Vaccine Vectors Harnessing the Power of Cytomegaloviruses
Abstract
1. Initial Considerations on Cytomegalovirus as a Viral Vector
2. Innate Immune Response to CMV is Relevant for Its Vector Properties
3. Adaptive Immune Response to CMV Relevant for Its Vector Properties
3.1. Humoral Response
3.2. Cellular Response—T cells
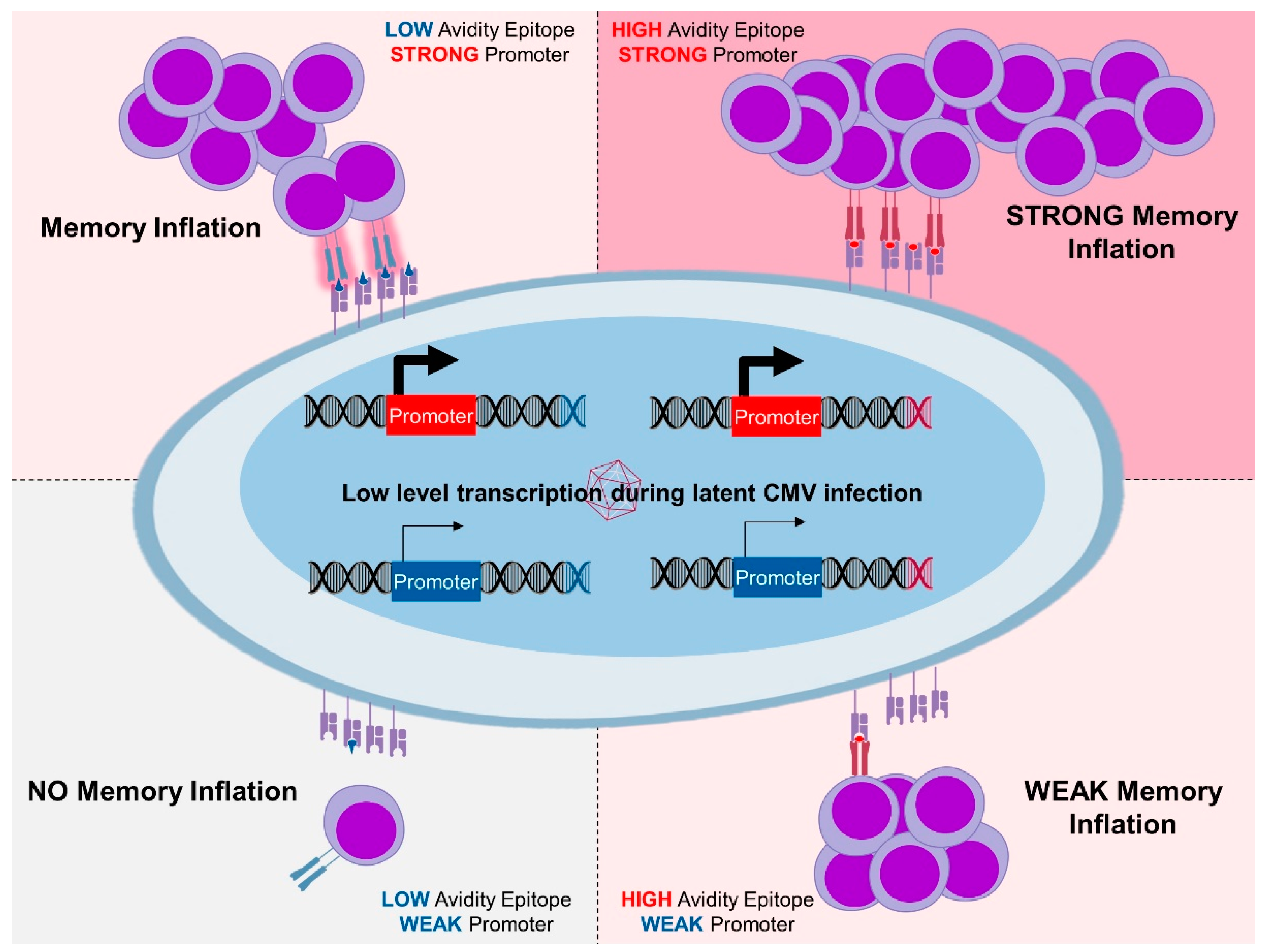
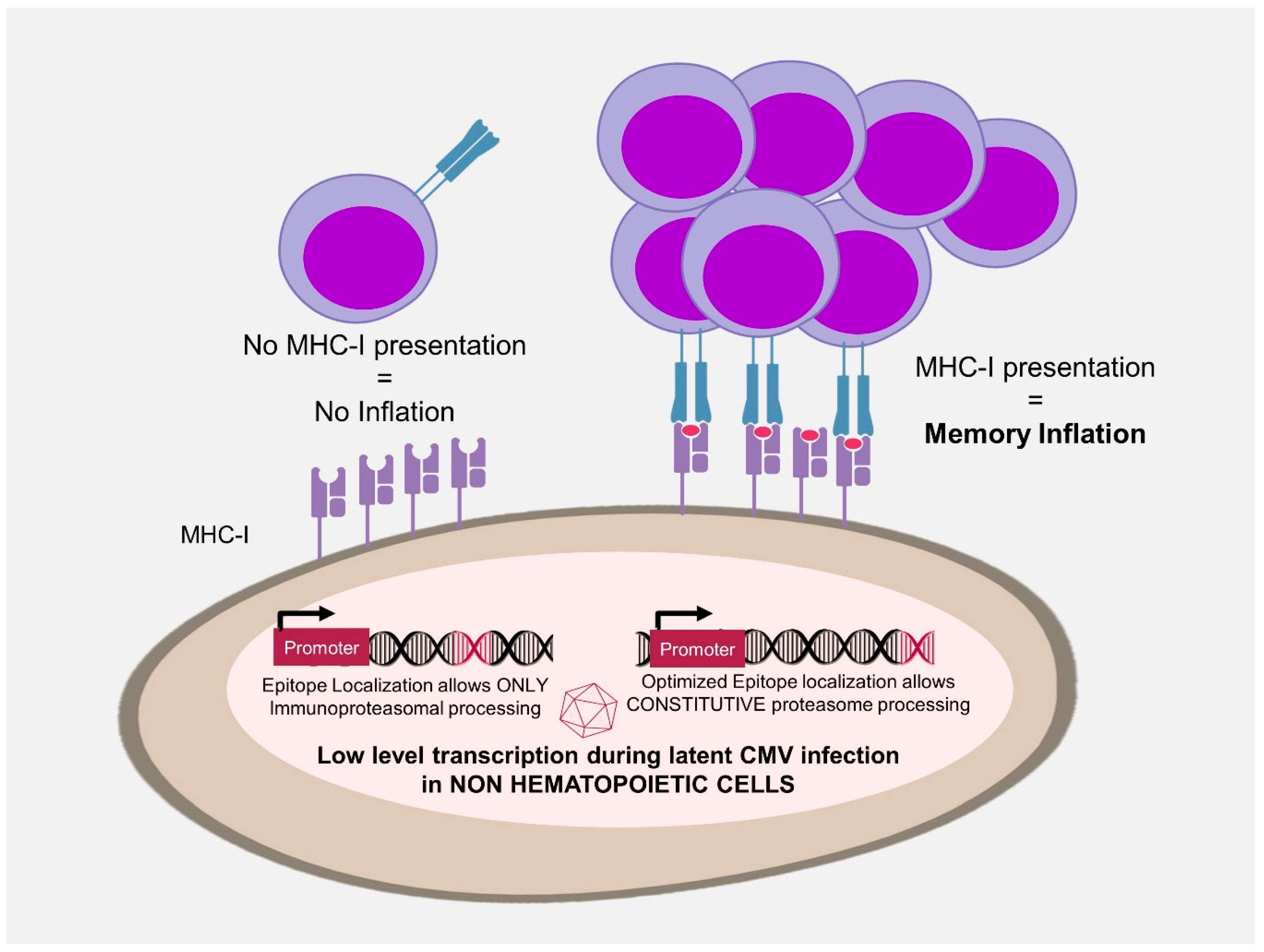
3.3. Antigen Processing in MI
3.4. Unconventional MHC Restriction
3.5. CD4 T Cells
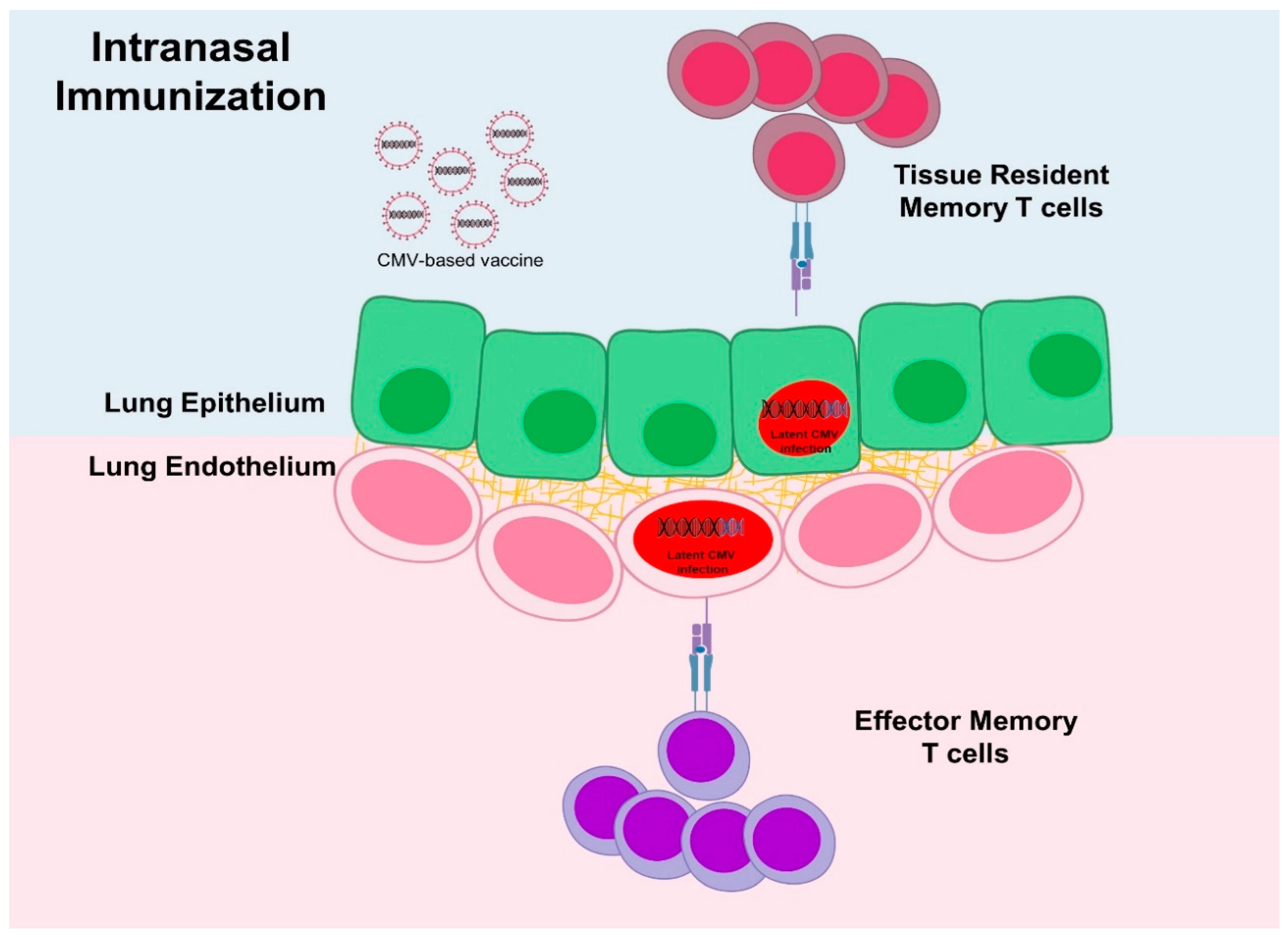
4. Factor Affecting Immunity—Dose and Administration Route
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boppana, S.B.; Britt, W.J. Synopsis of clinical aspects of human cytomegalovirus disease. In Cytomegaloviruses: From Molecular Pathogenesis to Intervention; Caister Academic Press: Poole, UK, 2013; Volume 2, pp. 1–25. [Google Scholar]
- Sylwester, A.W.; Mitchell, B.L.; Edgar, J.B.; Taormina, C.; Pelte, C.; Ruchti, F.; Sleath, P.R.; Grabstein, K.H.; Hosken, N.A.; Kern, F.; et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 2005, 202, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Brune, W. Molecular basis of cytomegalovirus host species specificity. In Cytomegaloviruses: From Molecular Pathogenesis to Intervention; Caister Academic Press: Poole, UK, 2013; Volume 1, pp. 322–329. [Google Scholar]
- Reddehase, M.J.; Lemmermann, N.A.W. Mouse Model of Cytomegalovirus Disease and Immunotherapy in the Immunocompromised Host: Predictions for Medical Translation that Survived the “Test of Time”. Viruses 2018, 10, 693. [Google Scholar] [CrossRef] [PubMed]
- Seckert, C.K.; Grießl, M.; Büttner, J.K.; Scheller, S.; Simon, C.O.; Kropp, K.A.; Renzaho, A.; Kühnapfel, B.; Grzimek, N.K.; Reddehase, M.J. Viral latency drives ‘memory inflation’: A unifying hypothesis linking two hallmarks of cytomegalovirus infection. Med. Microbiol. Immunol. 2012, 201, 551–566. [Google Scholar] [CrossRef] [PubMed]
- Kurz, S.; Steffens, H.P.; Mayer, A.; Harris, J.R.; Reddehase, M.J. Latency versus persistence or intermittent recurrences: Evidence for a latent state of murine cytomegalovirus in the lungs. J. Virol. 1997, 71, 2980–2987. [Google Scholar] [PubMed]
- Klenerman, P. The (gradual) rise of memory inflation. Immunol. Rev. 2018, 283, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Karrer, U.; Sierro, S.; Wagner, M.; Oxenius, A.; Hengel, H.; Koszinowski, U.H.; Phillips, R.E.; Klenerman, P. Memory inflation: Continuous accumulation of antiviral CD8+ T cells over time. J. Immunol. 2003, 170, 2022–2029. [Google Scholar] [CrossRef]
- Holtappels, R.; Pahl-Seibert, M.F.; Thomas, D.; Reddehase, M.J. Enrichment of immediate-early 1 (m123/pp89) peptide-specific CD8 T cells in a pulmonary CD62L(lo) memory-effector cell pool during latent murine cytomegalovirus infection of the lungs. J. Virol. 2000, 74, 11495–11503. [Google Scholar] [CrossRef]
- Klenerman, P.; Oxenius, A. T cell responses to cytomegalovirus. Nat. Rev. Immunol. 2016, 16, 367–377. [Google Scholar] [CrossRef]
- Dekhtiarenko, I.; Čičin-Šain, L.; Messerle, M. Use of recombinant approaches to construct human cytomegalovirus mutants. In Human Cytomegaloviruses; Humana Press: Totowa, NJ, USA, 2014; pp. 59–79. [Google Scholar]
- Humphreys, I.R.; Sebastian, S. Novel viral vectors in infectious diseases. Immunology 2018, 153, 1–9. [Google Scholar] [CrossRef]
- Liu, J.; Jaijyan, D.K.; Tang, Q.; Zhu, H. Promising Cytomegalovirus-Based Vaccine Vector Induces Robust CD8(+) T-Cell Response. Int. J. Mol. Sci. 2019, 20, 4457. [Google Scholar] [CrossRef]
- Dekhtiarenko, I.; Ratts, R.B.; Blatnik, R.; Lee, L.N.; Fischer, S.; Borkner, L.; Oduro, J.D.; Marandu, T.F.; Hoppe, S.; Ruzsics, Z. Peptide processing is critical for T-cell memory inflation and may be optimized to improve immune protection by CMV-based vaccine vectors. PLoS Pathog. 2016, 12, e1006072. [Google Scholar] [CrossRef] [PubMed]
- Borkner, L.; Sitnik, K.M.; Dekhtiarenko, I.; Pulm, A.K.; Tao, R.; Drexler, I.; Cicin-Sain, L. Immune Protection by a Cytomegalovirus Vaccine Vector Expressing a Single Low-Avidity Epitope. J. Immunol. 2017, 199, 1737–1747. [Google Scholar] [CrossRef] [PubMed]
- Cicin-Sain, L. Cytomegalovirus memory inflation and immune protection. Med. Microbiol. Immunol. 2019, 208, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.G.; Womack, J.; Scholz, I.; Renner, A.; Edgel, K.A.; Xu, G.; Ford, J.C.; Grey, M.; Laurent, B.S.; Turner, J.M. Cytomegalovirus vectors expressing Plasmodium knowlesi antigens induce immune responses that delay parasitemia upon sporozoite challenge. PLoS ONE 2019, 14, e0210252. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.G.; Zak, D.E.; Xu, G.; Ford, J.C.; Marshall, E.E.; Malouli, D.; Gilbride, R.M.; Hughes, C.M.; Ventura, A.B.; Ainslie, E.; et al. Prevention of tuberculosis in rhesus macaques by a cytomegalovirus-based vaccine. Nat. Med. 2018, 24, 130–143. [Google Scholar] [CrossRef]
- Hansen, S.G.; Ford, J.C.; Lewis, M.S.; Ventura, A.B.; Hughes, C.M.; Coyne-Johnson, L.; Whizin, N.; Oswald, K.; Shoemaker, R.; Swanson, T.; et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 2011, 473, 523–527. [Google Scholar] [CrossRef]
- Hansen, S.G.; Vieville, C.; Whizin, N.; Coyne-Johnson, L.; Siess, D.C.; Drummond, D.D.; Legasse, A.W.; Axthelm, M.K.; Oswald, K.; Trubey, C.M.; et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 2009, 15, 293–299. [Google Scholar] [CrossRef]
- Hansen, S.G.; Sacha, J.B.; Hughes, C.M.; Ford, J.C.; Burwitz, B.J.; Scholz, I.; Gilbride, R.M.; Lewis, M.S.; Gilliam, A.N.; Ventura, A.B.; et al. Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 2013, 340, 1237874. [Google Scholar] [CrossRef]
- Hansen, S.G.; Wu, H.L.; Burwitz, B.J.; Hughes, C.M.; Hammond, K.B.; Ventura, A.B.; Reed, J.S.; Gilbride, R.M.; Ainslie, E.; Morrow, D.W.; et al. Broadly targeted CD8(+) T cell responses restricted by major histocompatibility complex E. Science 2016, 351, 714–720. [Google Scholar] [CrossRef]
- Marshall, E.E.; Malouli, D.; Hansen, S.G.; Gilbride, R.M.; Hughes, C.M.; Ventura, A.B.; Ainslie, E.; Selseth, A.N.; Ford, J.C.; Burke, D.; et al. Enhancing safety of cytomegalovirus-based vaccine vectors by engaging host intrinsic immunity. Sci. Transl. Med. 2019, 11, eaaw2603. [Google Scholar] [CrossRef]
- Hansen, S.G.; Marshall, E.E.; Malouli, D.; Ventura, A.B.; Hughes, C.M.; Ainslie, E.; Ford, J.C.; Morrow, D.; Gilbride, R.M.; Bae, J.Y.; et al. A live-attenuated RhCMV/SIV vaccine shows long-term efficacy against heterologous SIV challenge. Sci. Transl. Med. 2019, 11, eaaw2607. [Google Scholar] [CrossRef] [PubMed]
