Palm Tocotrienol-Adjuvanted Dendritic Cells Decrease Expression of the SATB1 Gene in Murine Breast Cancer Cells and Tissues
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Generation of Bone Marrow-Derived Dendritic Cells
2.3. Cell Line
2.4. Freeze–Thaw 4T1 Tumor Lysate
2.5. Preparation of Tumor Lysate-Pulsed DC
2.6. Treatment of Mice
2.7. Analysis of Cell Surface Markers on in Blood Mice
2.8. RNA Extraction, Characterization and Integrity
2.9. Sample Preparation and Hybridization for Microarray Study
2.10. Real Time RT-PCR
2.11. Protein Expression
2.12. Statistical Analysis
3. Results
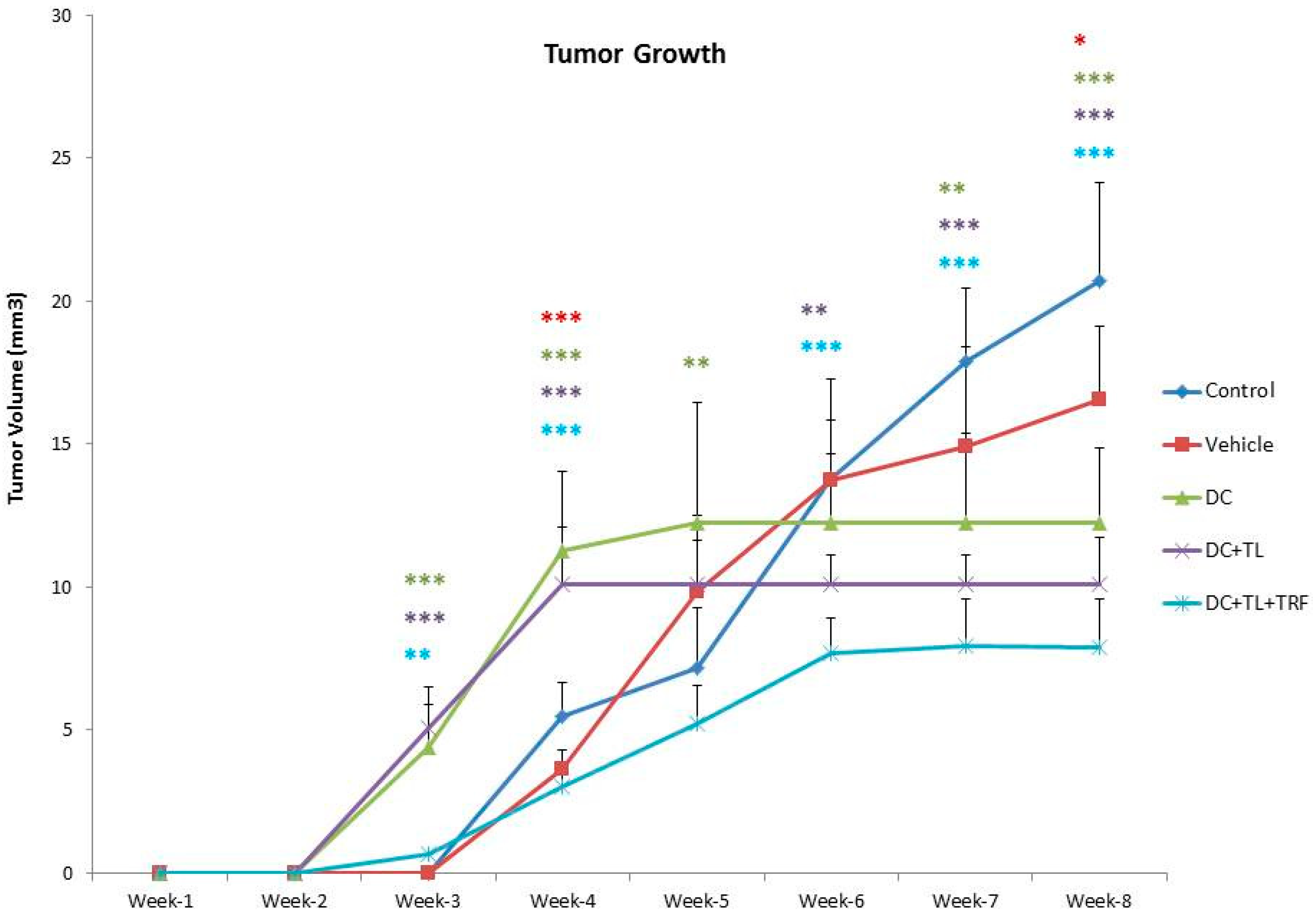
3.1. Anti-Tumor Effects of DC Pulsed with Tumor Lysate and Daily TRF Supplementation
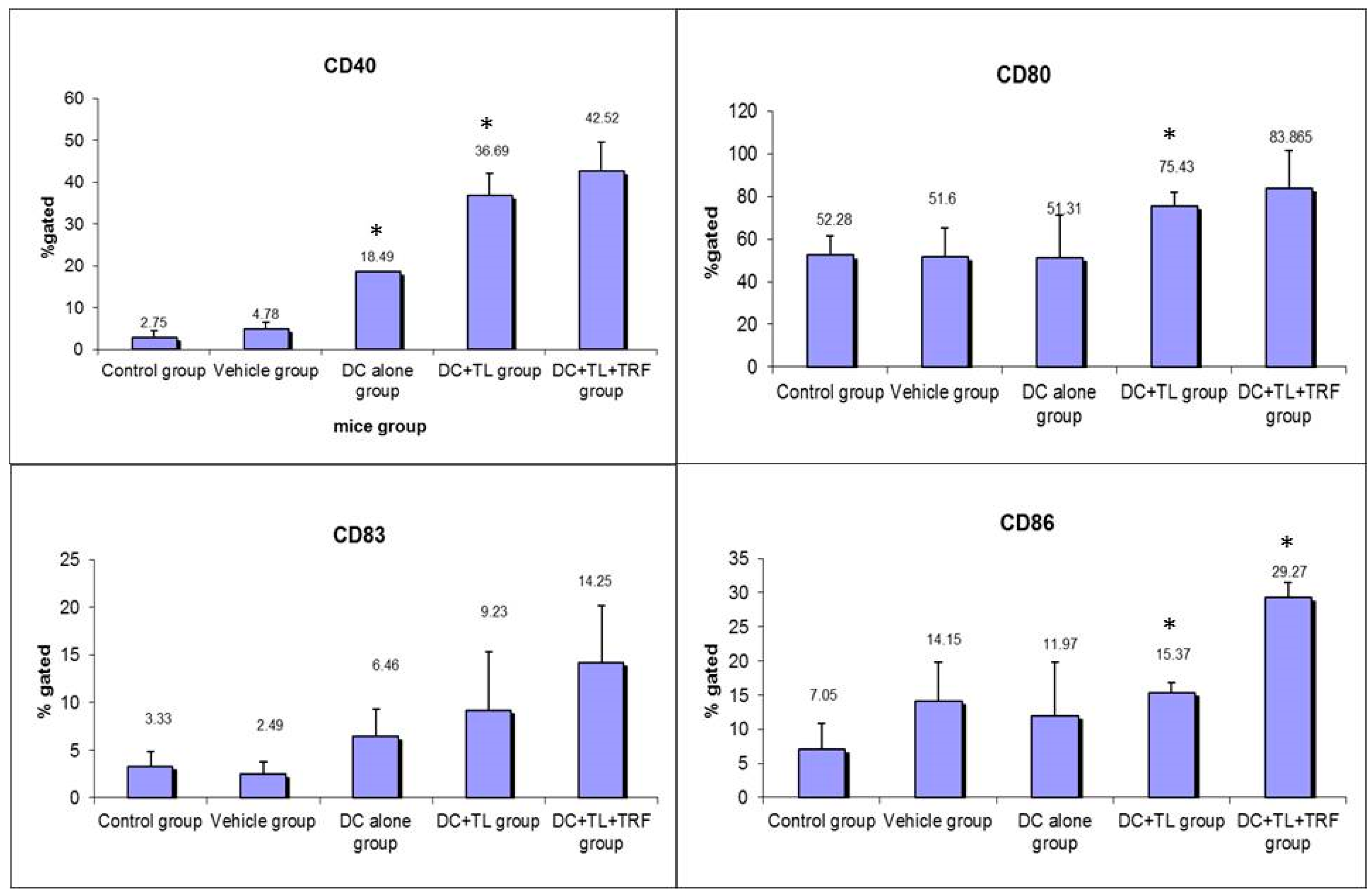
3.2. Expression of CD40, CD80, CD83, and CD86 in Peripheral Blood Leucocytes
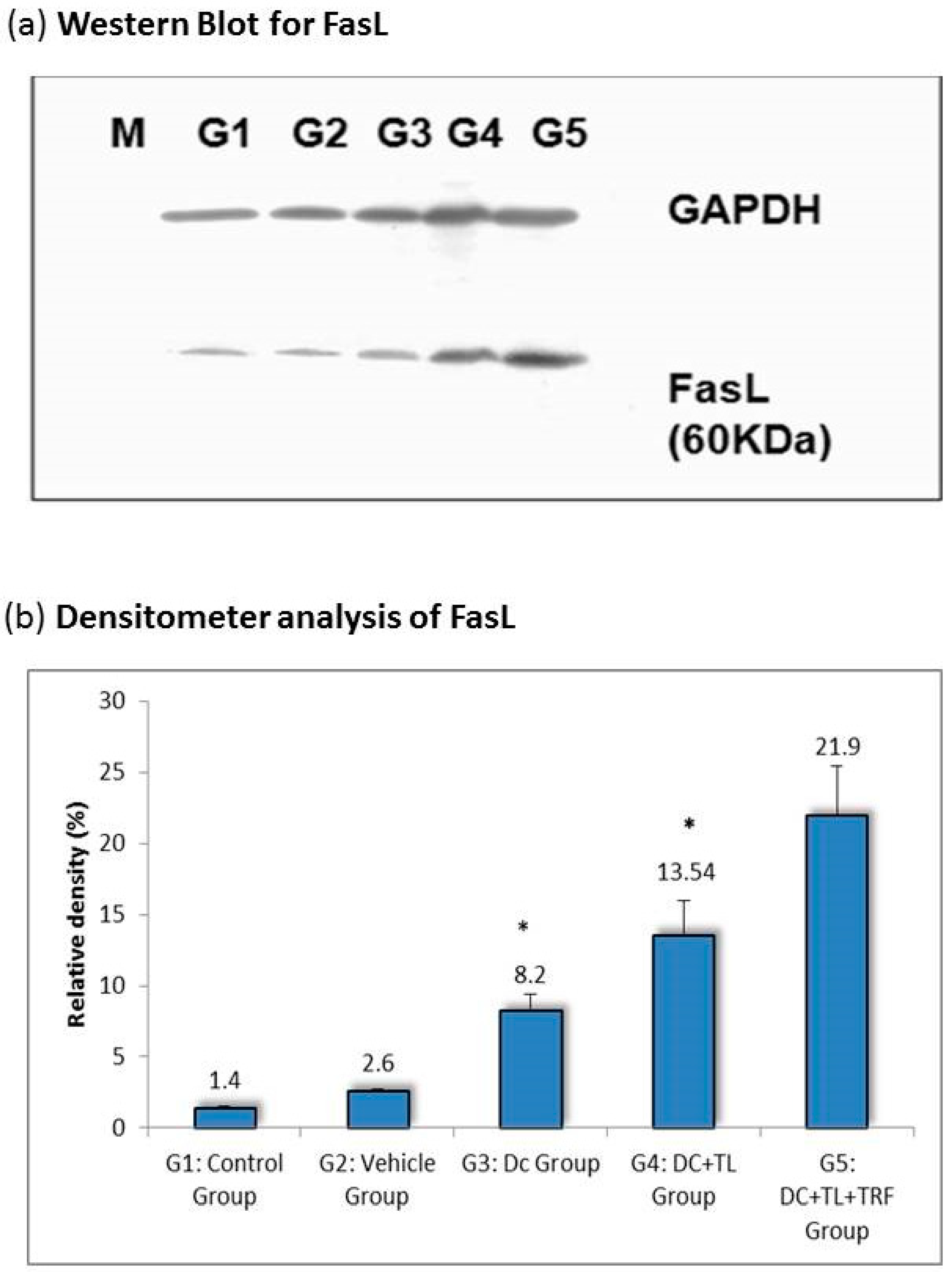
3.3. Expression of Fas Ligand in Tumor Tissues
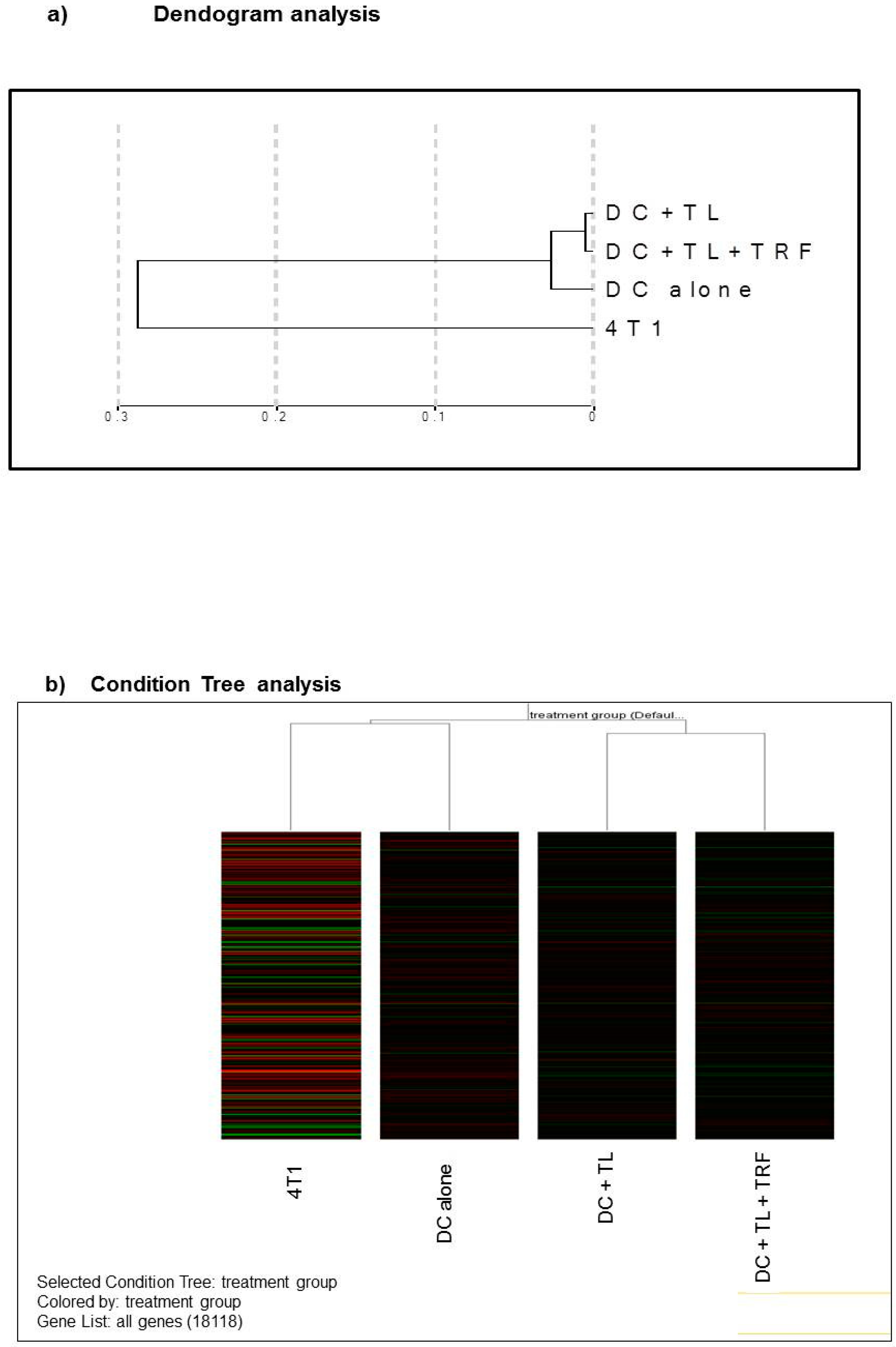
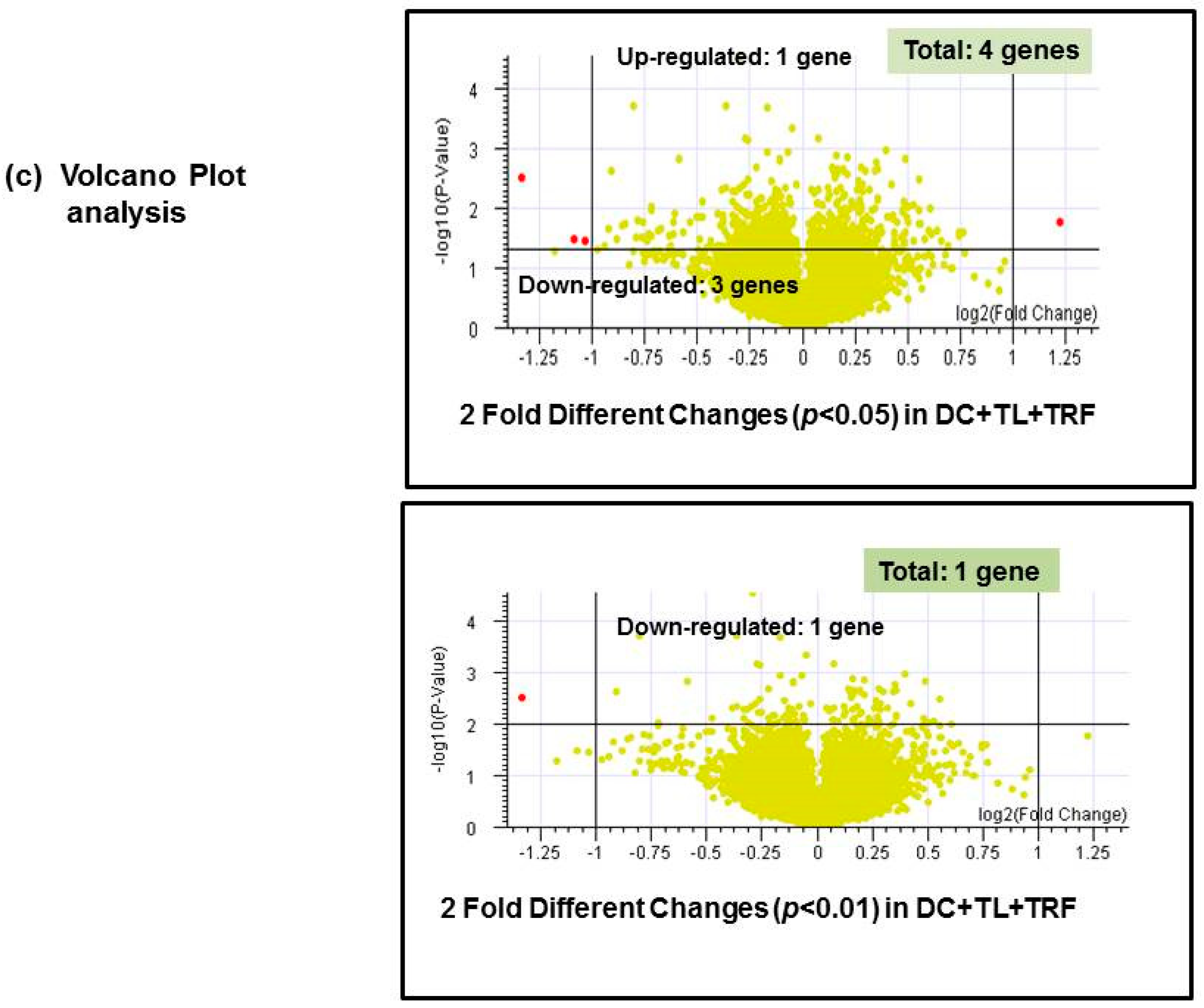
3.4. Microarray Analysis
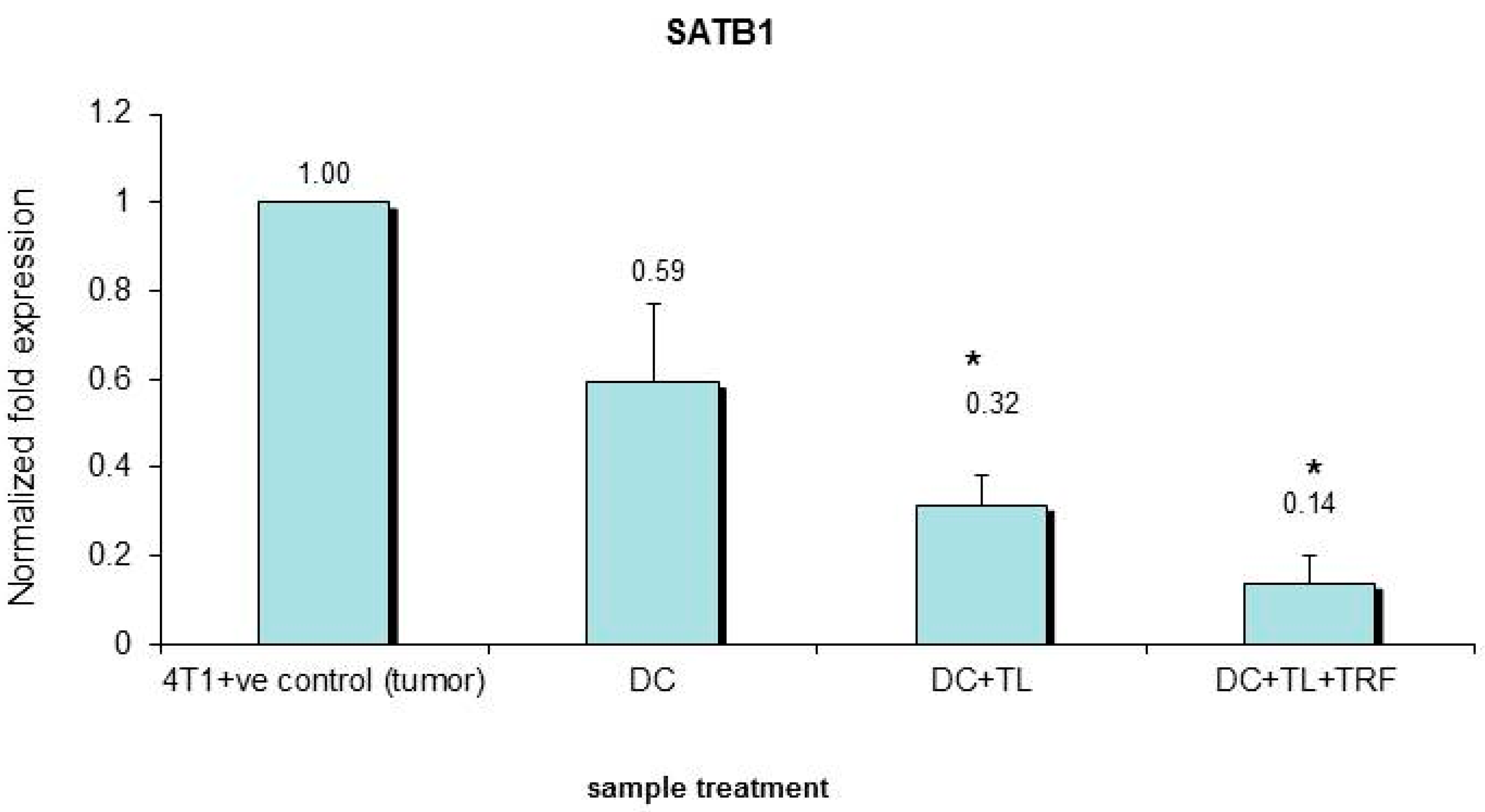
3.5. Verification of Microarray Results by Quantitative PCR
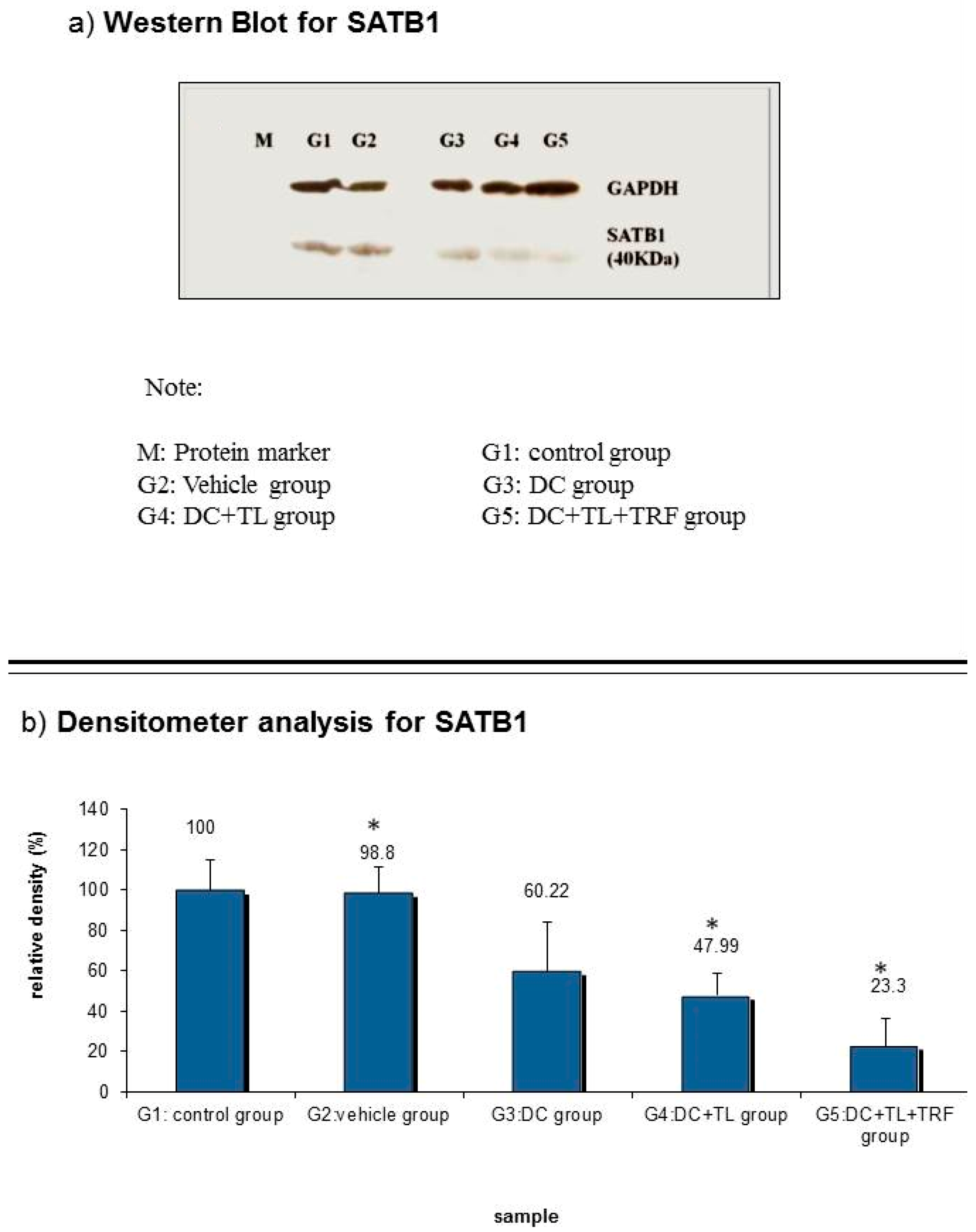
3.6. Expression of SATB1 Protein
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shuler, G.; Steinman, R. Dendritic cells as adjuvants for immune-mediated resistance to tumors. J. Exp. Med. 1997, 186, 1183–1187. [Google Scholar] [CrossRef] [PubMed]
