Nanoparticle Vaccines for Inducing HIV-1 Neutralizing Antibodies
Abstract
1. Introduction
2. Env as a Broadly Neutralizing Antibody Target
3. Nanoparticles and Immunity
4. Nanoparticle Platforms
4.1. Virus-Like Particles
4.2. Liposomes
4.3. Self-Assembling Protein Nanoparticles
5. Accessibility of Neutralizing Epitopes on Env Nanoparticles
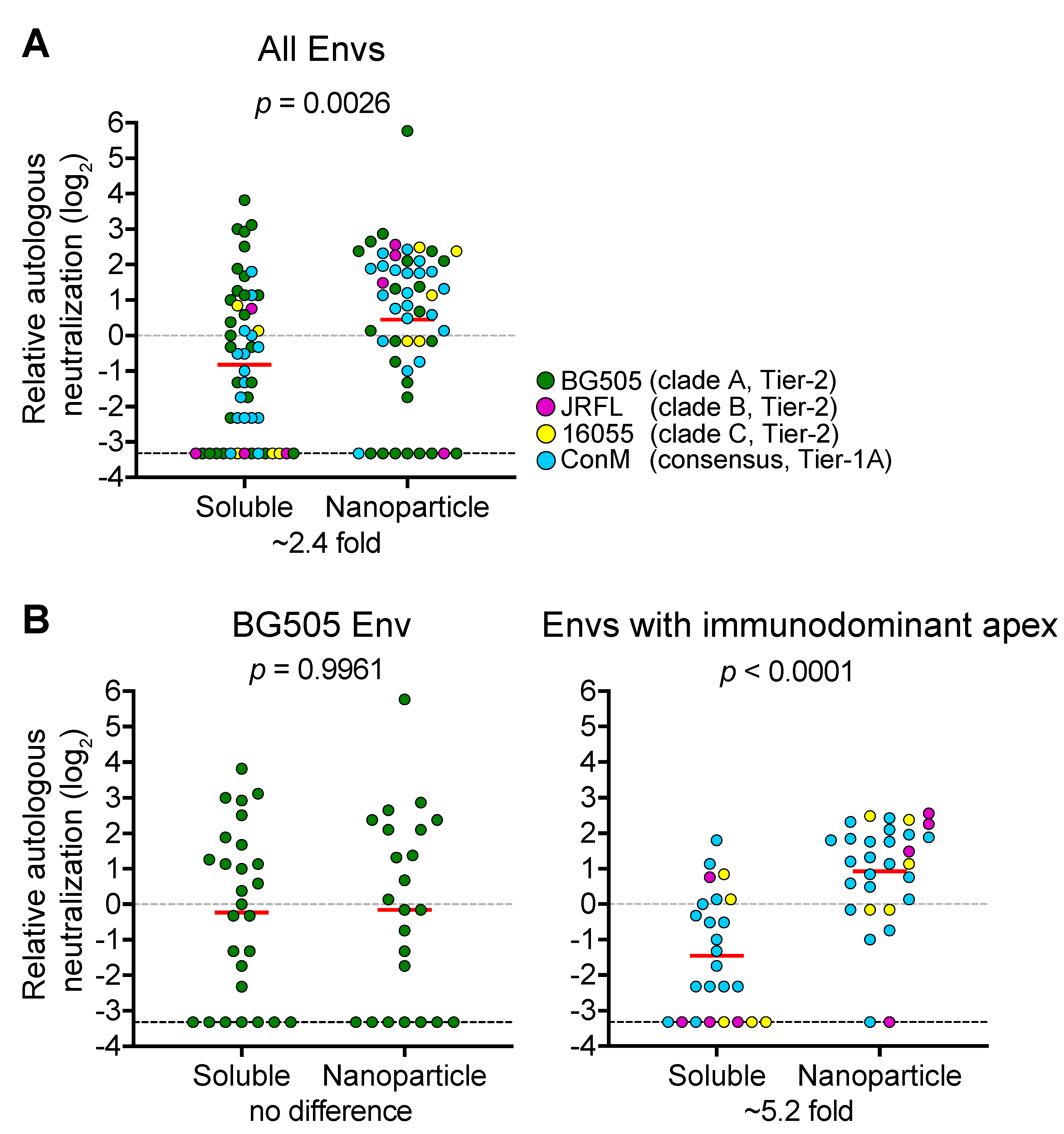
6. Epitope Focusing Using Nanoparticles
7. Anti-NP Responses
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- UNAIDS Fact Sheet—Latest Global and Regional Statistics on the Status of the AIDS Epidemic. Available online: http://www.unaids.org/sites/default/files/media_asset/UNAIDS_FactSheet_en.pdf (accessed on 19 March 2019).