- Marzi, A.; Murphy, A.A.; Feldmann, F.; Parkins, C.J.; Haddock, E.; Hanley, P.W.; Emery, M.J.; Engelmann, F.; Messaoudi, I.; Feldmann, H.; et al. Cytomegalovirus-based vaccine expressing Ebola virus glycoprotein protects nonhuman primates from Ebola virus infection. Sci. Rep. 2016, 6, 21674. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, Y.; Caposio, P.; Parkins, C.J.; Botto, S.; Messaoudi, I.; Cicin-Sain, L.; Feldmann, H.; Jarvis, M.A. A replicating cytomegalovirus-based vaccine encoding a single Ebola virus nucleoprotein CTL epitope confers protection against Ebola virus. PLoS Negl. Trop. Dis. 2011, 5, e1275. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, Y.; Parkins, C.J.; Caposio, P.; Feldmann, F.; Botto, S.; Ball, S.; Messaoudi, I.; Cicin-Sain, L.; Feldmann, H.; Jarvis, M.A. A cytomegalovirus-based vaccine provides long-lasting protection against lethal Ebola virus challenge after a single dose. Vaccine 2015, 33, 2261–2266. [Google Scholar] [CrossRef] [PubMed]
- Trsan, T.; Busche, A.; Abram, M.; Wensveen, F.M.; Lemmermann, N.A.; Arapovic, M.; Babic, M.; Tomic, A.; Golemac, M.; Brinkmann, M.M.; et al. Superior induction and maintenance of protective CD8 T cells in mice infected with mouse cytomegalovirus vector expressing RAE-1gamma. Proc. Natl. Acad. Sci. USA 2013, 110, 16550–16555. [Google Scholar] [CrossRef] [PubMed]
- Beverley, P.C.; Ruzsics, Z.; Hey, A.; Hutchings, C.; Boos, S.; Bolinger, B.; Marchi, E.; O’Hara, G.; Klenerman, P.; Koszinowski, U.H.; et al. A novel murine cytomegalovirus vaccine vector protects against Mycobacterium tuberculosis. J. Immunol. 2014, 193, 2306–2316. [Google Scholar] [CrossRef]
- Morabito, K.M.; Ruckwardt, T.R.; Redwood, A.J.; Moin, S.M.; Price, D.A.; Graham, B.S. Intranasal administration of RSV antigen-expressing MCMV elicits robust tissue-resident effector and effector memory CD8+ T cells in the lung. Mucosal Immunol. 2017, 10, 545–554. [Google Scholar] [CrossRef]
- Handke, W.; Luig, C.; Popovic, B.; Krmpotic, A.; Jonjic, S.; Brune, W. Viral inhibition of BAK promotes murine cytomegalovirus dissemination to salivary glands. J. Virol. 2013, 87, 3592–3596. [Google Scholar] [CrossRef]
- Morabito, K.M.; Ruckwardt, T.J.; Bar-Haim, E.; Nair, D.; Moin, S.M.; Redwood, A.J.; Price, D.A.; Graham, B.S. Memory Inflation Drives Tissue-Resident Memory CD8(+) T Cell Maintenance in the Lung After Intranasal Vaccination With Murine Cytomegalovirus. Front. Immunol. 2018, 9, 1861. [Google Scholar] [CrossRef]
- Dekhtiarenko, I.; Jarvis, M.A.; Ruzsics, Z.; Cicin-Sain, L. The context of gene expression defines the immunodominance hierarchy of cytomegalovirus antigens. J. Immunol. 2013, 190, 3399–3409. [Google Scholar] [CrossRef]
- Jordan, S.; Krause, J.; Prager, A.; Mitrovic, M.; Jonjic, S.; Koszinowski, U.H.; Adler, B. Virus progeny of murine cytomegalovirus bacterial artificial chromosome pSM3fr show reduced growth in salivary Glands due to a fixed mutation of MCK-2. J. Virol. 2011, 85, 10346–10353. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Oduro, J.D.; Boehme, J.D.; Borkner, L.; Ebensen, T.; Heise, U.; Gereke, M.; Pils, M.C.; Krmpotic, A.; Guzman, C.A.; et al. Mucosal CD8+ T cell responses induced by an MCMV based vaccine vector confer protection against influenza challenge. PLoS Pathog. 2019, 15, e1008036. [Google Scholar] [CrossRef] [PubMed]
- Nejad, E.B.; Ratts, R.B.; Panagioti, E.; Meyer, C.; Oduro, J.D.; Cicin-Sain, L.; Früh, K.; Van Der Burg, S.H.; Arens, R. Demarcated thresholds of tumor-specific CD8 T cells elicited by MCMV-based vaccine vectors provide robust correlates of protection. J. Immunother. Cancer 2019, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Van de Berg, P.J.; van Stijn, A.; Ten Berge, I.J.; van Lier, R.A. A fingerprint left by cytomegalovirus infection in the human T cell compartment. J. Clin. Virol. 2008, 41, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Messerle, M.; Crnkovic, I.; Hammerschmidt, W.; Ziegler, H.; Koszinowski, U.H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 1997, 94, 14759–14763. [Google Scholar] [CrossRef]
- Karrer, U.; Wagner, M.; Sierro, S.; Oxenius, A.; Hengel, H.; Dumrese, T.; Freigang, S.; Koszinowski, U.H.; Phillips, R.E.; Klenerman, P. Expansion of protective CD8+ T-cell responses driven by recombinant cytomegaloviruses. J. Virol. 2004, 78, 2255–2264. [Google Scholar] [CrossRef]
- Mohr, C.A.; Cicin-Sain, L.; Wagner, M.; Sacher, T.; Schnee, M.; Ruzsics, Z.; Koszinowski, U.H. Engineering of cytomegalovirus genomes for recombinant live herpesvirus vaccines. Int. J. Med. Microbiol. 2008, 298, 115–125. [Google Scholar] [CrossRef]
- Boppana, S.B.; Rivera, L.B.; Fowler, K.B.; Mach, M.; Britt, W.J. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N. Engl. J. Med. 2001, 344, 1366–1371. [Google Scholar] [CrossRef]
- Hansen, S.G.; Powers, C.J.; Richards, R.; Ventura, A.B.; Ford, J.C.; Siess, D.; Axthelm, M.K.; Nelson, J.A.; Jarvis, M.A.; Picker, L.J. Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science 2010, 328, 102–106. [Google Scholar] [CrossRef]
- Nogalski, M.T.; Collins-McMillen, D.; Yurochko, A.D. Overview of human cytomegalovirus pathogenesis. In Human Cytomegaloviruses; Humana Press: Totowa, NJ, USA, 2014; pp. 15–28. [Google Scholar]
- Snyder, C.M.; Cho, K.S.; Bonnett, E.L.; Allan, J.E.; Hill, A.B. Sustained CD8+ T cell memory inflation after infection with a single-cycle cytomegalovirus. PLoS Pathog 2011, 7, e1002295. [Google Scholar] [CrossRef]
- Čičin-Šain, L.; Bubić, I.; Schnee, M.; Ruzsics, Z.; Mohr, C.; Jonjić, S.; Koszinowski, U.H. Targeted deletion of regions rich in immune-evasive genes from the cytomegalovirus genome as a novel vaccine strategy. J. Virol. 2007, 81, 13825–13834. [Google Scholar] [CrossRef] [PubMed]
- Slavuljica, I.; Busche, A.; Babic, M.; Mitrovic, M.; Gasparovic, I.; Cekinovic, D.; Markova Car, E.; Pernjak Pugel, E.; Cikovic, A.; Lisnic, V.J.; et al. Recombinant mouse cytomegalovirus expressing a ligand for the NKG2D receptor is attenuated and has improved vaccine properties. J. Clin. Investig. 2010, 120, 4532–4545. [Google Scholar] [CrossRef] [PubMed]
- Hirsl, L.; Brizic, I.; Jenus, T.; Juranic Lisnic, V.; Reichel, J.J.; Jurkovic, S.; Krmpotic, A.; Jonjic, S. Murine CMV Expressing the High Affinity NKG2D Ligand MULT-1: A Model for the Development of Cytomegalovirus-Based Vaccines. Front. Immunol. 2018, 9, 991. [Google Scholar] [CrossRef] [PubMed]