- Ramanathapuram, L.V.; Kobie, J.J.; Bearss, D.; Payne, C.M.; Trevor, K.T.; Akporiaye, E.T. α-tocopheryl succinate sensitizes established tumors to vaccination with nonmatured dendritic cells. Cancer Immunol. Immunother. 2004, 53, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Steinman, R.M.; Dhodapkar, M. Active immunization against cancer with dendritic cells, the near future. Int. J. Cancer 2001, 94, 459. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Prasad, K.N.; Kumar, B.; Yan, X.D.; Hanson, A.J.; Cole, W.C. α-Tocopherol succinate, the most effective form of vitamin E adjuvant cancer treatment: A review. J. Am. Coll. Nutr. 2003, 22, 108–117. [Google Scholar] [CrossRef]
- Ramanathapuram, L.V. Chemo-Immunotherapy of Murine Cancer Using Alpha Tocopheryl Succinate and Non-Matured Dendritic Cells. Ph.D. Thesis, The University of Arizona, Tempe, AZ, USA, 2006. [Google Scholar]
- Sato, M.; Chamoto, K.; Nishimura, T. A novel tumor-vaccine cell therapy using bone marrow-derived dendritic cell type 1 and antigen-specific Th1 cells. Int. Immunol. 2003, 15, 837–843. [Google Scholar] [CrossRef]
- Gatza, E.; Okada, C.Y. Tumor cell lysate-pulsed dendritic cells are more effective than TCR Id protein vaccines for active immunotherapy of T cell lymphoma. J. Immunol. 2002, 169, 5227–5235. [Google Scholar] [CrossRef]
- Nagaraj, S.; Ziske, C.; Schmidt-Wolf, I.G.H. Dendritic cell, the immunotherapeutic cell for cancer. Indian J. Med. Res. 2004, 119, 133–138. [Google Scholar]
