Abstract
Tumor vaccine design requires prediction and validation of immunogenic MHC class I epitopes expressed by target cells as well as MHC class II epitopes expressed by antigen-presenting cells essential for the induction of optimal immune responses. Epitope prediction methods are based on different algorithms and are instrumental for a first screening of possible epitopes. However, their results do not reflect a one-to-one correlation with experimental data. We combined several in silico prediction methods to unravel the most promising C57BL/6 mouse-restricted Hepatitis C virus (HCV) MHC class I epitopes and validated these epitopes in vitro and in vivo. Cytotoxic T lymphocyte (CTL) epitopes within the HCV non-structural proteins were identified, and proteasomal cleavage sites and helper T cell (Th) epitopes at close proximity to these CTL epitopes were analyzed using multiple prediction algorithms. This combined in silico analysis enhances the precision of identification of functional HCV-specific CTL epitopes. This approach will be applicable to the design of human vaccines not only for HCV, but also for other antigens in which T-cell responses play a crucial role.
1. Introduction
Hepatitis C virus (HCV) primarily infects and persists in human hepatocytes. Clearance of HCV involves killing virus-infected cells through cellular immunity, particularly cytotoxic T lymphocytes (CTLs). However, 80% of HCV-infected patients are not able to clear the virus, meaning HCV infection persists. Development of chronic HCV results in liver cirrhosis, and 10% to 15% of chronically infected patients develop hepatocellular carcinoma in 10 to 15 years after the primary infection. During disease progression, viral mutants arise due to the high mutation rate of the HCV genome and immune pressure [1,2].
In 2011 two antiviral HCV protease inhibitors, telaprevir and boceprevir, were approved for treatment for chronic HCV infection next to/combined with the standard treatment with pegylated interferon alpha and ribavirin. Although the sustained antiviral response was improved when using these new drugs, two third of chronically HCV-infected patients are still non-responders to all available treatments [3,4]. Antiviral treatments inhibit HCV replication but do not induce HCV-specific CTLs, which is crucial for clearance of HCV-infected cells [5]. Therefore, to effectively control HCV infection, therapeutic vaccines that aim to induce robust cellular immunity should be developed and used in combination with current HCV therapies.
Spontaneous resolution of HCV infection is positively correlated to the activation of a wide T-cell repertoire recognizing multiple HCV epitopes [6,7]. CTL epitopes can be predicted by mathematical algorithms that compare the protein sequence of interest to a large database of known epitopes. However, for HCV, experimentally verified data on most of these predicted epitopes are limited. Some protective CTL epitopes have been identified in HCV-infected patients [8,9,10]. However, selection of epitopes to be included in a personalized vaccine is crucial and should take into account differences in virus genotypes and subtypes and the presence of escape viral mutants. Thus, to define high-potential candidates for a therapeutic HCV vaccine, it is crucial to identify virus sequences and their corresponding antigenic epitopes using high-throughput methods.
In this study, using a murine model, we aimed to set up a procedure to define and subsequently verify protective HCV CTL epitopes. We selected immunogenic epitopes within conserved HCV proteins based on the results from several mathematical algorithms available. Next, we measured the binding affinity of the predicted CTL peptides to MHC class I molecules in vitro. Furthermore, we determined the immunogenicity of the identified epitopes in vivo.
2. Materials and Methods
2.1. Prediction Algorithms and Nucleotide Sequences
HCV peptides associated with MHC class I molecules (H-2Db and H-2Kb) were predicted with SYFPEITHI, NetMHCpan 2.8 and Immune epitope database and analysis resource (IEDB). HCV peptides associated with MHC class II molecules (H-2-IAb) were predicted with IEDB. Proteasomal cleavage sites of the selected HCV peptides were predicted with MAPPP, PAProC I and Netchop. Protein analyzed: nsPs of HCV genotype 1 (H77) (Genomic sequence: NC_004102.1), ovalbumin (Genomic sequence: NC_006089.3) and E7 protein of Human papillomavirus type 16 (Genomic sequence: NC_001526.2).