- Fauci, A.S.; Folkers, G.K.; Marston, H.D. Ending the Global HIV/AIDS Pandemic: The Critical Role of an HIV Vaccine. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2014, 59, S80–S84. [Google Scholar] [CrossRef] [PubMed]
- Checkley, M.A.; Luttge, B.G.; Freed, E.O. HIV-1 Envelope Glycoprotein Biosynthesis, Trafficking, and Incorporation. J. Mol. Biol. 2011, 410, 582–608. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Bartesaghi, A.; Borgnia, M.J.; Sapiro, G.; Subramaniam, S. Molecular architecture of native HIV-1 gp 120 trimers. Nature 2008, 455, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Blumenthal, R.; Durell, S.; Viard, M. HIV Entry and Envelope Glycoprotein-mediated Fusion. J. Biol. Chem. 2012, 287, 40841–40849. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R.W.; Moore, J.P. HIV: A stamp on the envelope. Nature 2014, 514, 437–438. [Google Scholar] [CrossRef] [PubMed]
- Araújo, L.A.L.; Almeida, S.E.M. HIV-1 Diversity in the Envelope Glycoproteins: Implications for Viral Entry Inhibition. Viruses 2013, 5, 595–604. [Google Scholar] [CrossRef] [PubMed]
- McCoy, L.E.; van Gils, M.J.; Ozorowski, G.; Messmer, T.; Briney, B.; Voss, J.E.; Kulp, D.W.; Macauley, M.S.; Sok, D.; Pauthner, M.; et al. Holes in the glycan shield of the native HIV envelope are a target of trimer-elicited neutralizing antibodies. Cell Rep. 2016, 16, 2327–2338. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Doria-Rose, N.A.; Cheng, C.; Stewart-Jones, G.B.E.; Chuang, G.-Y.; Chambers, M.; Druz, A.; Geng, H.; McKee, K.; Kwon, Y.D.; et al. Quantification of the Impact of the HIV-1-Glycan Shield on Antibody Elicitation. Cell Rep. 2017, 19, 719–732. [Google Scholar] [CrossRef]
- Moore, P.L.; Crooks, E.T.; Porter, L.; Zhu, P.; Cayanan, C.S.; Grise, H.; Corcoran, P.; Zwick, M.B.; Franti, M.; Morris, L.; et al. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 2006, 80, 2515–2528. [Google Scholar] [CrossRef]
- Munro, J.B.; Gorman, J.; Ma, X.; Zhou, Z.; Arthos, J.; Burton, D.R.; Koff, W.C.; Courter, J.R.; Smith, A.B.; Kwong, P.D.; et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 2014, 346, 759–763. [Google Scholar] [CrossRef]
- Sok, D.; Burton, D.R. Recent progress in broadly neutralizing antibodies to HIV. Nat. Immunol. 2018, 19, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Van Gils, M.J.; Sanders, R.W. In vivo protection by broadly neutralizing HIV antibodies. Trends Microbiol. 2014, 22, 550–551. [Google Scholar] [CrossRef] [PubMed]
- Sliepen, K.; Sanders, R.W. HIV-1 envelope glycoprotein immunogens to induce broadly neutralizing antibodies. Expert Rev. Vaccines 2016, 15, 349–365. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R.W.; Moore, J.P. Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol. Rev. 2017, 275, 161–182. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Ozorowski, G.; Ward, A.B. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 2016, 351, 1043–1048. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R.W.; Derking, R.; Cupo, A.; Julien, J.-P.; Yasmeen, A.; de Val, N.; Kim, H.J.; Blattner, C.; de la Peña, A.T.; Korzun, J.; et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013, 9, e1003618. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R.W.; van Gils, M.J.; Derking, R.; Sok, D.; Ketas, T.J.; Burger, J.A.; Ozorowski, G.; Cupo, A.; Simonich, C.; Goo, L.; et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 2015, 349, aac4223. [Google Scholar] [CrossRef]
- De Taeye, S.W.; Moore, J.P.; Sanders, R.W. HIV-1 Envelope Trimer Design and Immunization Strategies To Induce Broadly Neutralizing Antibodies. Trends Immunol. 2016, 37, 221–232. [Google Scholar] [CrossRef]
- Crooks, E.T.; Tong, T.; Chakrabarti, B.; Narayan, K.; Georgiev, I.S.; Menis, S.; Huang, X.; Kulp, D.; Osawa, K.; Muranaka, J.; et al. Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site. PLoS Pathog. 2015, 11, e1004932. [Google Scholar] [CrossRef]
- del Moral-Sánchez, I.; Sliepen, K. Strategies for inducing broad, potent and long-lasting neutralizing antibody responses against HIV-1. Expert Rev. Vaccines. Submitted.