- Steinman, A.M. The dendritic cell system and its role in immunogenicity. Annu. Rev. Immunol. 1991, 9, 271. [Google Scholar] [CrossRef]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef]
- Lawrence, C.W.; Bracial, T.J. Activation, Differentiation, and Migration of Naive Virus-Specific CD8+ T Cells during Pulmonary Influenza Virus Infection. J. Immunol. 2004, 173, 1209–1218. [Google Scholar] [CrossRef]
- Brossart, P.; Bevan, M.J. Presentation of exogenous protein antigens on major histocompatability complex class I molecules by dendritic cells: Pathway of presentation and regulation by cytokines. Am. Soc. Hematol. 1997, 90, 1594–1599. [Google Scholar]
- Steinman, R.M. Decisions About Dendritic Cells: Past, Present, and Future. Annu. Rev. Immunol. 2012, 30, 1–22. [Google Scholar] [CrossRef]
- Herr, W.; Ranieri, E.; Olson, W.; Zarour, H.; Gesualdo, L.; Storkus, W.J. Mature dendritic cells pulsed with freeze-thaw lysates define an effective in vitro vaccine designed to elicit EBV-specific CD4+ and CD8+ T lymphocyte responses. Blood 2000, 96, 1857–1864. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.L.; Reizis, B. Dendritic Cells: Arbiters of Immunity and Immunological Tolerance. Cold Spring Harb. Perspect. Biol. 2012, 4, a007401. [Google Scholar] [CrossRef] [PubMed]
- Porgador, A.; Snyder, D.; Gilboa, E. Induction of antitumour immunity using bone marrow-generated dendritic cells. J. Immunol. 1996, 156, 2918–2926. [Google Scholar] [PubMed]