2.2. Synthetic Peptides
Long synthetic peptides (13 to 18-mers) were kindly received from BEI Resources, NIAID, NIH: Peptide Array, Hepatitis C Virus, H77, NS2 protein, NR-3751; NS3 protein, NR-3752; NS4A protein, NR-3753; NS4B protein, NR-3754; NS5A protein, NR-3755; NS5B protein, NR-3756. Short synthetic peptides (8 to 9-mers) were manufactured by the Leiden University Medical Center, the Netherlands. The purities of the synthetic peptide were analyzed with HPLC. All peptides had a purity of >90%.
2.3. MHC Class I Stabilization Assay
RMA-S cells were maintained in IMDM (Life Technologies, Bleiswijk, The Netherlands) supplemented with 10% FBS and 100 U/mL penicillin and 100 μg/mL streptomycin (Life Technologies). RMA-S cells were cultured at 26 °C for 48 h to induce expression of MHC class I molecules. Cells were then incubated with various concentrations of synthetic peptides at 26 °C for 4 h, followed by a 1-h culture at 37 °C. Cells were stained with APC-anti-H-2Kb Ab (clone: AF6-88.5.5.3) and FITC-anti-H-2Db Ab (clone: 28-14-8) (eBioscience, Vienna, Austria). The surface expression of MHC class I molecules were analyzed by FACSCalibur cytometer (BD Biosciences, Breda, The Netherlands) and data analyzed with FlowJo software (Treestar, Ashland, OR, USA). Fluorescence index was calculated by dividing the median fluorescence intensity (MFI) value of cells incubated with SLPs by the MFI value of cells incubated with equivalent concentration of DMSO.
2.4. Mice
Specific pathogen-free female inbred C57BL/6JOlaHsd (H-2b) mice expressing both MHC class I Kb and Db molecules were obtained from Harlan CPB (Zeist, The Netherlands) and kept under the institute guidelines. Mice were 8 to 10 weeks of age at the start of all experiments. Animal experiments were approved by the Institutional Animal Care and Use Committee of the University Medical Center of Groningen (DEC 5946).
2.5. Recombinant Semliki Forest Virus (rSFV) Particles Production and Immunizations
SFV plasmids (pSFVeNS2'-5B', pSFVeNS3/4A and pSFVeNS5A/B') containing HCV non-structure proteins were constructed by sub-cloning the HCV fragments from the full-length cDNA of HCV H77 genotype 1a consensus sequence (H/FL) into pSFVe (pSFV4.2 enclosed with translational enhancer, foot-and-mouth disease virus 2A auto-protease fragment) [11]. H/FL was provided by Charles M Rice via Apath, LLC (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: p90HCVconsensuslongpU) [12]. pSFV4.2 was provided by P Liljestrom (Karolinska Institute, Stockholm, Sweden). pSFVeNS2'-5B' (16,838 bps) contains part of the HCV NS2 and NS5B DNA and the complete HCV NS3, NS4A and NS5A DNA (nucleotide position 3106 to 9220 of H/FL). pSFVeNS3/4A (12,839 bps) contains the complete HCV NS3 and NS4A DNA (nucleotide position 3391 to 5543 of H/FL). pSFVeNS5A/B' (13,700 bps) contains the complete HCV NS5A DNA and part of the NS5B DNA (nucleotide position 6258 to 9220 of H/FL). Recombinant SFV particles (rSFVeNS2'-5B', rSFVeNS3/4A and rSFVeNS5A/B') were produced as previously described [11]. In brief, SFV particles were produced by the co-electroporation of BHK-21 cells with an RNA encoding the SFV replicase and the transgene and a helper RNA encoding the structural proteins of SFV. The rSFV replicon particles produced by the transfected BHK-21 cells were purified on a discontinuous sucrose density gradient. rSFV particles were titrated on BHK-21 cells using a polyclonal rabbit anti-replicase (nsP3) antibody (a kind gift from T. Ahola (Biocentre Viiki, Helsinki, Finland)). Mice were primed and boosted immunized intramuscularly with a 2-week interval with 5 × 106 rSFV in 50 μL (25 μL/thigh muscle) under anesthesia (isoflurane/O2). For negative controls, PBS was injected intramuscularly.