- Trsan, T.; Vukovic, K.; Filipovic, P.; Brizic, A.L.; Lemmermann, N.A.W.; Schober, K.; Busch, D.H.; Britt, W.J.; Messerle, M.; Krmpotic, A.; et al. Cytomegalovirus vector expressing RAE-1gamma induces enhanced anti-tumor capacity of murine CD8(+) T cells. Eur. J. Immunol. 2017, 47, 1354–1367. [Google Scholar] [CrossRef] [PubMed]
- Beswick, M.; Pachnio, A.; Al-Ali, A.; Sweet, C.; Moss, P.A. An attenuated temperature-sensitive strain of cytomegalovirus (tsm5) establishes immunity without development of CD8(+) T cell memory inflation. J. Med. Virol. 2013, 85, 1968–1974. [Google Scholar] [CrossRef] [PubMed]
- Mohr, C.A.; Arapovic, J.; Muhlbach, H.; Panzer, M.; Weyn, A.; Dolken, L.; Krmpotic, A.; Voehringer, D.; Ruzsics, Z.; Koszinowski, U.; et al. A spread-deficient cytomegalovirus for assessment of first-target cells in vaccination. J. Virol. 2010, 84, 7730–7742. [Google Scholar] [CrossRef]
- Snyder, C.M.; Allan, J.E.; Bonnett, E.L.; Doom, C.M.; Hill, A.B. Cross-presentation of a spread-defective MCMV is sufficient to prime the majority of virus-specific CD8+ T cells. PLoS ONE 2010, 5, e9681. [Google Scholar] [CrossRef]
- Valencia, S.; Gill, R.B.; Dowdell, K.C.; Wang, Y.; Hornung, R.; Bowman, J.J.; Lacayo, J.C.; Cohen, J.I. Comparison of vaccination with rhesus CMV (RhCMV) soluble gB with a RhCMV replication-defective virus deleted for MHC class I immune evasion genes in a RhCMV challenge model. Vaccine 2019, 37, 333–342. [Google Scholar] [CrossRef]
- Jordan, M.C. Adverse effects of cytomegalovirus vaccination in mice. J. Clin. Investig. 1980, 65, 798–803. [Google Scholar] [CrossRef]
- Kalejta, R.F.; Shenk, T. Manipulation of the cell cycle by human cytomegalovirus. Front. Biosci. 2002, 7, d295–d306. [Google Scholar] [CrossRef]
- Straat, K.; Liu, C.; Rahbar, A.; Zhu, Q.; Liu, L.; Wolmer-Solberg, N.; Lou, F.; Liu, Z.; Shen, J.; Jia, J.; et al. Activation of telomerase by human cytomegalovirus. J. Natl. Cancer Inst. 2009, 101, 488–497. [Google Scholar] [CrossRef]
- Albrecht, T.; Deng, C.Z.; Abdel-Rahman, S.Z.; Fons, M.; Cinciripini, P.; El-Zein, R.A. Differential mutagen sensitivity of peripheral blood lymphocytes from smokers and nonsmokers: effect of human cytomegalovirus infection. Environ. Mol. Mutagen. 2004, 43, 169–178. [Google Scholar] [CrossRef]
- Fortunato, E.A.; Spector, D.H. Viral induction of site-specific chromosome damage. Rev. Med. Virol. 2003, 13, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Tripathy, M.K.; Pasquereau, S.; Al Moussawi, F.; Abbas, W.; Coquard, L.; Khan, K.A.; Russo, L.; Algros, M.P.; Valmary-Degano, S.; et al. The Human Cytomegalovirus Strain DB Activates Oncogenic Pathways in Mammary Epithelial Cells. EBio. Med. 2018, 30, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.C.Y.; Pjechova, M.; Nightingale, K.; Vlahava, V.M.; Patel, M.; Ruckova, E.; Forbes, S.K.; Nobre, L.; Antrobus, R.; Roberts, D.; et al. Suppression of costimulation by human cytomegalovirus promotes evasion of cellular immune defenses. Proc. Natl. Acad. Sci. USA 2018, 115, 4998–5003. [Google Scholar] [CrossRef] [PubMed]
- Avdic, S.; McSharry, B.P.; Steain, M.; Poole, E.; Sinclair, J.; Abendroth, A.; Slobedman, B. Human Cytomegalovirus-Encoded Human Interleukin-10 (IL-10) Homolog Amplifies Its Immunomodulatory Potential by Upregulating Human IL-10 in Monocytes. J. Virol. 2016, 90, 3819–3827. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Slinger, E.; Maussang, D.; Schreiber, A.; Siderius, M.; Rahbar, A.; Fraile-Ramos, A.; Lira, S.A.; Soderberg-Naucler, C.; Smit, M.J. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci. Signal. 2010, 3, ra58. [Google Scholar] [CrossRef]
- Krenzlin, H.; Behera, P.; Lorenz, V.; Passaro, C.; Zdioruk, M.; Nowicki, M.O.; Grauwet, K.; Zhang, H.; Skubal, M.; Ito, H.; et al. Cytomegalovirus promotes murine glioblastoma growth via pericyte recruitment and angiogenesis. J. Clin. Invest. 2019, 130, 1671–1683. [Google Scholar] [CrossRef]
- Baumgarten, P.; Michaelis, M.; Rothweiler, F.; Starzetz, T.; Rabenau, H.F.; Berger, A.; Jennewein, L.; Braczynski, A.K.; Franz, K.; Seifert, V.; et al. Human cytomegalovirus infection in tumor cells of the nervous system is not detectable with standardized pathologico-virological diagnostics. Neuro. Oncol. 2014, 16, 1469–1477. [Google Scholar] [CrossRef]
- Lau, S.K.; Chen, Y.Y.; Chen, W.G.; Diamond, D.J.; Mamelak, A.N.; Zaia, J.A.; Weiss, L.M. Lack of association of cytomegalovirus with human brain tumors. Mod. Pathol. 2005, 18, 838–843. [Google Scholar] [CrossRef] [PubMed]
- Rahbar, A.; Orrego, A.; Peredo, I.; Dzabic, M.; Wolmer-Solberg, N.; Straat, K.; Stragliotto, G.; Soderberg-Naucler, C. Human cytomegalovirus infection levels in glioblastoma multiforme are of prognostic value for survival. J. Clin. Virol. 2013, 57, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.W.; Hellstrand, K.; Larsson, E. Absence of cytomegalovirus in high-coverage DNA sequencing of human glioblastoma multiforme. Int. J. Cancer 2015, 136, 977–981. [Google Scholar] [CrossRef]
- Yang, C.F.; Ho, H.L.; Lin, S.C.; Hsu, C.Y.; Ho, D.M. Detection of human cytomegalovirus in glioblastoma among Taiwanese subjects. PLoS ONE 2017, 12, e0179366. [Google Scholar] [CrossRef] [PubMed]
- Fredericks, D.N.; Relman, D.A. Sequence-based identification of microbial pathogens: A reconsideration of Koch’s postulates. Clin. Microbiol. Rev. 1996, 9, 18–33. [Google Scholar] [CrossRef]
- Soroceanu, L.; Cobbs, C.S. Is HCMV a tumor promoter? Virus Res. 2011, 157, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Herbein, G. The Human Cytomegalovirus, from Oncomodulation to Oncogenesis. Viruses 2018, 10, 408. [Google Scholar] [CrossRef] [PubMed]
- Erlach, K.C.; Reddehase, M.J.; Podlech, J. Mechanism of tumor remission by cytomegalovirus in a murine lymphoma model: Evidence for involvement of virally induced cellular interleukin-15. Med. Microbiol. Immunol. 2015, 204, 355–366. [Google Scholar] [CrossRef]
- Erkes, D.A.; Xu, G.; Daskalakis, C.; Zurbach, K.A.; Wilski, N.A.; Moghbeli, T.; Hill, A.B.; Snyder, C.M. Intratumoral Infection with Murine Cytomegalovirus Synergizes with PD-L1 Blockade to Clear Melanoma Lesions and Induce Long-term Immunity. Mol. Ther. 2016, 24, 1444–1455. [Google Scholar] [CrossRef]
- Wilski, N.A.; Snyder, C.M. From Vaccine Vector to Oncomodulation: Understanding the Complex Interplay between CMV and Cancer. Vaccines 2019, 7, 62. [Google Scholar] [CrossRef]