- Kokhaei, P.; Choudhury, A.; Mahdian, R.; Lundin, J.; Moshfegh, A.; Osterborg, A.; Mellstedt, H. Apoptotic tumor cells are superior to tumor cell lysate, and tumor cell RNA in induction of autologous T cell response in B-CLL. Leukemia 2004, 18, 1810–1815. [Google Scholar] [CrossRef]
- Ramanathapuram, L.V.; Hahn, T.; Dial, S.M.; Akporiaye, E.T. Chemo-Immunotherapy of Breast Cancer using vesiculated α-Tocophyrl succinate in combination with Dendritic Cell Vaccination. Nutr. Cancer 2005, 53, 177–193. [Google Scholar] [CrossRef]
- Abdul Hafid, S.R.; Radhakrishnan, A.K.; Nesaretnam, K. Tocotrienols are good adjuvant in developing cancer vaccines. BMC Cancer J. 2010, 10, 5. [Google Scholar] [CrossRef]
- Okamoto, M.; Kobayashi, M.; Yonemitsu, Y.; Koido, S.; Homma, V. Dendritic cell-based vaccine for pancreatic cancer in Japan. World J. Gastrointest. Pharmacol. Ther. 2016, 7, 133–138. [Google Scholar] [CrossRef]
- Shimizu, K.; Field, C.; Giedlin, M.; Mule, J.J. Systemic administration of interleukin 2 enhances the therapeutic efficacy of dendritic cell-based tumor vaccines. Proc. Natl. Acad. Sci. USA 1999, 96, 2268–2273. [Google Scholar] [CrossRef]
- Gapor, A.; Top, M. A value—added tocotrienols-rich fraction (TRF) from palm oil. Palm Oil Dev. 1995, 2, 7–17. [Google Scholar]
- Choo, Y.M.; Ma, A.N.; Yap, S.C. Carotenes, vitamin E and sterols in oils from Elaeis guineensis, Elaeis oleifera and their hybrids. Palm Oil Dev. 1997, 27. [Google Scholar]