- Zhao, Q.; Li, S.; Yu, H.; Xia, N.; Modis, Y. Virus-like particle-based human vaccines: Quality assessment based on structural and functional properties. Trends Biotechnol. 2013, 31, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Kelly, H.G.; Kent, S.J.; Wheatley, A.K. Immunological basis for enhanced immunity of nanoparticle vaccines. Expert Rev. Vaccines 2019, 18, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Schiller, J.; Chackerian, B. Why HIV virions have low numbers of envelope spikes: Implications for vaccine development. PLoS Pathog. 2014, 10, e1004254. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.F.; Jennings, G.T. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010, 10, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Victora, G.D.; Nussenzweig, M.C. Germinal centers. Annu. Rev. Immunol. 2012, 30, 429–457. [Google Scholar] [CrossRef] [PubMed]
- Abbott, R.K.; Lee, J.H.; Menis, S.; Skog, P.; Rossi, M.; Ota, T.; Kulp, D.W.; Bhullar, D.; Kalyuzhniy, O.; Havenar-Daughton, C.; et al. Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity 2018, 48, 133–146.e6. [Google Scholar] [CrossRef] [PubMed]
- Roozendaal, R.; Mempel, T.R.; Pitcher, L.A.; Gonzalez, S.F.; Verschoor, A.; Mebius, R.E.; von Andrian, U.H.; Carroll, M.C. Conduits mediate transport of low-molecular-weight antigen to lymph node follicles. Immunity 2009, 30, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Tokatlian, T.; Read, B.J.; Jones, C.A.; Kulp, D.W.; Menis, S.; Chang, J.Y.H.; Steichen, J.M.; Kumari, S.; Allen, J.D.; Dane, E.L.; et al. Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science 2019, 363, 649–654. [Google Scholar] [CrossRef]
- Dobrovolskaia, M.A.; Aggarwal, P.; Hall, J.B.; McNeil, S.E. Preclinical studies to understand nanoparticle interaction with the immune system and its potential effects on nanoparticle biodistribution. Mol. Pharm. 2008, 5, 487–495. [Google Scholar] [CrossRef]
- Manolova, V.; Flace, A.; Bauer, M.; Schwarz, K.; Saudan, P.; Bachmann, M.F. Nanoparticles target distinct dendritic cell populations according to their size. Eur. J. Immunol. 2008, 38, 1404–1413. [Google Scholar] [CrossRef]
- Nishioka, Y.; Yoshino, H. Lymphatic targeting with nanoparticulate system. Adv. Drug Deliv. Rev. 2001, 47, 55–64. [Google Scholar] [CrossRef]
- Joshi, V.B.; Geary, S.M.; Salem, A.K. Biodegradable Particles as Vaccine Delivery Systems: Size Matters. AAPS J. 2012, 15, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S.T.; van der Vlies, A.J.; Simeoni, E.; Angeli, V.; Randolph, G.J.; O’Neil, C.P.; Lee, L.K.; Swartz, M.A.; Hubbell, J.A. Exploiting lymphatic transport and complement activation in nanoparticle vaccines. Nat. Biotechnol. 2007, 25, 1159–1164. [Google Scholar] [CrossRef]
- Vogelstein, B.; Dintzis, R.Z.; Dintzis, H.M. Specific cellular stimulation in the primary immune response: A quantized model. Proc. Natl. Acad. Sci. USA 1982, 79, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.F.; Zinkernagel, R.M. Neutralizing antiviral B cell responses. Annu. Rev. Immunol. 1997, 15, 235–270. [Google Scholar] [CrossRef] [PubMed]
- Dintzis, H.M.; Dintzis, R.Z.; Vogelstein, B. Molecular determinants of immunogenicity: The immunon model of immune response. Proc. Natl. Acad. Sci. USA 1976, 73, 3671–3675. [Google Scholar] [CrossRef] [PubMed]
