Engineered Recombinant Single Chain Variable Fragment of Monoclonal Antibody Provides Protection to Chickens Infected with H9N2 Avian Influenza
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Viruses and Cells
2.3. Sequencing of Hybridoma Clones
2.4. Production and Purification of Recombinant Antibodies
2.5. ELISA
2.6. Virus Microneutralisation (MNT) Assay
2.7. Haemagglutination Inhibition (HI) Assay
2.8. Animals and AIV Challenge
2.9. Sample Collection and Tissue Homogenisation
2.10. Plaque Assay
2.11. qRT-PCR of Viral M Gene and Cytokine mRNAs
2.12. Virus Sequencing
2.13. Statistical Analysis
3. Results
3.1. scFvs can Bind to and Neutralize H9N2 Virus In Vitro
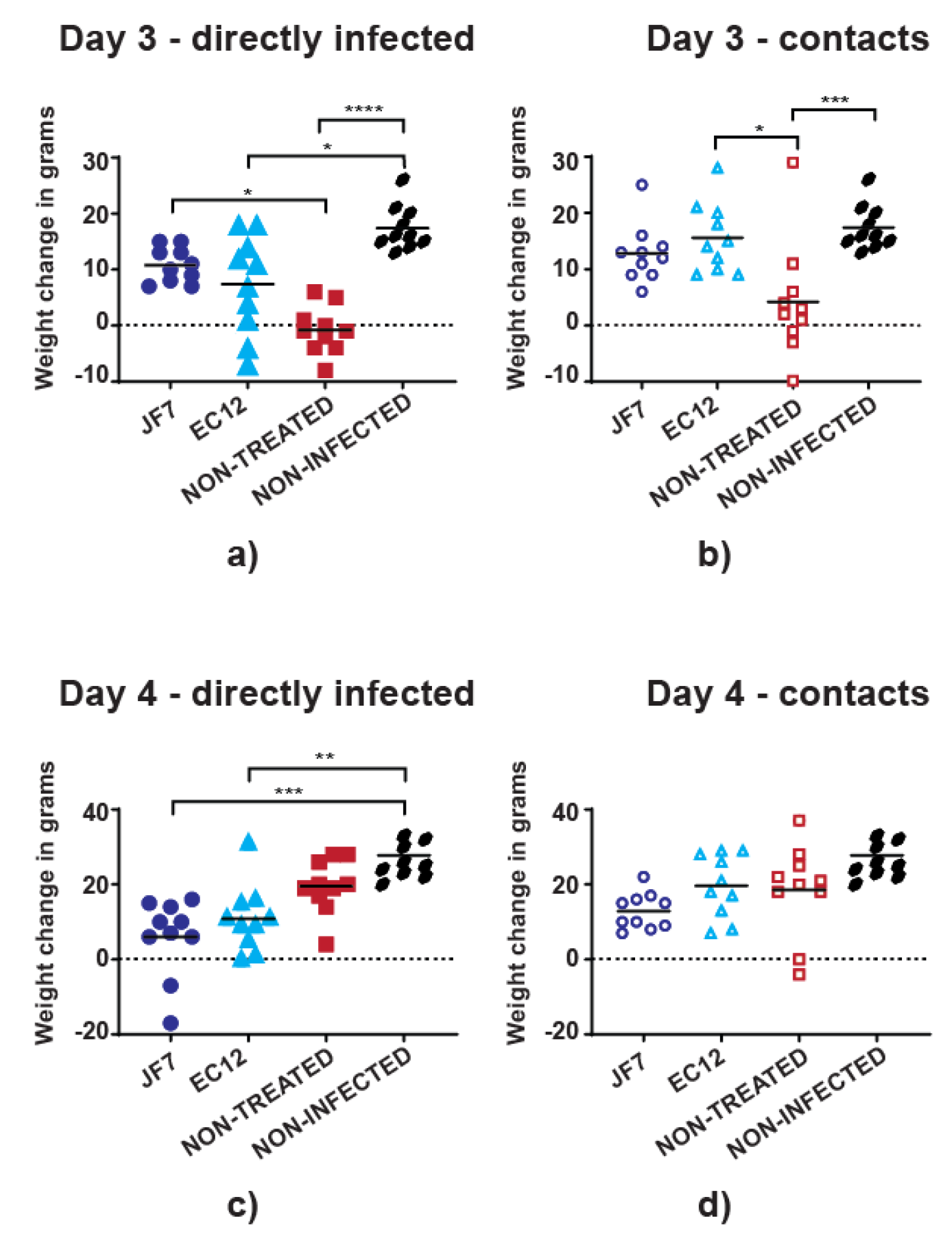
3.2. Antibody Therapy Results in Lower Morbidity Levels in Chickens
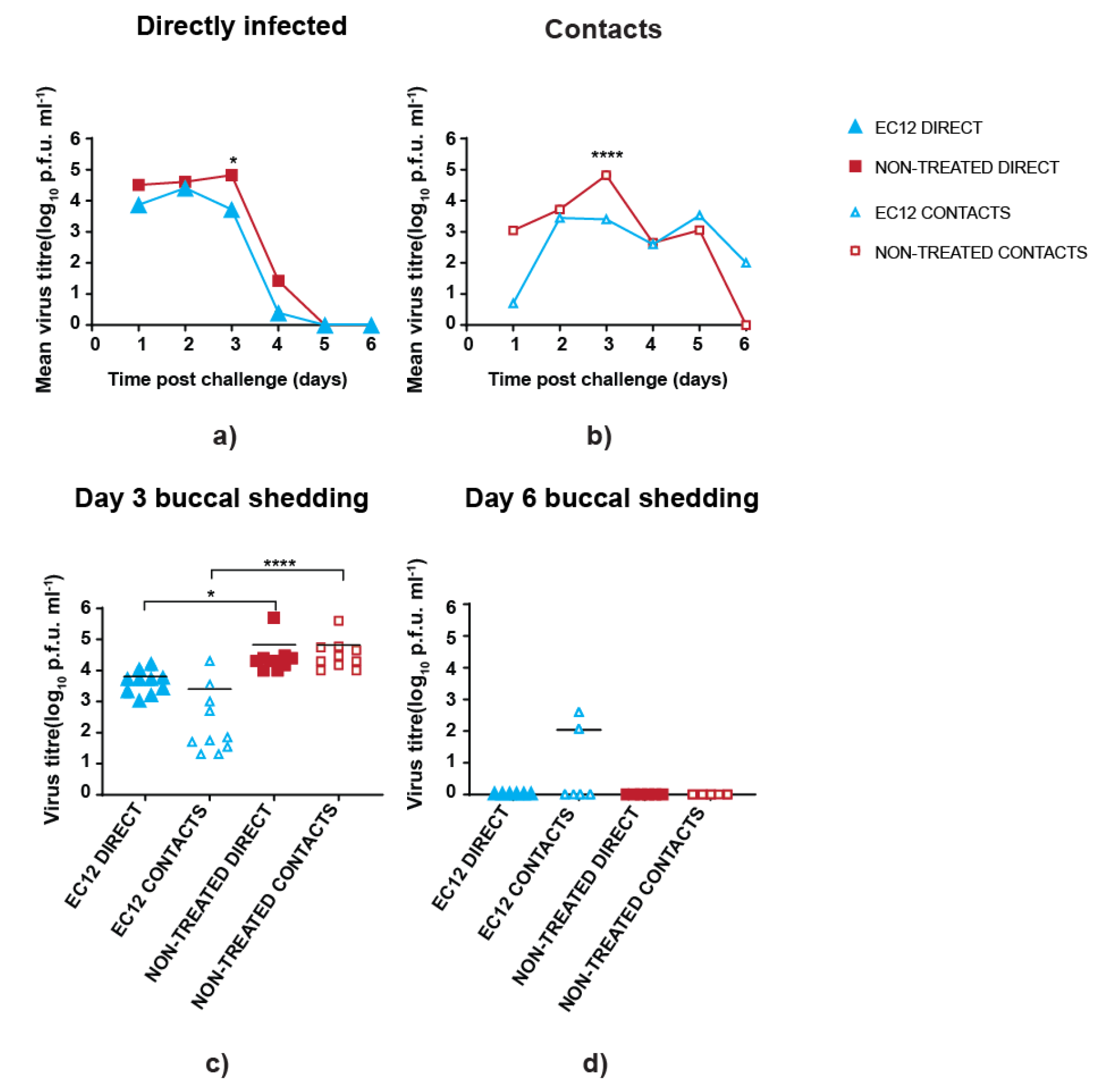
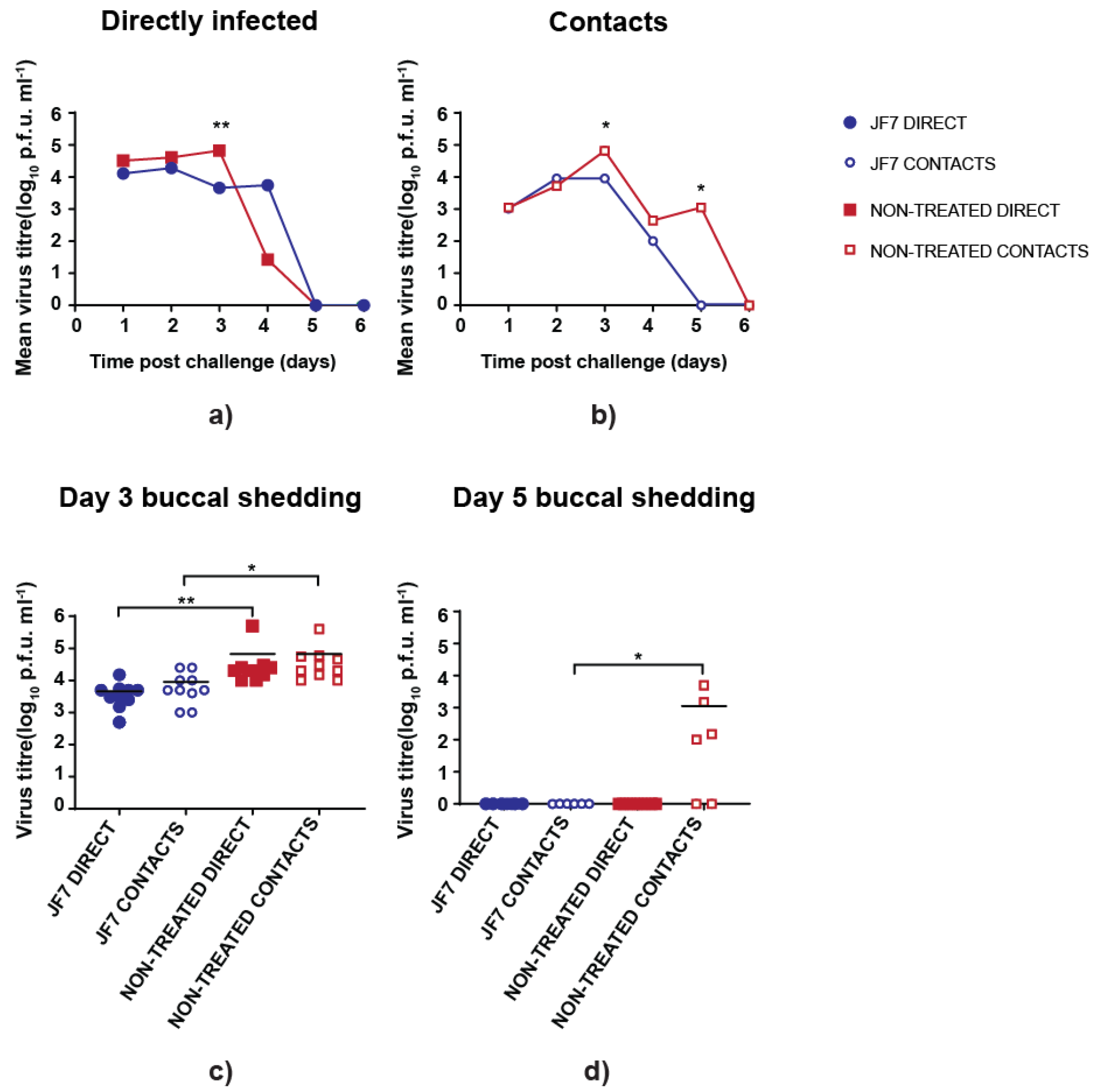
3.3. scFv Treatment Reduces Peak Virus Shedding
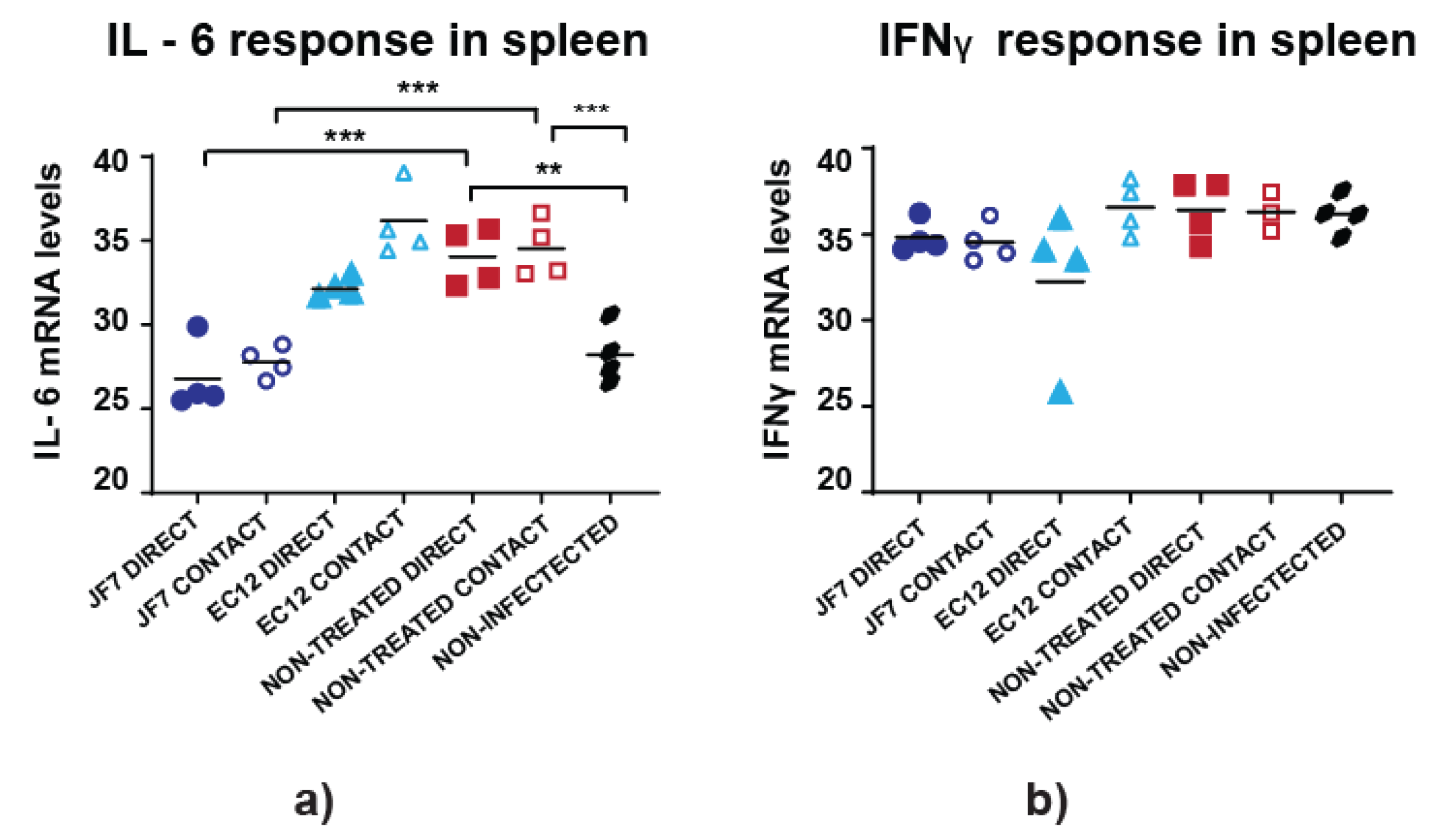
3.4. Non-Treated Birds Have Higher Inflammatory Responses in the Spleen
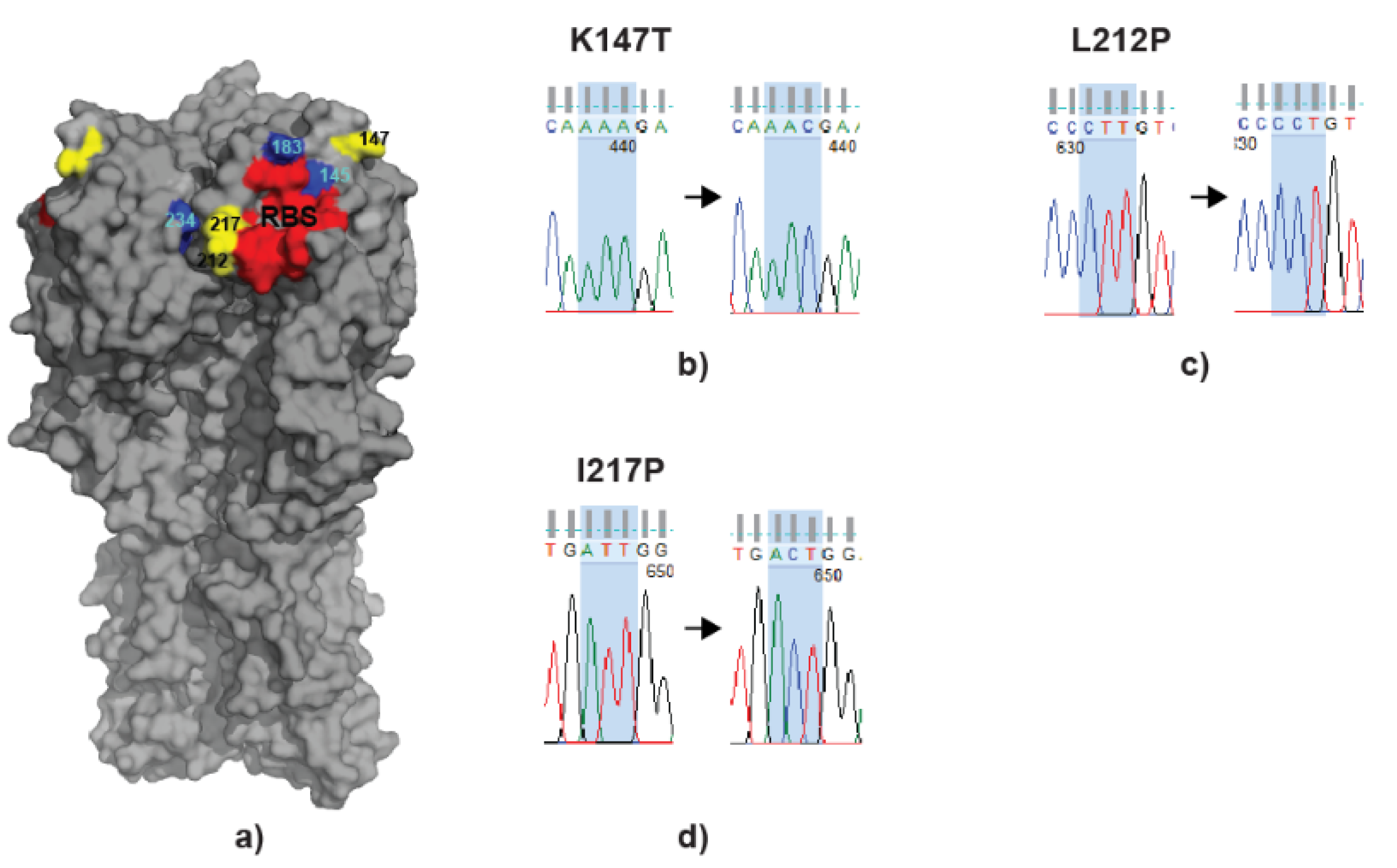
3.5. scFv Treatment Leads to Virus Escapes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Dunand, C.J.H.; Leon, P.E.; Huang, M.; Choi, A.; Chromikova, V.; Ho, I.Y.; Tan, G.S.; Cruz, J.; Hirsh, A.; Zheng, N.Y.; et al. Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host Microbe 2016, 19, 800–813. [Google Scholar] [CrossRef]