- Abdul Hafid, S.R.; Chakravarthi, S.; Nesaretnam, K.; Radhakrishnan, A.K. Tocotrienol-Adjuvanted Dendritic Cells Inhibit Tumor Growth and Metastasis: A murine Model of Breast Cancer. PLoS ONE 2013, 8, e74753. [Google Scholar] [CrossRef] [PubMed]
- Pulaski, B.A.; Terman, D.S.; Kabo, S.; Muller, E.; Ostrand-Resenberg, S. Cooperativity of staphylococcal aureus enterotoxin B superanti-gen, major histocompatibility complex class H, and CD80 for immunotherapy of advanced spontaneous metastases in a clinically relevant postoperative mouse breast cancer model. Cancer Res. 2000, 60, 2710–2715. [Google Scholar] [PubMed]
- DeMeyer, E.S.; Baar, J. Dendritic Cells: The Sentry Cells of the Immune System; Berlex Laboratories Inc.: Hanover, NJ, USA, 2007. [Google Scholar]
- Itano, A.A.; Jenkins, M.K. Antigen presentation to naive CD4T cels in the lymph node. Nat. Immunol. 2003, 4, 733. [Google Scholar] [CrossRef]
- Gregori, S. Dendritic cells in networks of immunological tolerance. Tissue Antigens 2011, 77, 89–99. [Google Scholar] [CrossRef]
- Kim, H.M.; Han, H.B.; Oh, G.T.; Kim, Y.H.; Hong, D.H.; Yoo, I.D. Stimulation oh humoral and cell mediated immunity by LPS from mushroom Phellinus linteus. Int. J. Immunopharmacol. 1996, 18, 295–303. [Google Scholar] [CrossRef]
- Nesaretnam, K.; Stephen, R.; Dils, R.; Darbre, P. Tocotrienols inhibit the growth of human breast cancer cells irrespective of estrogen receptor status. Lipids 1998, 33, 461. [Google Scholar] [CrossRef]
- Nesaretnam, K.; Guthrie, N.; Chambers, A.F.; Carroll, K.K. Effect of tocotrienols on the growth of a human breast cancer cell line in culture. Lipids 1995, 30, 1139–1143. [Google Scholar] [CrossRef]