- Francica, J.R.; Laga, R.; Lynn, G.M.; Mužíková, G.; Androvič, L.; Aussedat, B.; Walkowicz, W.E.; Padhan, K.; Ramirez-Valdez, R.A.; Parks, R.; et al. Star nanoparticles delivering HIV-1 peptide minimal immunogens elicit near-native envelope antibody responses in nonhuman primates. PLoS Biol. 2019, 17, e3000328. [Google Scholar] [CrossRef] [PubMed]
- Marcandalli, J.; Fiala, B.; Ols, S.; Perotti, M.; de van der Schueren, W.; Snijder, J.; Hodge, E.; Benhaim, M.; Ravichandran, R.; Carter, L.; et al. Induction of Potent Neutralizing Antibody Responses by a Designed Protein Nanoparticle Vaccine for Respiratory Syncytial Virus. Cell 2019, 176, 1420–1431.e17. [Google Scholar] [CrossRef]
- Naskalska, A.; Pyrć, K. Virus Like Particles as Immunogens and Universal Nanocarriers. Pol. J. Microbiol. 2015, 64, 3–13. [Google Scholar]
- Zhu, P.; Liu, J.; Bess, J.; Chertova, E.; Lifson, J.D.; Grisé, H.; Ofek, G.A.; Taylor, K.A.; Roux, K.H. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 2006, 441, 847–852. [Google Scholar] [CrossRef]
- Crooks, E.T.; Tong, T.; Osawa, K.; Binley, J.M. Enzyme digests eliminate nonfunctional Env from HIV-1 particle surfaces, leaving native Env trimers intact and viral infectivity unaffected. J. Virol. 2011, 85, 5825–5839. [Google Scholar] [CrossRef] [PubMed]
- Crooks, E.T.; Osawa, K.; Tong, T.; Grimley, S.L.; Dai, Y.D.; Whalen, R.G.; Kulp, D.W.; Menis, S.; Schief, W.R.; Binley, J.M. Effects of Partially Dismantling the CD4 Binding Site Glycan Fence of HIV-1 Envelope Glycoprotein Trimers on Neutralizing Antibody Induction. Virology 2017, 505, 193–209. [Google Scholar] [CrossRef] [PubMed]
- Stano, A.; Leaman, D.P.; Kim, A.S.; Zhang, L.; Autin, L.; Ingale, J.; Gift, S.K.; Truong, J.; Wyatt, R.T.; Olson, A.J.; et al. Dense Array of Spikes on HIV-1 Virion Particles. J. Virol. 2017, 91, e00415-17. [Google Scholar] [CrossRef] [PubMed]
- Bulbake, U.; Doppalapudi, S.; Kommineni, N.; Khan, W. Liposomal Formulations in Clinical Use: An Updated Review. Pharmaceutics 2017, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Kang, B.H.; Pancera, M.; Lee, J.H.; Tong, T.; Feng, Y.; Imamichi, H.; Georgiev, I.S.; Chuang, G.-Y.; Druz, A.; et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 2014, 515, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Muster, T.; Steindl, F.; Purtscher, M.; Trkola, A.; Klima, A.; Himmler, G.; Rüker, F.; Katinger, H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 1993, 67, 6642–6647. [Google Scholar] [PubMed]
- Zwick, M.B.; Labrijn, A.F.; Wang, M.; Spenlehauer, C.; Saphire, E.O.; Binley, J.M.; Moore, J.P.; Stiegler, G.; Katinger, H.; Burton, D.R.; et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 2001, 75, 10892–10905. [Google Scholar] [CrossRef]
- Matyas, G.R.; Wieczorek, L.; Beck, Z.; Ochsenbauer-Jambor, C.; Kappes, J.C.; Michael, N.L.; Polonis, V.R.; Alving, C.R. Neutralizing antibodies induced by liposomal HIV-1 glycoprotein 41 peptide simultaneously bind to both the 2F5 or 4E10 epitope and lipid epitopes. AIDS Lond. Engl. 2009, 23, 2069–2077. [Google Scholar] [CrossRef]
- Hanson, M.C.; Abraham, W.; Crespo, M.P.; Chen, S.H.; Liu, H.; Szeto, G.L.; Kim, M.; Reinherz, E.L.; Irvine, D.J. Liposomal vaccines incorporating molecular adjuvants and intrastructural T-cell help promote the immunogenicity of HIV membrane-proximal external region peptides. Vaccine 2015, 33, 861–868. [Google Scholar] [CrossRef]