- Koudstaal, W.; Koldijk, M.H.; Brakenhoff, J.P.J.; Cornelissen, L.A.H.M.; Weverling, G.J.; Friesen, R.H.E.; Goudsmit, J. Pre- and Postexposure Use of Human Monoclonal Antibody against H5N1 and H1N1 Influenza Virus in Mice: Viable Alternative to Oseltamivir. J. Infect. Dis. 2009, 200, 1870–1873. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Itoh, Y.; Yoshida, R.; Shichinohe, S.; Higuchi, M.; Ishigaki, H.; Nakayama, M.; Pham, V.L.; Ishida, H.; Kitano, M.; Arikata, M.; et al. Protective efficacy of passive immunization with monoclonal antibodies in animal models of H5N1 highly pathogenic avian influenza virus infection. PLoS Pathog. 2014, 10, e1004192. [Google Scholar] [CrossRef] [PubMed]
- Sparrow, E.; Friede, M.; Sheikh, M.; Torvaldsen, S.; Newall, A.T. Passive immunization for influenza through antibody therapies, a review of the pipeline, challenges and potential applications. Vaccine 2016, 34, 5442–5448. [Google Scholar] [CrossRef]
- Wilson, J.R.; Belser, J.A.; DaSilva, J.; Guo, Z.; Sun, X.; Gansebom, S.; Bai, Y.; Stark, T.J.; Chang, J.; Carney, P.; et al. An influenza A virus (H7N9) anti-neuraminidase monoclonal antibody protects mice from morbidity without interfering with the development of protective immunity to subsequent homologous challenge. Virology 2017, 511, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Murin, C.D.; Wilson, I.A.; Ward, A.B. Antibody responses to viral infections: A structural perspective across three different enveloped viruses. Nat. Microbiol. 2019, 4, 734–747. [Google Scholar] [CrossRef]
- Saphire, E.O.; Schendel, S.L.; Gunn, B.M.; Milligan, J.C.; Alter, G. Antibody-mediated protection against Ebola virus. Nat. Immunol. 2018, 19, 1169–1178. [Google Scholar] [CrossRef]