- Brodin, P.; Jojic, V.; Gao, T.; Bhattacharya, S.; Angel, C.J.; Furman, D.; Shen-Orr, S.; Dekker, C.L.; Swan, G.E.; Butte, A.J.; et al. Variation in the human immune system is largely driven by non-heritable influences. Cell 2015, 160, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Redeker, A.; Remmerswaal, E.B.M.; van der Gracht, E.T.I.; Welten, S.P.M.; Hollt, T.; Koning, F.; Cicin-Sain, L.; Nikolich-Zugich, J.; Ten Berge, I.J.M.; van Lier, R.A.W.; et al. The Contribution of Cytomegalovirus Infection to Immune Senescence Is Set by the Infectious Dose. Front. Immunol. 2017, 8, 1953. [Google Scholar] [CrossRef] [PubMed]
- Marandu, T.F.; Oduro, J.D.; Borkner, L.; Dekhtiarenko, I.; Uhrlaub, J.L.; Drabig, A.; Kroger, A.; Nikolich-Zugich, J.; Cicin-Sain, L. Immune Protection against Virus Challenge in Aging Mice Is Not Affected by Latent Herpesviral Infections. J. Virol. 2015, 89, 11715–11717. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barton, E.S.; White, D.W.; Cathelyn, J.S.; Brett-McClellan, K.A.; Engle, M.; Diamond, M.S.; Miller, V.L.; Virgin, H.W.t. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 2007, 447, 326–329. [Google Scholar] [CrossRef]
- Furman, D.; Jojic, V.; Sharma, S.; Shen-Orr, S.S.; Angel, C.J.; Onengut-Gumuscu, S.; Kidd, B.A.; Maecker, H.T.; Concannon, P.; Dekker, C.L.; et al. Cytomegalovirus infection enhances the immune response to influenza. Sci. Transl. Med. 2015, 7, 281ra243. [Google Scholar] [CrossRef]
- Pinschewer, D.D. Virally vectored vaccine delivery: Medical needs, mechanisms, advantages and challenges. Swiss Med. Wkly. 2017, 147, w14465. [Google Scholar]
- Klinman, D.M.; Shirota, H. Vaccine Adjuvants. In Inflammation: From Molecular and Cellular Mechanisms to the Clinic; Wiley-VCH: Weinheim, Germany, 2017; pp. 143–174. [Google Scholar] [CrossRef]
- Earl, P.L.; Americo, J.L.; Wyatt, L.S.; Eller, L.A.; Whitbeck, J.C.; Cohen, G.H.; Eisenberg, R.J.; Hartmann, C.J.; Jackson, D.L.; Kulesh, D.A.; et al. Immunogenicity of a highly attenuated MVA smallpox vaccine and protection against monkeypox. Nature 2004, 428, 182–185. [Google Scholar] [CrossRef]
- Waibler, Z.; Anzaghe, M.; Ludwig, H.; Akira, S.; Weiss, S.; Sutter, G.; Kalinke, U. Modified vaccinia virus Ankara induces Toll-like receptor-independent type I interferon responses. J. Virol. 2007, 81, 12102–12110. [Google Scholar] [CrossRef]
- Picarda, G.; Benedict, C.A. Cytomegalovirus: Shape-Shifting the Immune System. J. Immunol. 2018, 200, 3881–3889. [Google Scholar] [CrossRef]
- Stempel, M.; Chan, B.; Brinkmann, M.M. Coevolution pays off: Herpesviruses have the license to escape the DNA sensing pathway. Med. Microbiol. Immunol. 2019, 208, 495–512. [Google Scholar] [CrossRef]
- Verma, S.; Benedict, C.A. Sources and signals regulating type I interferon production: lessons learned from cytomegalovirus. J. Interferon Cytokine Res. 2011, 31, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Tegtmeyer, P.K.; Spanier, J.; Borst, K.; Becker, J.; Riedl, A.; Hirche, C.; Ghita, L.; Skerra, J.; Baumann, K.; Lienenklaus, S.; et al. STING induces early IFN-beta in the liver and constrains myeloid cell-mediated dissemination of murine cytomegalovirus. Nat. Commun. 2019, 10, 2830. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.; Loewendorf, A.; De Trez, C.; Fulton, J.; Rhode, A.; Shumway, H.; Ha, S.; Patterson, G.; Pfeffer, K.; Nedospasov, S.A.; et al. Lymphotoxin-mediated crosstalk between B cells and splenic stroma promotes the initial type I interferon response to cytomegalovirus. Cell Host Microbe 2008, 3, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Wang, Q.; Chodaczek, G.; Benedict, C.A. Lymphoid-tissue stromal cells coordinate innate defense to cytomegalovirus. J. Virol. 2013, 87, 6201–6210. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reddehase, M.J. Antigens and immunoevasins: Opponents in cytomegalovirus immune surveillance. Nat. Rev. Immunol. 2002, 2, 831–844. [Google Scholar] [CrossRef]
- Brinkmann, M.M.; Dag, F.; Hengel, H.; Messerle, M.; Kalinke, U.; Cicin-Sain, L. Cytomegalovirus immune evasion of myeloid lineage cells. Med. Microbiol. Immunol. 2015, 204, 367–382. [Google Scholar] [CrossRef]
- Loewendorf, A.; Kruger, C.; Borst, E.M.; Wagner, M.; Just, U.; Messerle, M. Identification of a mouse cytomegalovirus gene selectively targeting CD86 expression on antigen-presenting cells. J. Virol. 2004, 78, 13062–13071. [Google Scholar] [CrossRef]
- Jonjic, S.; Polic, B.; Krmpotic, A. Viral inhibitors of NKG2D ligands: Friends or foes of immune surveillance? Eur J. Immunol. 2008, 38, 2952–2956. [Google Scholar] [CrossRef]
- Goodier, M.R.; Jonjic, S.; Riley, E.M.; Juranic Lisnic, V. CMV and natural killer cells: Shaping the response to vaccination. Eur J. Immunol. 2018, 48, 50–65. [Google Scholar] [CrossRef]
- Slavuljica, I.; Krmpotic, A.; Jonjic, S. Manipulation of NKG2D ligands by cytomegaloviruses: Impact on innate and adaptive immune response. Front. Immunol. 2011, 2, 85. [Google Scholar]
- Snyder, C.M.; Cho, K.S.; Bonnett, E.L.; van Dommelen, S.; Shellam, G.R.; Hill, A.B. Memory inflation during chronic viral infection is maintained by continuous production of short-lived, functional T cells. Immunity 2008, 29, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Tomic, A.; Varanasi, P.R.; Golemac, M.; Malic, S.; Riese, P.; Borst, E.M.; Mischak-Weissinger, E.; Guzman, C.A.; Krmpotic, A.; Jonjic, S.; et al. Activation of Innate and Adaptive Immunity by a Recombinant Human Cytomegalovirus Strain Expressing an NKG2D Ligand. PLoS Pathog. 2016, 12, e1006015. [Google Scholar] [CrossRef] [PubMed]
- Venkatasubramanian, S.; Cheekatla, S.; Paidipally, P.; Tripathi, D.; Welch, E.; Tvinnereim, A.R.; Nurieva, R.; Vankayalapati, R. IL-21-dependent expansion of memory-like NK cells enhances protective immune responses against Mycobacterium tuberculosis. Mucosal Immunol. 2017, 10, 1031–1042. [Google Scholar] [CrossRef] [PubMed]
- Nigro, G.; Adler, S.P. Hyperimmunoglobulin for prevention of congenital cytomegalovirus disease. Clin. Infect. Dis. 2013, 57, S193–S195. [Google Scholar] [CrossRef]
- Revello, M.G.; Lazzarotto, T.; Guerra, B.; Spinillo, A.; Ferrazzi, E.; Kustermann, A.; Guaschino, S.; Vergani, P.; Todros, T.; Frusca, T.; et al. A randomized trial of hyperimmune globulin to prevent congenital cytomegalovirus. N. Engl. J. Med. 2014, 370, 1316–1326. [Google Scholar] [CrossRef]
- Raanani, P.; Gafter-Gvili, A.; Paul, M.; Ben-Bassat, I.; Leibovici, L.; Shpilberg, O. Immunoglobulin prophylaxis in hematopoietic stem cell transplantation: systematic review and meta-analysis. J. Clin. Oncol. 2009, 27, 770–781. [Google Scholar] [CrossRef]