- Moon, J.J.; Suh, H.; Bershteyn, A.; Stephan, M.T.; Liu, H.; Huang, B.; Sohail, M.; Luo, S.; Um, S.H.; Khant, H.; et al. Interbilayer-crosslinked multilamellar vesicles as synthetic vaccines for potent humoral and cellular immune responses. Nat. Mater. 2011, 10, 243–251. [Google Scholar] [CrossRef]
- Moon, J.J.; Suh, H.; Li, A.V.; Ockenhouse, C.F.; Yadava, A.; Irvine, D.J. Enhancing humoral responses to a malaria antigen with nanoparticle vaccines that expand Tfh cells and promote germinal center induction. Proc. Natl. Acad. Sci. USA 2012, 109, 1080–1085. [Google Scholar] [CrossRef] [PubMed]
- Pejawar-Gaddy, S.; Kovacs, J.M.; Barouch, D.H.; Chen, B.; Irvine, D.J. Design of Lipid Nanocapsule Delivery Vehicles for Multivalent Display of Recombinant Env Trimers in HIV Vaccination. Bioconjug. Chem. 2014, 25, 1470–1478. [Google Scholar] [CrossRef] [PubMed]
- Steichen, J.M.; Kulp, D.W.; Tokatlian, T.; Escolano, A.; Dosenovic, P.; Stanfield, R.L.; McCoy, L.E.; Ozorowski, G.; Hu, X.; Kalyuzhniy, O.; et al. HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies. Immunity 2016, 45, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Bale, S.; Goebrecht, G.; Stano, A.; Wilson, R.; Ota, T.; Tran, K.; Ingale, J.; Zwick, M.B.; Wyatt, R.T. Covalent Linkage of HIV-1 Trimers to Synthetic Liposomes Elicits Improved B Cell and Antibody Responses. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Ingale, J.; Stano, A.; Guenaga, J.; Sharma, S.K.; Nemazee, D.; Zwick, M.B.; Wyatt, R.T. High-density array of well-ordered HIV-1 spikes on synthetic liposomal nanoparticles efficiently activate B cells. Cell Rep. 2016, 15, 1986–1999. [Google Scholar] [CrossRef]
- Tokatlian, T.; Kulp, D.W.; Mutafyan, A.A.; Jones, C.A.; Menis, S.; Georgeson, E.; Kubitz, M.; Zhang, M.H.; Melo, M.B.; Silva, M.; et al. Enhancing Humoral Responses Against HIV Envelope Trimers via Nanoparticle Delivery with Stabilized Synthetic Liposomes. Sci. Rep. 2018, 8, 16527. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.K.; Crampton, J.C.; Cupo, A.; Ketas, T.; van Gils, M.J.; Sliepen, K.; de Taeye, S.W.; Sok, D.; Ozorowski, G.; Deresa, I.; et al. Murine Antibody Responses to Cleaved Soluble HIV-1 Envelope Trimers Are Highly Restricted in Specificity. J. Virol. 2015, 89, 10383–10398. [Google Scholar] [CrossRef]
- Martinez-Murillo, P.; Tran, K.; Guenaga, J.; Lindgren, G.; Àdori, M.; Feng, Y.; Phad, G.E.; Vázquez Bernat, N.; Bale, S.; Ingale, J.; et al. Particulate Array of Well-Ordered HIV Clade C Env Trimers Elicits Neutralizing Antibodies that Display a Unique V2 Cap Approach. Immunity 2017, 46, 804–817.e7. [Google Scholar] [CrossRef]
- Pauthner, M.; Havenar-Daughton, C.; Sok, D.; Nkolola, J.P.; Bastidas, R.; Boopathy, A.V.; Carnathan, D.G.; Chandrashekar, A.; Cirelli, K.M.; Cottrell, C.A.; et al. Elicitation of Robust Tier 2 Neutralizing Antibody Responses in Nonhuman Primates by HIV Envelope Trimer Immunization Using Optimized Approaches. Immunity 2017, 46, 1073–1088.e6. [Google Scholar] [CrossRef]
- López-Sagaseta, J.; Malito, E.; Rappuoli, R.; Bottomley, M.J. Self-assembling protein nanoparticles in the design of vaccines. Comput. Struct. Biotechnol. J. 2015, 14, 58–68. [Google Scholar] [CrossRef]