- Jefferis, R.; Lowe, J.; Ling, N.R.; Porter, P.; Senior, S. Immunogenic and antigenic epitopes of immunoglobulins I. Cross-reactivity of murine monoclonal antibodies to human IgG with the immunoglobulins of certain animal species. Immunology 1982, 45, 71–77. [Google Scholar]
- Hwang, W.Y.; Foote, J. Immunogenicity of engineered antibodies. Methods 2005, 36, 3–10. [Google Scholar] [CrossRef]
- Taylor, A.I.; Gould, H.J.; Sutton, B.J.; Calvert, R.A. The first avian Ig-like Fc receptor family member combines features of mammalian FcR and FCRL. Immunogenetics 2007, 59, 323–328. [Google Scholar] [CrossRef]
- Swayne, D.E.; Suarez, D.L. Highly pathogenic avian influenza. Rev. Sci. Tech. 2000, 19, 463–482. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.H.P.; James, J.; Sealy, J.E.; Iqbal, M. A Global Perspective on H9N2 Avian Influenza Virus. Viruses 2019, 11, 620. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.P.; Su, C.H.; Wang, D.Y.; Peng, Y.S.; Liu, M.; Hua, S.; Li, T.X.; Gao, G.F.; Tang, H.; Chen, J.Z.; et al. Sequential Reassortments Underlie Diverse Influenza H7N9 Genotypes in China. Cell Host Microbe 2013, 14, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Cheng, V.C.; Chan, J.F.; Wen, X.; Wu, W.L.; Que, T.L.; Chen, H.; Chan, K.H.; Yuen, K.Y. Infection of immunocompromised patients by avian H9N2 influenza A virus. J. Infect. 2011, 62, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, J. H9N2 influenza virus in China: A cause of concern. Protein Cell 2015, 6, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Swayne, D.E.; Sharma, P.; Rehmani, S.F.; Wajid, A.; Suarez, D.L.; Afonso, C. H9N2 low pathogenic avian influenza in Pakistan (2012–2015). Vet. Rec. Open 2016, 3, e000171. [Google Scholar] [CrossRef]
