
| - | HIV-1 | HIV-1-Infected | [30,72,101,102,104] | |
| IGHV1-3 | - | HBV2 | HBV-Associated Acute Liver Failure Patients | [108] |
| IGHV1-18 | - | Influenza A | Participants of Influenza Vaccine Trial | [32] |
| - | DENV | Acute Dengue Infection | [107] | |
| IGHV1-46 | - | RV3 | RV-Infected Adults/Infants | [109,110,111] |
| - | HIV-1 | HIV-1-Infected | [72,112] | |
| IGHV1-69 | - | Influenza A | Hu. Non-Immune Antibody Phage-Display Library | [113] |
| Seasonal Influenza Vaccinees | [114] | |||
| Human Donor with Influenza A Broadly Neutralizing Serum | [115] | |||
| - | HIV-1 | HIV-1-Infected | [72,101,116] | |
| IGHV3-7 | - | HBV | HBV Vaccinees | [117] |
| - | Influenza A | Vaccinated Healthy Individuals | [118] | |
| IGHV3-15 | IGLV1-40 | EBOV4 | rVSV-ZEBOV5 Vaccinees | [34] |
| ChAD3 EBOV6 Vaccinees | [119] | |||
| Survivor of 2014 EBOV Outbreak in Zaire | [120] | |||
| IGHV3-23 | IGKV2D-29 | Hib7 | Hib-PS8 Conjugate-Vaccinated Infants | [121] |
| IGKV1-5 | ZIKV9 | Brazilian/Mexican DENV- and ZIKV-Infected Individuals | [33] | |
| IGHV3-30 | IGKV3-11 | HCMV10 | HCMV-Infected Individuals | [122] |
| Streptococcus pneumonieae | N/A | [123] | ||
| - | ZIKV | ZIKV-Infected Donors | [124,125] | |
| Phage-Display Naive Antibody Library | [126] | |||
| IGHV4-30-4 | RV | RV-Infected Adults/Infants | [109] | |
| IGHV4-1 | RV | RV-Infected Adults/Infants | [109] | |
| IGHV4-39 | RV | RV-Infected Adults/Infants | [109] | |
| IGHV4-61 | RV | RV-Infected Adults/Infants | [109] | |
| IGHV5-51 | - | HIV-1 | HIV-1-Infected | [127] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kreer, C.; Gruell, H.; Mora, T.; Walczak, A.M.; Klein, F. Exploiting B Cell Receptor Analyses to Inform on HIV-1 Vaccination Strategies. Vaccines 2020, 8, 13. https://doi.org/10.3390/vaccines8010013
Kreer C, Gruell H, Mora T, Walczak AM, Klein F. Exploiting B Cell Receptor Analyses to Inform on HIV-1 Vaccination Strategies. Vaccines. 2020; 8(1):13. https://doi.org/10.3390/vaccines8010013
Chicago/Turabian StyleKreer, Christoph, Henning Gruell, Thierry Mora, Aleksandra M. Walczak, and Florian Klein. 2020. "Exploiting B Cell Receptor Analyses to Inform on HIV-1 Vaccination Strategies" Vaccines 8, no. 1: 13. https://doi.org/10.3390/vaccines8010013
APA StyleKreer, C., Gruell, H., Mora, T., Walczak, A. M., & Klein, F. (2020). Exploiting B Cell Receptor Analyses to Inform on HIV-1 Vaccination Strategies. Vaccines, 8(1), 13. https://doi.org/10.3390/vaccines8010013