- Nesaretnam, K. Antioxidant and anti-cancer properties of tocotrienols. Palm Oil Dev. 2000, 32, 5–10. [Google Scholar]
- Selvaduray, K.R.; Radhakrishnan, A.K.; Kannan, M.; Nesaretnam, K. Palm tocotrienols inhibit proliferation of murine mammary cancer cells and induce expression of IL-24 mRNA. J. Interferon Cytokine Res. 2010, 30, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Loo, G. Disruption of mitochondria during tocotrienol-induced apoptosis in MDA-MB-231 human breast cancer cells. Biochem. Pharmacol. 2004, 67, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Nesaretnam, K.; Dorasamy, S.; Darbre, P.D. Tocotrienols inhibit growth of ZR-75-1 breast cancer cells. Int. J. Food Sci. Nutr. 2000, 51, S95–S103. [Google Scholar] [CrossRef] [PubMed]
- Nesaretnam, K.; Ambra, R.; Selvaduray, K.R.; Radhakrishnan, A.; Canali, R.; Virgili, F. Tocotrienol-rich fraction from palm oil and gene expression in human breast cancer cells. Ann. N. Y. Acad. Sci. 2004, 1031, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Nesaretnam, K.; Ambra, R.; Selvaduray, K.R.; Radhakrishnan, A.; Reimann, K.; Razak, G.; Virgili, F. Tocotrienol-rich fraction from palm oil affects gene expression in tumors resulting from MCF-7 cell inoculation in athymic mice. Lipids 2004, 39, 459–467. [Google Scholar] [CrossRef]