- Toroghi, R.; Momayez, R. Biological and molecular characterization of Avian influenza virus (H9N2) isolates from Iran. Acta Virol. 2006, 50, 163–168. [Google Scholar]
- Shehata, A.A.; Parvin, R.; Sultan, H.; Halami, M.Y.; Talaat, S.; Abd Elrazek, A.; Ibrahim, M.; Heenemann, K.; Vahlenkamp, T. Isolation and full genome characterization of avian influenza subtype H9N2 from poultry respiratory disease outbreak in Egypt. Virus Genes 2015, 50, 389–400. [Google Scholar] [CrossRef]
- Astill, J.; Alkie, T.; Yitbarek, A.; Taha-Abdelaziz, K.; Bavananthasivam, J.; Nagy, E.; Petrik, J.J.; Sharif, S. Examination of the effects of virus inactivation methods on the induction of antibody- and cell-mediated immune responses against whole inactivated H9N2 avian influenza virus vaccines in chickens. Vaccine 2018, 36, 3908–3916. [Google Scholar] [CrossRef]
- Sun, Y.; Pu, J.; Fan, L.; Sun, H.; Wang, J.; Zhang, Y.; Liu, L.; Liu, J. Evaluation of the protective efficacy of a commercial vaccine against different antigenic groups of H9N2 influenza viruses in chickens. Vet. Microbiol. 2012, 156, 193–199. [Google Scholar] [CrossRef]
- Park, K.J.; Kwon, H.I.; Song, M.S.; Pascua, P.N.; Baek, Y.H.; Lee, J.H.; Jang, H.L.; Lim, J.Y.; Mo, I.P.; Moon, H.J.; et al. Rapid evolution of low-pathogenic H9N2 avian influenza viruses following poultry vaccination programmes. J. Gen. Virol. 2011, 92, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Sealy, J.E.; Fournie, G.; Trang, P.H.; Dang, N.H.; Sadeyen, J.R.; Thanh, T.L.; van Doorn, H.R.; Bryant, J.E.; Iqbal, M. Poultry trading behaviours in Vietnamese live bird markets as risk factors for avian influenza infection in chickens. Transbound. Emerg. Dis. 2019. [Google Scholar] [CrossRef] [PubMed]