- Ljungman, P.; Hakki, M.; Boeckh, M. Cytomegalovirus in hematopoietic stem cell transplant recipients. Hematol. Oncol. Clin. North Am. 2011, 25, 151–169. [Google Scholar] [CrossRef]
- Martins, J.P.; Andoniou, C.E.; Fleming, P.; Kuns, R.D.; Schuster, I.S.; Voigt, V.; Daly, S.; Varelias, A.; Tey, S.K.; Degli-Esposti, M.A.; et al. Strain-specific antibody therapy prevents cytomegalovirus reactivation after transplantation. Science 2019, 363, 288–293. [Google Scholar] [CrossRef]
- Meyer-König, U.; Ebert, K.; Schrage, B.; Pollak, S.; Hufert, F.T. Simultaneous infection of healthy people with multiple human cytomegalovirus strains. Lancet. 1998, 352, 1280–1281. [Google Scholar] [CrossRef]
- Chou, S.W. Acquisition of donor strains of cytomegalovirus by renal-transplant recipients. N. Engl. J. Med. 1986, 314, 1418–1423. [Google Scholar] [CrossRef]
- Welten, S.P.; Redeker, A.; Toes, R.E.; Arens, R. Viral persistence induces antibody inflation without altering antibody avidity. J. Virol. 2016, 90, 4402–4411. [Google Scholar] [CrossRef] [PubMed]
- Vescovini, R.; Telera, A.R.; Pedrazzoni, M.; Abbate, B.; Rossetti, P.; Verzicco, I.; Arcangeletti, M.C.; Medici, M.C.; Calderaro, A.; Volpi, R.; et al. Impact of Persistent Cytomegalovirus Infection on Dynamic Changes in Human Immune System Profile. PLoS ONE 2016, 11, e0151965. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, M.L.; Shellam, G.R.; Papadimitriou, J.M.; Lawson, M.A. Immunocontraception is induced in BALB/c mice inoculated with murine cytomegalovirus expressing mouse zona pellucida 3. Biol. Reprod. 2003, 68, 2024–2032. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, M.L.; Papadimitriou, J.M.; O’Leary, S.; Robertson, S.A.; Shellam, G.R. Immunoglobulin to zona pellucida 3 mediates ovarian damage and infertility after contraceptive vaccination in mice. J. Autoimmun. 2010, 35, 77–85. [Google Scholar] [CrossRef]
- Tierney, R.; Nakai, T.; Parkins, C.J.; Caposio, P.; Fairweather, N.F.; Sesardic, D.; Jarvis, M.A. A single-dose cytomegalovirus-based vaccine encoding tetanus toxin fragment C induces sustained levels of protective tetanus toxin antibodies in mice. Vaccine 2012, 30, 3047–3052. [Google Scholar] [CrossRef]
- Xu, G.; Smith, T.; Grey, F.; Hill, A.B. Cytomegalovirus-based cancer vaccines expressing TRP2 induce rejection of melanoma in mice. Biochem. Biophys. Res. Commun. 2013, 437, 287–291. [Google Scholar] [CrossRef]
- Cichon, G.; Boeckh-Herwig, S.; Schmidt, H.H.; Wehnes, E.; Muller, T.; Pring-Akerblom, P.; Burger, R. Complement activation by recombinant adenoviruses. Gene Ther. 2001, 8, 1794–1800. [Google Scholar] [CrossRef]
- Buchbinder, S.P.; Mehrotra, D.V.; Duerr, A.; Fitzgerald, D.W.; Mogg, R.; Li, D.; Gilbert, P.B.; Lama, J.R.; Marmor, M.; Del Rio, C.; et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): A double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 2008, 372, 1881–1893. [Google Scholar] [CrossRef]
- Liu, Y.; Mu, R.; Gao, Y.-P.; Dong, J.; Zhu, L.; Ma, Y.; Li, Y.-H.; Zhang, H.-Q.; Han, D.; Zhang, Y. A cytomegalovirus peptide-specific antibody alters natural killer cell homeostasis and is shared in several autoimmune diseases. Cell Host Microbe 2016, 19, 400–408. [Google Scholar] [CrossRef]
- Pardieck, I.N.; Beyrend, G.; Redeker, A.; Arens, R. Cytomegalovirus infection and progressive differentiation of effector-memory T cells. F1000Res 2018, 7. [Google Scholar] [CrossRef]
- Martin, M.D.; Badovinac, V.P. Defining Memory CD8 T Cell. Front. Immunol. 2018, 9, 2692. [Google Scholar] [CrossRef] [PubMed]
- Arens, R.; Wang, P.; Sidney, J.; Loewendorf, A.; Sette, A.; Schoenberger, S.P.; Peters, B.; Benedict, C.A. Cutting edge: Murine cytomegalovirus induces a polyfunctional CD4 T cell response. J. Immunol. 2008, 180, 6472–6476. [Google Scholar] [CrossRef] [PubMed]
- Saeidi, A.; Zandi, K.; Cheok, Y.Y.; Saeidi, H.; Wong, W.F.; Lee, C.Y.Q.; Cheong, H.C.; Yong, Y.K.; Larsson, M.; Shankar, E.M. T-Cell Exhaustion in Chronic Infections: Reversing the State of Exhaustion and Reinvigorating Optimal Protective Immune Responses. Front. Immunol. 2018, 9, 2569. [Google Scholar] [CrossRef] [PubMed]
- Cicin-Sain, L.; Arens, R. Exhaustion and inflation at antipodes of T cell responses to chronic virus infection. Trends Microbiol. 2018, 26, 498–509. [Google Scholar] [CrossRef] [PubMed]
- Baumann, N.S.; Torti, N.; Welten, S.P.M.; Barnstorf, I.; Borsa, M.; Pallmer, K.; Oduro, J.D.; Cicin-Sain, L.; Ikuta, K.; Ludewig, B.; et al. Tissue maintenance of CMV-specific inflationary memory T cells by IL-15. PLoS Pathog. 2018, 14, e1006993. [Google Scholar] [CrossRef] [PubMed]
- Loewendorf, A.I.; Arens, R.; Purton, J.F.; Surh, C.D.; Benedict, C.A. Dissecting the requirements for maintenance of the CMV-specific memory T-cell pool. Viral Immunol. 2011, 24, 351–355. [Google Scholar] [CrossRef]
- Welten, S.P.M.; Baumann, N.S.; Oxenius, A. Fuel and brake of memory T cell inflation. Med. Microbiol. Immunol. 2019, 208, 329–338. [Google Scholar] [CrossRef]
- Torti, N.; Walton, S.M.; Brocker, T.; Rulicke, T.; Oxenius, A. Non-hematopoietic cells in lymph nodes drive memory CD8 T cell inflation during murine cytomegalovirus infection. PLoS Pathog. 2011, 7, e1002313. [Google Scholar] [CrossRef]
- Smith, C.J.; Turula, H.; Snyder, C.M. Systemic hematogenous maintenance of memory inflation by MCMV infection. PLoS Pathog. 2014, 10, e1004233. [Google Scholar] [CrossRef]
- Seckert, C.K.; Schader, S.I.; Ebert, S.; Thomas, D.; Freitag, K.; Renzaho, A.; Podlech, J.; Reddehase, M.J.; Holtappels, R. Antigen-presenting cells of haematopoietic origin prime cytomegalovirus-specific CD8 T-cells but are not sufficient for driving memory inflation during viral latency. J. Gen. Virol. 2011, 92, 1994–2005. [Google Scholar] [CrossRef]
- Hutchinson, S.; Sims, S.; O’Hara, G.; Silk, J.; Gileadi, U.; Cerundolo, V.; Klenerman, P. A dominant role for the immunoproteasome in CD8+ T cell responses to murine cytomegalovirus. PloS One 2011, 6, e14646. [Google Scholar] [CrossRef] [PubMed]
- Waller, E.C.; McKinney, N.; Hicks, R.; Carmichael, A.J.; Sissons, J.G.; Wills, M.R. Differential costimulation through CD137 (4-1BB) restores proliferation of human virus-specific “effector memory” (CD28(-) CD45RA(HI)) CD8(+) T cells. Blood 2007, 110, 4360–4366. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Weiskopf, D.; Gupta, A.; McDonald, B.; Peters, B.; Sette, A.; Benedict, C.A. Cytomegalovirus-Specific CD4 T Cells Are Cytolytic and Mediate Vaccine Protection. J. Virol. 2016, 90, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, E.M.; Remmerswaal, E.B.; Vossen, M.T.; Rowshani, A.T.; Wertheim-van Dillen, P.M.; van Lier, R.A.; ten Berge, I.J. Emergence of a CD4+CD28- granzyme B+, cytomegalovirus-specific T cell subset after recovery of primary cytomegalovirus infection. J. Immunol. 2004, 173, 1834–1841. [Google Scholar] [CrossRef] [PubMed]