- Bale, J.B.; Gonen, S.; Liu, Y.; Sheffler, W.; Ellis, D.; Thomas, C.; Cascio, D.; Yeates, T.O.; Gonen, T.; King, N.P.; et al. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 2016, 353, 389–394. [Google Scholar] [CrossRef] [PubMed]
- King, N.P.; Lai, Y.-T. Practical approaches to designing novel protein assemblies. Curr. Opin. Struct. Biol. 2013, 23, 632–638. [Google Scholar] [CrossRef] [PubMed]
- King, N.P.; Sheffler, W.; Sawaya, M.R.; Vollmar, B.S.; Sumida, J.P.; André, I.; Gonen, T.; Yeates, T.O.; Baker, D. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science 2012, 336, 1171–1174. [Google Scholar] [CrossRef] [PubMed]
- Malay, A.D.; Miyazaki, N.; Biela, A.; Chakraborti, S.; Majsterkiewicz, K.; Stupka, I.; Kaplan, C.S.; Kowalczyk, A.; Piette, B.M.A.G.; Hochberg, G.K.A.; et al. An ultra-stable gold-coordinated protein cage displaying reversible assembly. Nature 2019, 569, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Kanekiyo, M.; Wei, C.-J.; Yassine, H.M.; McTamney, P.M.; Boyington, J.C.; Whittle, J.R.R.; Rao, S.S.; Kong, W.-P.; Wang, L.; Nabel, G.J. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 2013, 499, 102–106. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Cheng, Y.; Kong, L.; Azadnia, P.; Giang, E.; Kim, J.; Wood, M.R.; Wilson, I.A.; Law, M.; Zhu, J. Approaching rational epitope vaccine design for hepatitis C virus with meta-server and multivalent scaffolding. Sci. Rep. 2015, 5, 12501. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wang, X.; Lou, P.; Hu, Z.; Qu, P.; Li, D.; Li, Q.; Xu, Y.; Niu, J.; He, Y.; et al. A nanoparticle-based HCV vaccine with enhanced potency. J. Infect. Dis. 2019. [Google Scholar] [CrossRef] [PubMed]
- Kanekiyo, M.; Bu, W.; Joyce, M.G.; Meng, G.; Whittle, J.R.R.; Baxa, U.; Yamamoto, T.; Narpala, S.; Todd, J.-P.; Rao, S.S.; et al. Rational Design of an Epstein-Barr Virus Vaccine Targeting the Receptor-Binding Site. Cell 2015, 162, 1090–1100. [Google Scholar] [CrossRef] [PubMed]
- Georgiev, I.S.; Joyce, M.G.; Chen, R.E.; Leung, K.; McKee, K.; Druz, A.; Van Galen, J.G.; Kanekiyo, M.; Tsybovsky, Y.; Yang, E.S.; et al. Two-Component Ferritin Nanoparticles for Multimerization of Diverse Trimeric Antigens. ACS Infect. Dis. 2018, 4, 788–796. [Google Scholar] [CrossRef]
- He, L.; Kumar, S.; Allen, J.D.; Huang, D.; Lin, X.; Mann, C.J.; Saye-Francisco, K.L.; Copps, J.; Sarkar, A.; Blizard, G.S.; et al. HIV-1 vaccine design through minimizing envelope metastability. Sci. Adv. 2018, 4, eaau6769. [Google Scholar] [CrossRef]
- Sliepen, K.; Ozorowski, G.; Burger, J.A.; van Montfort, T.; Stunnenberg, M.; LaBranche, C.; Montefiori, D.C.; Moore, J.P.; Ward, A.B.; Sanders, R.W. Presenting native-like HIV-1 envelope trimers on ferritin nanoparticles improves their immunogenicity. Retrovirology 2015, 12, 82. [Google Scholar] [CrossRef] [PubMed]
- He, L.; de Val, N.; Morris, C.D.; Vora, N.; Thinnes, T.C.; Kong, L.; Azadnia, P.; Sok, D.; Zhou, B.; Burton, D.R.; et al. Presenting native-like trimeric HIV-1 antigens with self-assembling nanoparticles. Nat. Commun. 2016, 7, 12041. [Google Scholar] [CrossRef] [PubMed]