- Sealy, J.E.; Yaqub, T.; Peacock, T.P.; Chang, P.; Ermetal, B.; Clements, A.; Sadeyen, J.R.; Mehboob, A.; Shelton, H.; Bryant, J.E.; et al. Association of Increased Receptor-Binding Avidity of Influenza A(H9N2) Viruses with Escape from Antibody-Based Immunity and Enhanced Zoonotic Potential. Emerg. Infect. Dis. 2018, 25, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.; Reddy, K.; James, J.; Adamiak, B.; Barclay, W.; Shelton, H.; Iqbal, M. Antigenic mapping of an H9N2 avian influenza virus reveals two discrete antigenic sites and a novel mechanism of immune escape. Sci. Rep. 2016, 6, 18745. [Google Scholar] [CrossRef]
- Doenecke, A.; Winnacker, E.L.; Hallek, M. Rapid amplification of cDNA ends (RACE) improves the PCR-based isolation of immunoglobulin variable region genes from murine and human lymphoma cells and cell lines. Leukemia 1997, 11, 1787–1792. [Google Scholar] [CrossRef]
- Leboeuf, R.D.; Galin, F.S.; Hollinger, S.K.; Peiper, S.C.; Blalock, J.E. Cloning and Sequencing of Immunoglobulin Variable-Region Genes Using Degenerate Oligodeoxy-Ribonucleotides and Polymerase Chain-Reaction. Gene 1989, 82, 371–377. [Google Scholar] [CrossRef]
- Webster, R.C.N.; Stöhr , K. WHO Manual on Animal Influenza Diagnosis and Surveillance. WHO Global Influenza Programme 2002, WHO/CDS/CSR/NCS/2002.5 Rev. 1. Available online: https://www.who.int/csr/resources/publications/influenza/whocdscsrncs20025rev.pdf (accessed on 2 March 2020).