- Baumann, N.S.; Welten, S.P.M.; Torti, N.; Pallmer, K.; Borsa, M.; Barnstorf, I.; Oduro, J.D.; Cicin-Sain, L.; Oxenius, A. Early primed KLRG1- CMV-specific T cells determine the size of the inflationary T cell pool. PLoS Pathog. 2019, 15, e1007785. [Google Scholar] [CrossRef]
- Goodrum, F. Human Cytomegalovirus Latency: Approaching the Gordian Knot. Annu. Rev. Virol. 2016, 3, 333–357. [Google Scholar] [CrossRef]
- Reddehase, M.J.; Lemmermann, N.A.W. Cellular reservoirs of latent cytomegaloviruses. Med. Microbiol. Immunol. 2019, 208, 391–403. [Google Scholar] [CrossRef]
- Pomeroy, C.; Hilleren, P.J.; Jordan, M.C. Latent murine cytomegalovirus DNA in splenic stromal cells of mice. J. Virol. 1991, 65, 3330–3334. [Google Scholar]
- Koffron, A.J.; Hummel, M.; Patterson, B.K.; Yan, S.; Kaufman, D.B.; Fryer, J.P.; Stuart, F.P.; Abecassis, M.I. Cellular localization of latent murine cytomegalovirus. J. Virol. 1998, 72, 95–103. [Google Scholar]
- Elder, E.; Sinclair, J. HCMV latency: What regulates the regulators? Med. Microbiol. Immunol. 2019, 1–8. [Google Scholar] [CrossRef]
- Reddehase, M.; Simon, C.; Seckert, C.; Lemmermann, N.; Grzimek, N. Murine model of cytomegalovirus latency and reactivation. In Human Cytomegalovirus; Springer: Berlin, Germany, 2008; pp. 315–331. [Google Scholar]
- Simon, C.O.; Holtappels, R.; Tervo, H.M.; Bohm, V.; Daubner, T.; Oehrlein-Karpi, S.A.; Kuhnapfel, B.; Renzaho, A.; Strand, D.; Podlech, J.; et al. CD8 T cells control cytomegalovirus latency by epitope-specific sensing of transcriptional reactivation. J. Virol. 2006, 80, 10436–10456. [Google Scholar] [CrossRef] [PubMed]
- Shnayder, M.; Nachshon, A.; Krishna, B.; Poole, E.; Boshkov, A.; Binyamin, A.; Maza, I.; Sinclair, J.; Schwartz, M.; Stern-Ginossar, N. Defining the Transcriptional Landscape during Cytomegalovirus Latency with Single-Cell RNA Sequencing. MBio. 2018, 9, e00013–e00018. [Google Scholar] [CrossRef] [PubMed]
- Sims, S.; Bolinger, B.; Klenerman, P. Increasing inflationary T-cell responses following transient depletion of MCMV-specific memory T cells. Eur. J. Immunol. 2015, 45, 113–118. [Google Scholar] [CrossRef] [PubMed]
- DeMarchi, J.M.; Schmidt, C.A.; Kaplan, A.S. Patterns of transcription of human cytomegalovirus in permissively infected cells. J. Virol. 1980, 35, 277–286. [Google Scholar] [PubMed]
- Malone, C.L.; Vesole, D.H.; Stinski, M.F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: Mutational analysis of the viral proteins. J. Virol. 1990, 64, 1498–1506. [Google Scholar]
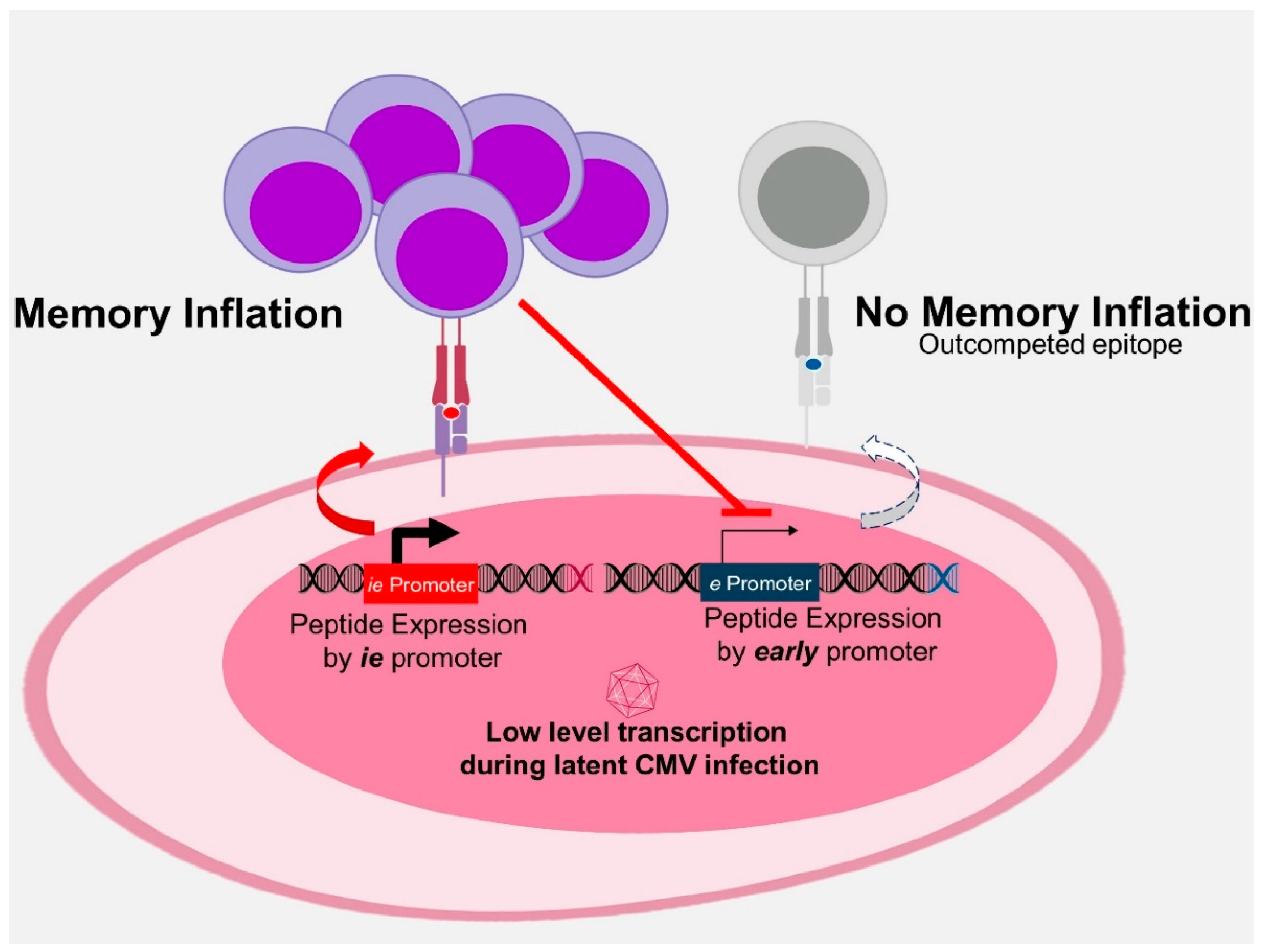
- Grzimek, N.K.; Dreis, D.; Schmalz, S.; Reddehase, M.J. Random, Asynchronous, and Asymmetric Transcriptional Activity of Enhancer-Flanking Major Immediate-Early Genes ie1/3 andie2 during Murine Cytomegalovirus Latency in the Lungs. J. Virol. 2001, 75, 2692–2705. [Google Scholar] [CrossRef]
- Munks, M.W.; Cho, K.S.; Pinto, A.K.; Sierro, S.; Klenerman, P.; Hill, A.B. Four distinct patterns of memory CD8 T cell responses to chronic murine cytomegalovirus infection. J. Immunol. 2006, 177, 450–458. [Google Scholar] [CrossRef]
- Kern, F.; Faulhaber, N.; Frommel, C.; Khatamzas, E.; Prosch, S.; Schonemann, C.; Kretzschmar, I.; Volkmer-Engert, R.; Volk, H.D.; Reinke, P. Analysis of CD8 T cell reactivity to cytomegalovirus using protein-spanning pools of overlapping pentadecapeptides. Eur. J. Immunol. 2000, 30, 1676–1682. [Google Scholar] [CrossRef]
- Jones, S.M.; Ströher, U.; Fernando, L.; Qiu, X.; Alimonti, J.; Melito, P.; Bray, M.; Klenk, H.-D.; Feldmann, H. Assessment of a vesicular stomatitis virus-based vaccine by use of the mouse model of Ebola virus hemorrhagic fever. J. Infect. Dis. 2007, 196, S404–S412. [Google Scholar] [CrossRef]
- Utting, O.; Teh, S.-J.; Teh, H.-S. T Cells Expressing Receptors of Different Affinity for Antigen Ligands Reveal a Unique Role for p59fyn in T Cell Development and Optimal Stimulation of T Cells by Antigen. J. Immunol. 1998, 160, 5410. [Google Scholar]
- Qiu, Z.; Huang, H.; Grenier, J.M.; Perez, O.A.; Smilowitz, H.M.; Adler, B.; Khanna, K.M. Cytomegalovirus-Based Vaccine Expressing a Modified Tumor Antigen Induces Potent Tumor-Specific CD8(+) T-cell Response and Protects Mice from Melanoma. Cancer Immunol. Res. 2015, 3, 536–546. [Google Scholar] [CrossRef] [PubMed]
- Van Stipdonk, M.J.; Badia-Martinez, D.; Sluijter, M.; Offringa, R.; van Hall, T.; Achour, A. Design of agonistic altered peptides for the robust induction of CTL directed towards H-2Db in complex with the melanoma-associated epitope gp100. Cancer Res. 2009, 69, 7784–7792. [Google Scholar] [CrossRef] [PubMed]
- Oscherwitz, J. The promise and challenge of epitope-focused vaccines. Hum. Vaccin. Immunother. 2016, 12, 2113–2116. [Google Scholar] [CrossRef] [PubMed]
- Klyushnenkova, E.N.; Kouiavskaia, D.V.; Parkins, C.J.; Caposio, P.; Botto, S.; Alexander, R.B.; Jarvis, M.A. A cytomegalovirus-based vaccine expressing a single tumor-specific CD8+ T-cell epitope delays tumor growth in a murine model of prostate cancer. J. Immunother. 2012, 35, 390–399. [Google Scholar] [CrossRef]
- McCarthy, M.K.; Weinberg, J.B. The immunoproteasome and viral infection: A complex regulator of inflammation. Front. Microbiol. 2015, 6, 21. [Google Scholar] [CrossRef]
- Del Val, M.; Schlicht, H.J.; Ruppert, T.; Reddehase, M.J.; Koszinowski, U.H. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell 1991, 66, 1145–1153. [Google Scholar] [CrossRef]
- Kincaid, E.Z.; Che, J.W.; York, I.; Escobar, H.; Reyes-Vargas, E.; Delgado, J.C.; Welsh, R.M.; Karow, M.L.; Murphy, A.J.; Valenzuela, D.M. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat. Immunol. 2012, 13, 129. [Google Scholar] [CrossRef]
- Farrington, L.A.; Smith, T.A.; Grey, F.; Hill, A.B.; Snyder, C.M. Competition for antigen at the level of the APC is a major determinant of immunodominance during memory inflation in murine cytomegalovirus infection. J. Immunol. 2013, 190, 3410–3416. [Google Scholar] [CrossRef]
- Holtappels, R.; Simon, C.O.; Munks, M.W.; Thomas, D.; Deegen, P.; Kuhnapfel, B.; Daubner, T.; Emde, S.F.; Podlech, J.; Grzimek, N.K.; et al. Subdominant CD8 T-cell epitopes account for protection against cytomegalovirus independent of immunodomination. J. Virol. 2008, 82, 5781–5796. [Google Scholar] [CrossRef]