- Sliepen, K.; Han, B.W.; Bontjer, I.; Mooij, P.; Garces, F.; Behrens, A.-J.; Rantalainen, K.; Kumar, S.; Sarkar, A.; Brouwer, P.J.M.; et al. Structure and immunogenicity of a stabilized HIV-1 envelope trimer based on a group-M consensus sequence. Nat. Commun. 2019, 10, 2355. [Google Scholar] [CrossRef] [PubMed]
- Jardine, J.; Julien, J.-P.; Menis, S.; Ota, T.; Kalyuzhniy, O.; McGuire, A.; Sok, D.; Huang, P.-S.; MacPherson, S.; Jones, M.; et al. Rational HIV immunogen design to target specific germline B cell receptors. Science 2013, 340, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Jardine, J.G.; Ota, T.; Sok, D.; Pauthner, M.; Kulp, D.W.; Kalyuzhniy, O.; Skog, P.D.; Thinnes, T.C.; Bhullar, D.; Briney, B.; et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 2015, 349, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Havenar-Daughton, C.; Sarkar, A.; Kulp, D.W.; Toy, L.; Hu, X.; Deresa, I.; Kalyuzhniy, O.; Kaushik, K.; Upadhyay, A.A.; Menis, S.; et al. The human naive B cell repertoire contains distinct subclasses for a germline-targeting HIV-1 vaccine immunogen. Sci. Transl. Med. 2018, 10, eaat0381. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, P.J.M.; Sanders, R.W. Presentation of HIV-1 envelope glycoprotein trimers on diverse nanoparticle platforms. Curr. Opin. HIV AIDS 2019, 14, 302–308. [Google Scholar] [CrossRef]
- Brune, K.D.; Leneghan, D.B.; Brian, I.J.; Ishizuka, A.S.; Bachmann, M.F.; Draper, S.J.; Biswas, S.; Howarth, M. Plug-and-Display: Decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci. Rep. 2016, 6, 19234. [Google Scholar] [CrossRef]
- Escolano, A.; Gristick, H.B.; Abernathy, M.E.; Merkenschlager, J.; Gautam, R.; Oliveira, T.Y.; Pai, J.; West, A.P.; Barnes, C.O.; Cohen, A.A.; et al. Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques. Nature 2019, 570, 468–473. [Google Scholar] [CrossRef]
- Brouwer, P.J.M.; Antanasijevic, A.; Berndsen, Z.; Yasmeen, A.; Fiala, B.; Bijl, T.P.L.; Bontjer, I.; Bale, J.B.; Sheffler, W.; Allen, J.D.; et al. Enhancing and shaping the immunogenicity of native-like HIV-1 envelope trimers by presentation on a designed two-component protein nanoparticle. Nat. Commun. Submitted.
- Bianchi, M.; Turner, H.L.; Nogal, B.; Cottrell, C.A.; Oyen, D.; Pauthner, M.; Bastidas, R.; Nedellec, R.; McCoy, L.E.; Wilson, I.A.; et al. Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization. Immunity 2018, 49, 288–300.e8. [Google Scholar] [CrossRef] [PubMed]
- Klasse, P.J.; Ketas, T.J.; Cottrell, C.A.; Ozorowski, G.; Debnath, G.; Camara, D.; Francomano, E.; Pugach, P.; Ringe, R.P.; LaBranche, C.C.; et al. Epitopes for neutralizing antibodies induced by HIV-1 envelope glycoprotein BG505 SOSIP trimers in rabbits and macaques. PLoS Pathog. 2018, 14, e1006913. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Yang, Y.R.; Tran, K.; Wang, Y.; Chiang, C.-I.; Ozorowski, G.; Xiao, Y.; Ward, A.B.; Wyatt, R.T.; Li, Y. The HIV-1 Envelope Glycoprotein C3/V4 Region Defines a Prevalent Neutralization Epitope following Immunization. Cell Rep. 2019, 27, 586–598.e6. [Google Scholar] [CrossRef] [PubMed]
- Hoot, S.; McGuire, A.T.; Cohen, K.W.; Strong, R.K.; Hangartner, L.; Klein, F.; Diskin, R.; Scheid, J.F.; Sather, D.N.; Burton, D.R.; et al. Recombinant HIV Envelope Proteins Fail to Engage Germline Versions of Anti-CD4bs bNAbs. PLoS Pathog. 2013, 9, e1003106. [Google Scholar] [CrossRef]