- Collecting, preserving and shipping specimens for the diagnosis of avian influenza A(H5N1) virus infection. Guide for Field Operations. Available online: https://www.who.int/csr/resources/publications/surveillance/WHO_CDS_EPR_ARO_2006_1/en/ (accessed on 2 March 2020).
- Spackman, E.; Senne, D.A.; Myers, T.J.; Bulaga, L.L.; Garber, L.P.; Perdue, M.L.; Lohman, K.; Daum, L.T.; Suarez, D.L. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 2002, 40, 3256–3260. [Google Scholar] [CrossRef]
- Kapczynski, D.R.; Jiang, H.J.; Kogut, M.H. Characterization of cytokine expression induced by avian influenza virus infection with real-time RT-PCR. Methods Mol. Biol. 2014, 1161, 217–233. [Google Scholar]
- Morgan, S.B.; Holzer, B.; Hemmink, J.D.; Salguero, F.J.; Schwartz, J.C.; Agatic, G.; Cameroni, E.; Guarino, B.; Porter, E.; Rijal, P.; et al. Therapeutic Administration of Broadly Neutralizing FI6 Antibody Reveals Lack of Interaction Between Human IgG1 and Pig Fc Receptors. Front. Immunol. 2018, 9, 865. [Google Scholar] [CrossRef]
- Hoffmann, E.; Stech, J.; Guan, Y.; Webster, R.G.; Perez, D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001, 146, 2275–2289. [Google Scholar] [CrossRef]
- Bangaru, S.; Zhang, H.; Gilchuk, I.M.; Voss, T.G.; Irving, R.P.; Gilchuk, P.; Matta, P.; Zhu, X.Y.; Lang, S.S.; Nieusma, T.; et al. A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA. Nat. Commun. 2018, 9, 2669. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.O.; Takas, T.; Nyborg, A.; Shoemaker, K.; Kallewaard, N.L.; Chiong, R.; Dubovsky, F.; Mallory, R.M. Evaluation of MEDI8852, an Anti-Influenza A Monoclonal Antibody, in Treating Acute Uncomplicated Influenza. Antimicrob. Agents Chemother. 2018, 62, e00694-18. [Google Scholar] [CrossRef] [PubMed]
- Kolpe, A.; Schepens, B.; Ye, L.; Staeheli, P.; Saelens, X. Passively transferred M2e-specific monoclonal antibody reduces influenza A virus transmission in mice. Antivir. Res. 2018, 158, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Kallewaard, N.L.; Corti, D.; Collins, P.J.; Neu, U.; McAuliffe, J.M.; Benjamin, E.; Wachter-Rosati, L.; Palmer-Hill, F.J.; Yuan, A.Q.; Walker, P.A.; et al. Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes. Cell 2016, 166, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al. A Neutralizing Antibody Selected from Plasma Cells That Binds to Group 1 and Group 2 Influenza A Hemagglutinins. Science 2011, 333, 850–856. [Google Scholar] [CrossRef]
- Throsby, M.; van den Brink, E.; Jongeneelen, M.; Poon, L.L.M.; Alard, P.; Cornelissen, L.; Bakker, A.; Cox, F.; van Deventer, E.; Guan, Y.; et al. Heterosubtypic Neutralizing Monoclonal Antibodies Cross-Protective against H5N1 and H1N1 Recovered from Human IgM(+) Memory B Cells. PLoS ONE 2008, 3, e3942. [Google Scholar] [CrossRef]
- Hu, H.; Voss, J.; Zhang, G.; Buchy, P.; Zuo, T.; Wang, L.; Wang, F.; Zhou, F.; Wang, G.; Tsai, C.; et al. A human antibody recognizing a conserved epitope of H5 hemagglutinin broadly neutralizes highly pathogenic avian influenza H5N1 viruses. J. Virol. 2012, 86, 2978–2989. [Google Scholar] [CrossRef]
- Nogales, A.; Piepenbrink, M.S.; Wang, J.; Ortega, S.; Basu, M.; Fucile, C.F.; Treanor, J.J.; Rosenberg, A.F.; Zand, M.S.; Keefer, M.C.; et al. A Highly Potent and Broadly Neutralizing H1 Influenza-Specific Human Monoclonal Antibody. Sci. Rep. 2018, 8, 4374. [Google Scholar] [CrossRef]
- DiLillo, D.J.; Palese, P.; Wilson, P.C.; Ravetch, J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Investig. 2016, 126, 605–610. [Google Scholar] [CrossRef]