- Früh, K. (Vaccine and Gene Therapy Institute, Oregon National Primate Research Center, Oregon Health and Science University, Beaverton, OR, USA). Personal Communication, 2018.
- Murray, S.E.; Nesterenko, P.A.; Vanarsdall, A.L.; Munks, M.W.; Smart, S.M.; Veziroglu, E.M.; Sagario, L.C.; Lee, R.; Claas, F.H.J.; Doxiadis, I.I.N.; et al. Fibroblast-adapted human CMV vaccines elicit predominantly conventional CD8 T cell responses in humans. J. Exp. Med. 2017, 214, 1889–1899. [Google Scholar] [CrossRef]
- Otting, N.; Heijmans, C.M.; Noort, R.C.; de Groot, N.G.; Doxiadis, G.G.; van Rood, J.J.; Watkins, D.I.; Bontrop, R.E. Unparalleled complexity of the MHC class I region in rhesus macaques. Proc. Natl. Acad. Sci. USA 2005, 102, 1626–1631. [Google Scholar] [CrossRef] [PubMed]
- Ranasinghe, S.; Lamothe, P.A.; Soghoian, D.Z.; Kazer, S.W.; Cole, M.B.; Shalek, A.K.; Yosef, N.; Jones, R.B.; Donaghey, F.; Nwonu, C.; et al. Antiviral CD8(+) T Cells Restricted by Human Leukocyte Antigen Class II Exist during Natural HIV Infection and Exhibit Clonal Expansion. Immunity 2016, 45, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Joosten, S.A.; Sullivan, L.C.; Ottenhoff, T.H. Characteristics of HLA-E restricted T-cell responses and their role in infectious diseases. J. Immunol. Res. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, H.R.; Bowyer, G.; Brackenridge, S.; Lambe, T. HLA-E: exploiting pathogen-host interactions for vaccine development. Clin. Exp. Immunol. 2019, 196, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Davis, Z.B.; Cogswell, A.; Scott, H.; Mertsching, A.; Boucau, J.; Wambua, D.; Le Gall, S.; Planelles, V.; Campbell, K.S.; Barker, E. A Conserved HIV-1-Derived Peptide Presented by HLA-E Renders Infected T-cells Highly Susceptible to Attack by NKG2A/CD94-Bearing Natural Killer Cells. PLoS Pathog. 2016, 12, e1005421. [Google Scholar] [CrossRef]
- Walters, L.C.; Harlos, K.; Brackenridge, S.; Rozbesky, D.; Barrett, J.R.; Jain, V.; Walter, T.S.; O’Callaghan, C.A.; Borrow, P.; Toebes, M. Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding. Nat. Commun. 2018, 9, 3137. [Google Scholar] [CrossRef]
- Gamadia, L.E.; Remmerswaal, E.B.; Weel, J.F.; Bemelman, F.; van Lier, R.A.; Ten Berge, I.J. Primary immune responses to human CMV: A critical role for IFN-γ–producing CD4+ T cells in protection against CMV disease. Blood 2003, 101, 2686–2692. [Google Scholar] [CrossRef]
- Komanduri, K.V.; Viswanathan, M.N.; Wieder, E.D.; Schmidt, D.K.; Bredt, B.M.; Jacobson, M.A.; McCune, J.M. Restoration of cytomegalovirus-specific CD4+ T-lymphocyte responses after ganciclovir and highly active antiretroviral therapy in individuals infected with HIV-1. Nat. Med. 1998, 4, 953–956. [Google Scholar] [CrossRef]
- Lilleri, D.; Fornara, C.; Revello, M.G.; Gerna, G. Human cytomegalovirus-specific memory CD8+ and CD4+ T cell differentiation after primary infection. J. Infect. Dis. 2008, 198, 536–543. [Google Scholar] [CrossRef]
- Tu, W.; Chen, S.; Sharp, M.; Dekker, C.; Manganello, A.M.; Tongson, E.C.; Maecker, H.T.; Holmes, T.H.; Wang, Z.; Kemble, G.; et al. Persistent and selective deficiency of CD4+ T cell immunity to cytomegalovirus in immunocompetent young children. J. Immunol. 2004, 172, 3260–3267. [Google Scholar] [CrossRef]
- Walter, E.A.; Greenberg, P.D.; Gilbert, M.J.; Finch, R.J.; Watanabe, K.S.; Thomas, E.D.; Riddell, S.R. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N. Engl. J. Med. 1995, 333, 1038–1044. [Google Scholar] [CrossRef] [PubMed]
- Jonjić, S.; Mutter, W.; Weiland, F.; Reddehase, M.J.; Koszinowski, U.H. Site-restricted persistent cytomegalovirus infection after selective long-term depletion of CD4+ T lymphocytes. J. Exp. Med. 1989, 169, 1199–1212. [Google Scholar] [CrossRef] [PubMed]
- Seder, R.A.; Ahmed, R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat. Immunol. 2003, 4, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, J.R.; Petersen, H.E.; Frederick, D.R.; Morici, L.A.; McLachlan, J.B. Vaccination with a single CD4 T cell peptide epitope from a Salmonella type III-secreted effector protein provides protection against lethal infection. Infect. Immun. 2014, 82, 2424–2433. [Google Scholar] [CrossRef]
- Juno, J.A.; van Bockel, D.; Kent, S.J.; Kelleher, A.D.; Zaunders, J.J.; Munier, C. Cytotoxic CD4 T cells—Friend or foe during viral infection? Front. Immunol. 2017, 8, 19. [Google Scholar] [CrossRef]
- Yao, S.; Huang, D.; Chen, C.Y.; Halliday, L.; Wang, R.C.; Chen, Z.W. CD4+ T cells contain early extrapulmonary tuberculosis (TB) dissemination and rapid TB progression and sustain multieffector functions of CD8+ T and CD3- lymphocytes: Mechanisms of CD4+ T cell immunity. J. Immunol. 2014, 192, 2120–2132. [Google Scholar] [CrossRef]
- Jost, N.H.; Abel, S.; Hutzler, M.; Sparwasser, T.; Zimmermann, A.; Roers, A.; Muller, W.; Klopfleisch, R.; Hengel, H.; Westendorf, A.M.; et al. Regulatory T cells and T-cell-derived IL-10 interfere with effective anti-cytomegalovirus immune response. Immunol. Cell Biol. 2014, 92, 860–871. [Google Scholar] [CrossRef]
- Clement, M.; Marsden, M.; Stacey, M.A.; Abdul-Karim, J.; Gimeno Brias, S.; Costa Bento, D.; Scurr, M.J.; Ghazal, P.; Weaver, C.T.; Carlesso, G.; et al. Cytomegalovirus-Specific IL-10-Producing CD4+ T Cells Are Governed by Type-I IFN-Induced IL-27 and Promote Virus Persistence. PLoS Pathog. 2016, 12, e1006050. [Google Scholar] [CrossRef]
- Jackson, S.E.; Sedikides, G.X.; Okecha, G.; Wills, M.R. Generation, maintenance and tissue distribution of T cell responses to human cytomegalovirus in lytic and latent infection. Med. Microbiol. Immunol. 2019, 208, 375–389. [Google Scholar] [CrossRef]
- Abana, C.O.; Pilkinton, M.A.; Gaudieri, S.; Chopra, A.; McDonnell, W.J.; Wanjalla, C.; Barnett, L.; Gangula, R.; Hager, C.; Jung, D.K. Cytomegalovirus (CMV) Epitope–Specific CD4+ T Cells Are Inflated in HIV+ CMV+ Subjects. J. Immunol. 2017, 199, 3187–3201. [Google Scholar] [CrossRef]
- Pachnio, A.; Ciaurriz, M.; Begum, J.; Lal, N.; Zuo, J.; Beggs, A.; Moss, P. Cytomegalovirus infection leads to development of high frequencies of cytotoxic virus-specific CD4+ T cells targeted to vascular endothelium. PLoS Pathog. 2016, 12, e1005832. [Google Scholar] [CrossRef] [PubMed]
- Redeker, A.; Welten, S.P.; Arens, R. Viral inoculum dose impacts memory T-cell inflation. Eur. J. Immunol. 2014, 44, 1046–1057. [Google Scholar] [CrossRef] [PubMed]