- Terumi Kohwi, S.; Mass, K.; Bode, J. A Thymocyte Factor SATB1 Suppresses Transcription of Stably Integrated Matrix-Attachment Region-Linked Reporter Genes. Biochemistry 1997, 36, 12005–12010. [Google Scholar] [CrossRef]
- Han, H.J.; Russo, J.; Kohwi, Y.; Kohwi-Shigematsu, T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature 2008, 452, 187–193. [Google Scholar] [CrossRef]
- Alvarez, J.D.; Yasui, D.H.; Niida, H.; Joh, T.; Loh, D.Y.; Kohwi-Shigematsu, T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000, 14, 521–535. [Google Scholar] [CrossRef]
| Groups | Name | Treatment |
|---|---|---|
| 1 | Control | Injected with 4T1 cells |
| 2 | Vehicle | Fed with vehicle daily Injected with 4T1 cells |
| 3 | DC | Injected with DC (three times) Injected with 4T1 cells a week after last DC injection |
| 4 | DC + TL | Injected with DC + TL (three times) Injected with 4T1 cells a week after last the DC + TL injection |
| 5 | DC + TL + TRF | Injected with DC + TL (three times) Fed with 1 mg TRF daily Injected with 4T1 cells a week after the last DC + TL injection |
| No. | Primary Antibody | Secondary Antibody |
|---|---|---|
| (1) | Mouse FAS ligand/TNFSF6 antibody (AF526) | Goat IgG HRP-conjugated antibody (HAF017) |
| (2) | Human/mouse/rat GAPDH/G3PDH antibody, antigen affinity-purified polyclonal goat IgG (AF5718) | Goat IgG HRP-conjugated antibody (HAF017) |
| No. | Gene Name | Description | Regulation in DC + TL + TRF | Fold * Change |
|---|---|---|---|---|
| 1. | CRIP2 | Mus musculus cysteine-rich protein 2 (Crip2), mRNA | Up-regulated | 2.337 |