- Sliepen, K.; Medina-Ramírez, M.; Yasmeen, A.; Moore, J.P.; Klasse, P.J.; Sanders, R.W. Binding of inferred germline precursors of broadly neutralizing HIV-1 antibodies to native-like envelope trimers. Virology 2015, 486, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Medina-Ramírez, M.; Garces, F.; Escolano, A.; Skog, P.; de Taeye, S.W.; Del Moral-Sanchez, I.; McGuire, A.T.; Yasmeen, A.; Behrens, A.-J.; Ozorowski, G.; et al. Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo. J. Exp. Med. 2017, 214, 2573–2590. [Google Scholar] [CrossRef] [PubMed]
- Escolano, A.; Steichen, J.M.; Dosenovic, P.; Kulp, D.W.; Golijanin, J.; Sok, D.; Freund, N.T.; Gitlin, A.D.; Oliveira, T.; Araki, T.; et al. Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice. Cell 2016, 166, 1445–1458.e12. [Google Scholar] [CrossRef]
- Haynes, B.F.; Kelsoe, G.; Harrison, S.C.; Kepler, T.B. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotechnol. 2012, 30, 423–433. [Google Scholar] [CrossRef]
- Xiao, X.; Chen, W.; Feng, Y.; Zhu, Z.; Prabakaran, P.; Wang, Y.; Zhang, M.-Y.; Longo, N.S.; Dimitrov, D.S. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: Implications for evasion of immune responses and design of vaccine immunogens. Biochem. Biophys. Res. Commun. 2009, 390, 404–409. [Google Scholar] [CrossRef]
- McGuire, A.T.; Gray, M.D.; Dosenovic, P.; Gitlin, A.D.; Freund, N.T.; Petersen, J.; Correnti, C.; Johnsen, W.; Kegel, R.; Stuart, A.B.; et al. Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nat. Commun. 2016, 7, 10618. [Google Scholar] [CrossRef]
- Kanekiyo, M.; Joyce, M.G.; Gillespie, R.A.; Gallagher, J.R.; Andrews, S.F.; Yassine, H.M.; Wheatley, A.K.; Fisher, B.E.; Ambrozak, D.R.; Creanga, A.; et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 2019, 20, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.M.; Nanda, A.; Havenga, M.J.E.; Abbink, P.; Lynch, D.M.; Ewald, B.A.; Liu, J.; Thorner, A.R.; Swanson, P.E.; Gorgone, D.A.; et al. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature 2006, 441, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Buchbinder, S.P.; Mehrotra, D.V.; Duerr, A.; Fitzgerald, D.W.; Mogg, R.; Li, D.; Gilbert, P.B.; Lama, J.R.; Marmor, M.; Del Rio, C.; et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): A double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet Lond. Engl. 2008, 372, 1881–1893. [Google Scholar] [CrossRef]
- Sliepen, K.; van Montfort, T.; Melchers, M.; Isik, G.; Sanders, R.W. Immunosilencing a highly immunogenic protein trimerization domain. J. Biol. Chem. 2015, 290, 7436–7442. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brinkkemper, M.; Sliepen, K. Nanoparticle Vaccines for Inducing HIV-1 Neutralizing Antibodies. Vaccines 2019, 7, 76. https://doi.org/10.3390/vaccines7030076
Brinkkemper M, Sliepen K. Nanoparticle Vaccines for Inducing HIV-1 Neutralizing Antibodies. Vaccines. 2019; 7(3):76. https://doi.org/10.3390/vaccines7030076
Chicago/Turabian StyleBrinkkemper, Mitch, and Kwinten Sliepen. 2019. "Nanoparticle Vaccines for Inducing HIV-1 Neutralizing Antibodies" Vaccines 7, no. 3: 76. https://doi.org/10.3390/vaccines7030076
APA StyleBrinkkemper, M., & Sliepen, K. (2019). Nanoparticle Vaccines for Inducing HIV-1 Neutralizing Antibodies. Vaccines, 7(3), 76. https://doi.org/10.3390/vaccines7030076