- Laursen, N.S.; Friesen, R.H.E.; Zhu, X.; Jongeneelen, M.; Blokland, S.; Vermond, J.; van Eijgen, A.; Tang, C.; van Diepen, H.; Obmolova, G.; et al. Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science 2018, 362, 598–602. [Google Scholar] [CrossRef]
- Bangaru, S.; Lang, S.; Schotsaert, M.; Vanderven, H.A.; Zhu, X.; Kose, N.; Bombardi, R.; Finn, J.A.; Kent, S.J.; Gilchuk, P.; et al. A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface. Cell 2019, 177, 1136–1152. [Google Scholar] [CrossRef] [PubMed]
- Nachbagauer, R.; Shore, D.; Yang, H.; Johnson, S.K.; Gabbard, J.D.; Tompkins, S.M.; Wrammert, J.; Wilson, P.C.; Stevens, J.; Ahmed, R.; et al. Broadly Reactive Human Monoclonal Antibodies Elicited following Pandemic H1N1 Influenza Virus Exposure Protect Mice against Highly Pathogenic H5N1 Challenge. J. Virol. 2018, 92, e00949-18. [Google Scholar] [CrossRef] [PubMed]
- Kapczynski, D.R.; Tumpey, T.M.; Hidajat, R.; Zsak, A.; Chrzastek, K.; Tretyakova, I.; Pushko, P. Vaccination with virus-like particles containing H5 antigens from three H5N1 clades protects chickens from H5N1 and H5N8 influenza viruses. Vaccine 2016, 34, 1575–1581. [Google Scholar] [CrossRef] [PubMed]
- Lone, N.A.; Spackman, E.; Kapczynski, D. Immunologic evaluation of 10 different adjuvants for use in vaccines for chickens against highly pathogenic avian influenza virus. Vaccine 2017, 35, 3401–3408. [Google Scholar] [CrossRef] [PubMed]
- Kapczynski, D.R.; Pantin-Jackwood, M.J.; Spackman, E.; Chrzastek, K.; Suarez, D.L.; Swayne, D.E. Homologous and heterologous antigenic matched vaccines containing different H5 hemagglutinins provide variable protection of chickens from the 2014 U.S. H5N8 and H5N2 clade 2.3.4.4 highly pathogenic avian influenza viruses. Vaccine 2017, 35, 6345–6353. [Google Scholar] [CrossRef] [PubMed]
- Leyva-Grado, V.H.; Tan, G.S.; Leon, P.E.; Yondola, M.; Palese, P. Direct administration in the respiratory tract improves efficacy of broadly neutralizing anti-influenza virus monoclonal antibodies. Antimicrob. Agents Chemother. 2015, 59, 4162–4172. [Google Scholar] [CrossRef]
- Stevens, N.E.; Hatjopolous, A.; Fraser, C.K.; Alsharifi, M.; Diener, K.R.; Hayball, J.D. Preserved antiviral adaptive immunity following polyclonal antibody immunotherapy for severe murine influenza infection. Sci. Rep. 2016, 6, 29154. [Google Scholar] [CrossRef]
- Iqbal, M.; Yaqub, T.; Mukhtar, N.; Shabbir, M.Z.; McCauley, J.W. Infectivity and transmissibility of H9N2 avian influenza virus in chickens and wild terrestrial birds. Vet. Res. 2013, 44, 100. [Google Scholar] [CrossRef]
- Post, J.; Burt, D.W.; Cornelissen, J.B.; Broks, V.; van Zoelen, D.; Peeters, B.; Rebel, J.M. Systemic virus distribution and host responses in brain and intestine of chickens infected with low pathogenic or high pathogenic avian influenza virus. Virol. J. 2012, 9, 61. [Google Scholar] [CrossRef]
- Zhu, R.; Xu, D.; Yang, X.; Zhang, J.; Wang, S.; Shi, H.; Liu, X. Genetic and biological characterization of H9N2 avian influenza viruses isolated in China from 2011 to 2014. PLoS ONE 2018, 13, e0199260. [Google Scholar] [CrossRef]
- Corbanie, E.A.; Matthijs, M.G.; van Eck, J.H.; Remon, J.P.; Landman, W.J.; Vervaet, C. Deposition of differently sized airborne microspheres in the respiratory tract of chickens. Avian Pathol. 2006, 35, 475–485. [Google Scholar] [CrossRef]
- van den Berg, F.T.; Makoah, N.A.; Ali, S.A.; Scott, T.A.; Mapengo, R.E.; Mutsvunguma, L.Z.; Mkhize, N.N.; Lambson, B.E.; Kgagudi, P.D.; Crowther, C.; et al. AAV-Mediated Expression of Broadly Neutralizing and Vaccine-like Antibodies Targeting the HIV-1 Envelope V2 Region. Mol. Ther. Methods Clin. Dev. 2019, 14, 100–112. [Google Scholar] [CrossRef]