| 2. | IFITM1 | Mus musculus interferon-induced transmembrane protein 1 (Ifitm1) | Down-regulated | 0.488 |
| 3. | MRC1 | Mus musculus mannose receptor, C type 1 (Mrc1), mRNA | Down-regulated | 0.471 |
| 4. | SATB1 | Mus musculus special AT-rich binding protein 1 | Down-regulated | 0.396 |
| No. | Gene Name | Description | Regulation in DC + TL + TRF | Fold * Change |
|---|---|---|---|---|
| 1. | SATB1 | Special AT-rich binding protein 1 | Down-regulated | 0.396 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdul Hafid, S.R.; Radhakrishnan, A.K. Palm Tocotrienol-Adjuvanted Dendritic Cells Decrease Expression of the SATB1 Gene in Murine Breast Cancer Cells and Tissues. Vaccines 2019, 7, 198. https://doi.org/10.3390/vaccines7040198
Abdul Hafid SR, Radhakrishnan AK. Palm Tocotrienol-Adjuvanted Dendritic Cells Decrease Expression of the SATB1 Gene in Murine Breast Cancer Cells and Tissues. Vaccines. 2019; 7(4):198. https://doi.org/10.3390/vaccines7040198
Chicago/Turabian StyleAbdul Hafid, Sitti Rahma, and Ammu Kutty Radhakrishnan. 2019. "Palm Tocotrienol-Adjuvanted Dendritic Cells Decrease Expression of the SATB1 Gene in Murine Breast Cancer Cells and Tissues" Vaccines 7, no. 4: 198. https://doi.org/10.3390/vaccines7040198
APA StyleAbdul Hafid, S. R., & Radhakrishnan, A. K. (2019). Palm Tocotrienol-Adjuvanted Dendritic Cells Decrease Expression of the SATB1 Gene in Murine Breast Cancer Cells and Tissues. Vaccines, 7(4), 198. https://doi.org/10.3390/vaccines7040198