- Oduro, J.D.; Redeker, A.; Lemmermann, N.A.; Ebermann, L.; Marandu, T.F.; Dekhtiarenko, I.; Holzki, J.K.; Busch, D.; Arens, R.; Cicin-Sain, L. Murine cytomegalovirus infection via the intranasal route offers a robust model of immunity upon mucosal CMV infection. J. Gen. Virol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Muruganandah, V.; Sathkumara, H.D.; Navarro, S.; Kupz, A. A systematic review: The role of resident memory T cells in infectious diseases and their relevance for vaccine development. Front. Immunol. 2018, 9, 1574. [Google Scholar] [CrossRef]
- Zens, K.D.; Chen, J.K.; Farber, D.L. Vaccine-generated lung tissue–resident memory T cells provide heterosubtypic protection to influenza infection. JCI Insight 2016, 1, pii: 85832. [Google Scholar] [CrossRef]
- Blanc, C.; Hans, S.; Tran, T.; Granier, C.; Saldman, A.; Anson, M.; Oudard, S.; Tartour, E. Targeting Resident Memory T Cells for Cancer Immunotherapy. Front. Immunol. 2018, 9, 1722. [Google Scholar] [CrossRef]
- Enamorado, M.; Iborra, S.; Priego, E.; Cueto, F.J.; Quintana, J.A.; Martínez-Cano, S.; Mejías-Pérez, E.; Esteban, M.; Melero, I.; Hidalgo, A. Enhanced anti-tumour immunity requires the interplay between resident and circulating memory CD8+ T cells. Nat. Commun. 2017, 8, 16073. [Google Scholar] [CrossRef]
- Szabo, P.A.; Miron, M.; Farber, D.L. Location, location, location: Tissue resident memory T cells in mice and humans. Sci. Immunol. 2019, 4, eaas9673. [Google Scholar] [CrossRef]
- McMichael, A.J.; Picker, L.J. Unusual antigen presentation offers new insight into HIV vaccine design. Curr. Opin. Immunol. 2017, 46, 75–81. [Google Scholar] [CrossRef]
- Fatima, K.; Syed, N.I. Dengvaxia controversy: impact on vaccine hesitancy. J. Glob. Health 2018, 8, 010312. [Google Scholar] [CrossRef]
| Vaccine Candidates | Vector Backbone | Immune Response | Disease Model | Antigen | Design Context | Protection |
|---|---|---|---|---|---|---|
| RhCMV/Pk [17] | RhCMV strain 68-1 | Polyfunctional TEM CD8 T cells. Response to non-canonical epitopes with atypical MHC restriction. | Plasmodium knowlesi sporozoite challenge | Codon-optimized AMA1, CSP, MSP1c or SSP2 under HCMV gH promoter | Inserted in Rh211 | 3/16 were blood-stage free until day 10 post challenge. |
| RhCMV/TB set [18] | Erdman strain Mtb intrabronchial challenge | Polyproteins containing Ag85A, Ag85B, Rv3407, Rv2626, Rv1733, ESAT-6, Ag85B, RpfA, RpfC and RpD under MCMV ie promoter, human EF1a promoter or Rh107 promoter., Rpf A and Rpf D polyprotein under the Rh107 promoter | In place of Rh211 or Rh107 | 14/34 did not have detectable granulomatous disease. | ||
| RhCMV/SIV set [19,20,21,22] | Highly pathogenic SIV intra-rectal challenge | Env, gag, rev-tat-nef (fusion construct), pol1 and pol2 under human EF1a promoter or HCMV gH promoter. | Inserted between rh213 and Rh214 | 12/24 showed rapid control after infection and long-term protection. | ||
| ΔRh110 RhCMV/SIV set [23,24] | Attenuated RhCMV strain 68-1 | 7/13 showed long-lasting absence of SIV viremia | ||||
| RhCMV/EBOV-GP [25] | RhCMV strain 68-1 | IgG responses correlated to protection with no neutralizing antibodies. | EBOV with lethal dose challenge | Codon-optimized EBOV GP | In place of Rh112 | 3/4 survived EBOV challenge with undetectable viremia |
| MCMV/ZEBOV-NPCTL [26,27] | MCMV Smith strain with m157 deletion to avoid NK control | Polyfunctional TEM-biased ‘inflationary’ CD8 responses | Mouse-adapted ZEBOV IP challenge | H2b-restricted T cell epitope from ZEBOV NP | Fused to the C- terminus of ie2 | All were protected against lethal challenge |
| RAE-1γMCMVList [28] | Attenuated RAE-1γMCMV | Protective CD8 T cells | Listeria monocytogenesEGD strain (serovar1/2a) | Listeriolysin O 91–99 CD8 T-cell epitope | In place of the immunodominant m164167–175 | All vaccinated mice survived after challenge. |
| MCMV-85A [29] | Δm1-16-FRT-MCMV: deletion of MHC class I downregulators | CD8 T cell response after AdV vector booster. | Erdman strain Mtb intranasal challenge | Mtb 85A expressed under HCMV ie promoter | In place of backbone deletion | Lung mycobacterial load reduction |
| MCMV-M [30] | MCMV K181 strain with m157 deletion [31] | IN route induced inflationary CD8 TEM and TRM in the lungs. | RSV IN challenge | RSV M protein | Inserted in ie2 | IN route had lower lung viral loads than IP. MCMV-M2 failed to mediate early RSV control. |
| MCMV-M2 [32] | IN route induced a non-inflationary response. | RSV M2 protein | ||||
| MCMVie2SL MCMVM45SL [33] | MCK2-repaired MCMV Smith Strain [34] | MCMVie2SL Inflationary CD8 T cell response MCMVM45SL Non-inflationary CD8 T cell response | HSV-1 challenge on susceptible 129/Sv mice | Kb-restricted peptide from HSV-1 glycoprotein B498–505 | MCMVie2SL in-frame fusion to the C-terminus of ie2 MCMVM45SL in the 3′ end of M45 | No viral detection by day 7 post-challenge in brains and lungs |
| (HA)-MCMVIVL [35] | IN route induced lung CD8 TRM accumulation | IAV PR8M variant IN challenge | MHC-I restricted peptide IVL533–541 | Inserted into the C-terminus of the ie2 | IN reduced lung viral load and weight loss at a higher magnitude than IP route | |
| MCMVie2E6/E7Full [14] | Protective CD8 T cells | Heterotopic (subcutaneous) administration of TC-1 cells transformed with HPV16 E6 and E7-oncogenes | Full-length HPV16 E6 and E7 | Antigen fused to the C-terminus of ie2 | Transient limitation of tumour cell growth | |
| MCMVIE2E7 [14] | MHC-I restricted HPV16 E749–57 epitope | Epitope fused to the C-terminus of ie2 | No tumour cell growth upon challenge | |||
| MCMV-M79-FKBP-E7 [36] | MCMV Smith Strain with FKBP-mediated destabilization of the essential M79 gene |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ynga-Durand, M.A.; Dekhtiarenko, I.; Cicin-Sain, L. Vaccine Vectors Harnessing the Power of Cytomegaloviruses. Vaccines 2019, 7, 152. https://doi.org/10.3390/vaccines7040152
Ynga-Durand MA, Dekhtiarenko I, Cicin-Sain L. Vaccine Vectors Harnessing the Power of Cytomegaloviruses. Vaccines. 2019; 7(4):152. https://doi.org/10.3390/vaccines7040152
Chicago/Turabian StyleYnga-Durand, Mario Alberto, Iryna Dekhtiarenko, and Luka Cicin-Sain. 2019. "Vaccine Vectors Harnessing the Power of Cytomegaloviruses" Vaccines 7, no. 4: 152. https://doi.org/10.3390/vaccines7040152
APA StyleYnga-Durand, M. A., Dekhtiarenko, I., & Cicin-Sain, L. (2019). Vaccine Vectors Harnessing the Power of Cytomegaloviruses. Vaccines, 7(4), 152. https://doi.org/10.3390/vaccines7040152