- van Lieshout, L.P.; Soule, G.; Sorensen, D.; Frost, K.L.; He, S.; Tierney, K.; Safronetz, D.; Booth, S.A.; Kobinger, G.P.; Qiu, X.; et al. Intramuscular Adeno-Associated Virus-Mediated Expression of Monoclonal Antibodies Provides 100% Protection Against Ebola Virus Infection in Mice. J. Infect. Dis. 2018, 217, 916–925. [Google Scholar] [CrossRef] [PubMed]
- Paules, C.I.; Lakdawala, S.; McAuliffe, J.M.; Paskel, M.; Vogel, L.; Kallewaard, N.L.; Zhu, Q.; Subbarao, K. The Hemagglutinin A Stem Antibody MEDI8852 Prevents and Controls Disease and Limits Transmission of Pandemic Influenza Viruses. J. Infect. Dis. 2017, 216, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.Y.; Fang, D.; Lv, P.; Bian, X.B.; Ruan, X.Z.; Yan, Y.; Zhou, J.Y. Differential cellular immune responses between chickens and ducks to H9N2 avian influenza virus infection. Vet. Immunol. Immunopathol. 2012, 150, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Cardona, C.J.; Li, J.; Dao, N.; Tran, T.; Andrada, J. Modulation of the immune responses in chickens by low-pathogenicity avian influenza virus H9N2. J. Gen. Virol. 2008, 89, 1288–1299. [Google Scholar] [CrossRef] [PubMed]
- Cyranoski, D. China’s chicken farmers under fire for antiviral abuse. Nature 2005, 435, 1009. [Google Scholar] [CrossRef]
- Dong, G.; Peng, C.; Luo, J.; Wang, C.; Han, L.; Wu, B.; Ji, G.; He, H. Adamantane-resistant influenza a viruses in the world (1902–2013): Frequency and distribution of M2 gene mutations. PLoS ONE 2015, 10, e0119115. [Google Scholar] [CrossRef]
- Takashita, E.; Meijer, A.; Lackenby, A.; Gubareva, L.; Rebelo-de-Andrade, H.; Besselaar, T.; Fry, A.; Gregory, V.; Leang, S.K.; Huang, W.; et al. Global update on the susceptibility of human influenza viruses to neuraminidase inhibitors, 2013-2014. Antiviral Res. 2015, 117, 27–38. [Google Scholar] [CrossRef]
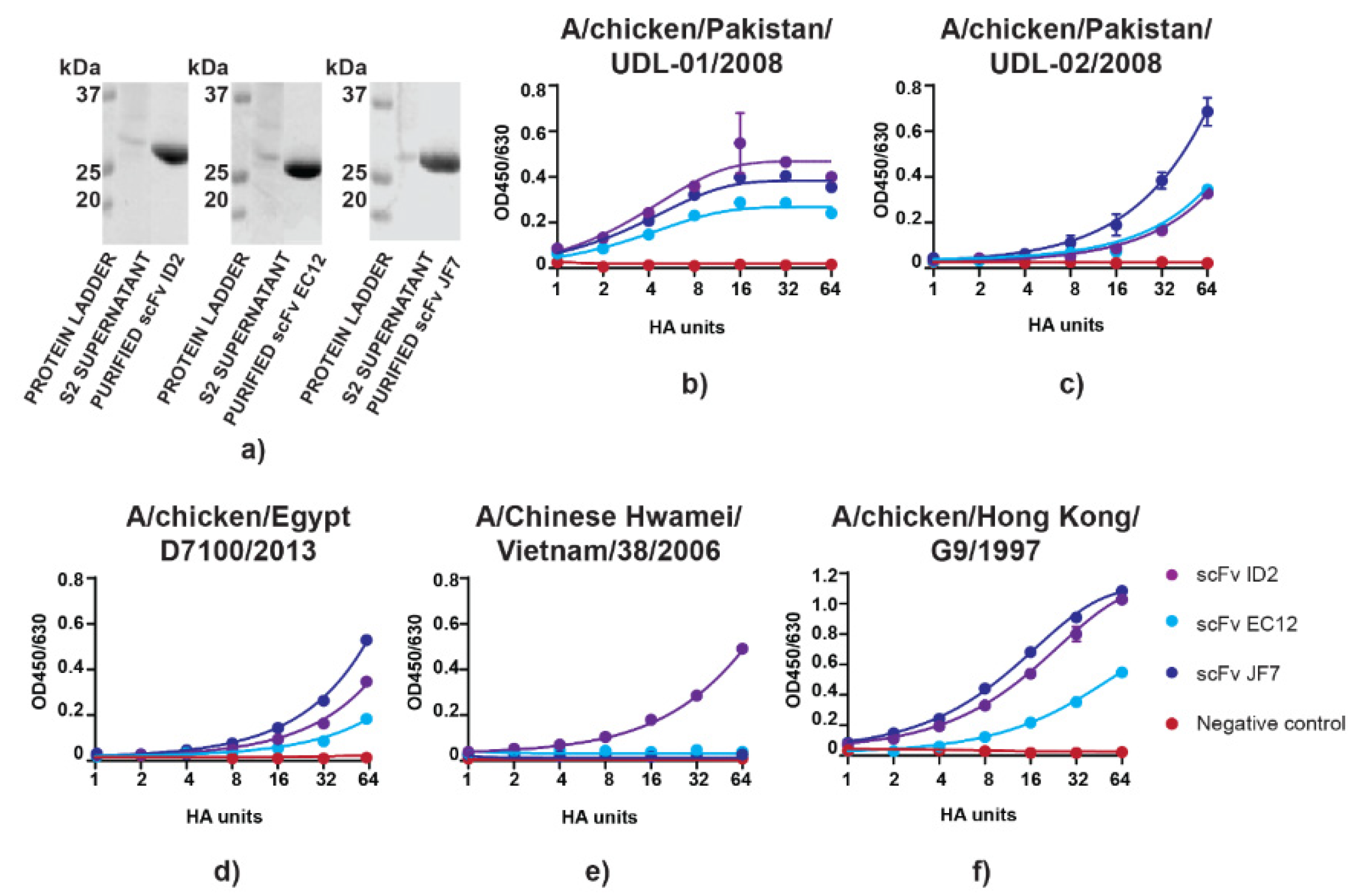
| scFv | MNT Titre/mg | HI Titre/mg |
|---|---|---|
| ID2 | 59 (±61) | 30,720 |
| EC12 | 384 (±222) | 2560 |
| JF7 | 4096 | 160 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lukosaityte, D.; Sadeyen, J.-R.; Shrestha, A.; Sealy, J.E.; Bhat, S.; Chang, P.; Digard, P.; Iqbal, M. Engineered Recombinant Single Chain Variable Fragment of Monoclonal Antibody Provides Protection to Chickens Infected with H9N2 Avian Influenza. Vaccines 2020, 8, 118. https://doi.org/10.3390/vaccines8010118
Lukosaityte D, Sadeyen J-R, Shrestha A, Sealy JE, Bhat S, Chang P, Digard P, Iqbal M. Engineered Recombinant Single Chain Variable Fragment of Monoclonal Antibody Provides Protection to Chickens Infected with H9N2 Avian Influenza. Vaccines. 2020; 8(1):118. https://doi.org/10.3390/vaccines8010118
Chicago/Turabian StyleLukosaityte, Deimante, Jean-Remy Sadeyen, Angita Shrestha, Joshua E. Sealy, Sushant Bhat, Pengxiang Chang, Paul Digard, and Munir Iqbal. 2020. "Engineered Recombinant Single Chain Variable Fragment of Monoclonal Antibody Provides Protection to Chickens Infected with H9N2 Avian Influenza" Vaccines 8, no. 1: 118. https://doi.org/10.3390/vaccines8010118
APA StyleLukosaityte, D., Sadeyen, J.-R., Shrestha, A., Sealy, J. E., Bhat, S., Chang, P., Digard, P., & Iqbal, M. (2020). Engineered Recombinant Single Chain Variable Fragment of Monoclonal Antibody Provides Protection to Chickens Infected with H9N2 Avian Influenza. Vaccines, 8(1), 118. https://doi.org/10.3390/vaccines8010118