2.6. Degranulation and IFN-γ Staining
Splenocytes isolated from immunized mice were cultured with 10 μg/mL of synthetic peptides in the presence of anti-CD28 Ab (clone: PV-1, Bioceros B.V., Utrecht, The Netherlands), eFluor® 660-anti-CD107a Ab (clone: eBio1D4B) and eFluor® 660-anti-CD107b Ab (clone: eBioABL-93) in a 96-well plate at 37 °C with 5% CO2. As a negative control, splenocytes were cultured without peptides but in the presence of anti-CD28 Ab. One hour after culture, brefeldin A (10 μg/mL) was added and the cultures were incubated for another 4 h. Cells were harvested, washed and stained with LIVE/DEAD® fixable violet dead cell stain kit (Life Technologies), followed by surface staining with PE-Cy7-anti-CD8a Ab (clone: 53-6.7) and intracellular staining with PerCP-Cyanine5.5-anti-IFN-γ Ab (clone: XMG1.2). Unless otherwise indicated, antibodies were purchased from eBioscience. FACS analysis was conducted with LSR-II flow cytometer (BD Biosciences), and data was analyzed with FlowJo software (Treestar, Ashland, OR, USA).
3. Results and Discussion
3.1. Selection and Characteristics of HCV Synthetic Long Peptides that May Contain CTL Epitopes
The MHC class I epitope prediction algorithms SYFPEITHI [13], NetMHCpan 2.8 [14] and Immune Epitope Database and Analysis Resource (IEDB) [15] were used to predict CTL epitopes within HCV nonstructural proteins (nsPs) that bind H-2Kb/Db. The first round of HCV CTL epitope selection was based on predictions from SYFPEITHI (Table 1). Eighteen CTL epitopes that scored higher than 20 in the SYFPEITHI prediction were selected. In addition, NS5B1–16 (H-2d) [16] and NS5B46–63 (multiple human MHC alleles) [17], which are known binders for other MHC alleles, were selected as negative controls. As a result, twenty CTL epitopes were further analyzed with the NetMHCpan 2.8 and IEDB prediction algorithms. The threshold for a strong binder is <0.5% rank in both NetMHCpan 2.8 and IEDB. Seven out of eighteen SYFPEITHI predicted CTL epitopes were predicted by either NetMHCpan 2.8 or IEDB (Table 1). Surprisingly, the negative controls, NS5B1–16 and NS5B46–63, were predicted as strong binders by both NetMHCpan 2.8 and IEDB.

Table 1.
Selection of synthetic long peptides containing CTL epitopes from HCV nsPs by prediction algorithms. Sequences of the long synthetic peptides, for which MHC binding affinity was shown in Figure 1, are shown. Strong binders are depicted in bold and highlighted in grey. The cut off score of SYFPEITHI is set at 20; a high score indicates a strong binder. The cut off score of NetMHCpan 2.8 and IEDB is set at 0.5 (% rank); a low score indicates a strong binder.
| Protein/Position | Sequence (CTL Epitopes Are Underlined) | MHC I Peptides Prediction | MHC I Stabilization Assay (Figure 1) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SYFPEITHI (>20 strong binding) | NetMHCpan 2.8 (<0.5 strong binding) | IEDB (<0.5 strong binding) | Peptide Concentration = 10 μM 1,2 | Selected for Short Peptides Synthesis (v) | ||||||
| H-2Db | H-2Kb | H-2Db | H-2Kb | H-2Db | H-2Kb | H-2Db | H-2Kb | |||
| NS372-87 | IQMYTNVDQDLVGWPA | 24 | 10 | 0.8 | 3 | 2.05 | 2.25 | + | - | |
| NS3165-180 | KAVDFIPVENLGTTMR | 30 | 8 | 2 | 32 | 2.5 | 18.5 | - | + | |
| NS3214-228 | VPAAYAAQGYKVLVL | 0 | 22 | 10 | 15 | 24 | 6.8 | ++ | + | |
| NS3323-340 | ATPPGSVTVSHPNIEEVA | 23 | 9 | 0.08 | 8 | 0.3 | 9.45 | + | - | v |
| NS3383-400 | ALGINAVAYYRGLDVSVI | 0 | 22 | 32 | 1.5 | 19.1 | 1.15 | ++ | - | |
| NS3507-524 | AETTVRLRAYMNTPGLPV | 22 | 11 | 0.1 | 0.25 | 0.7 | 0.7 | +++ | - | v |
| NS3525-542 | CQDHLEFWEGVFTGLTHI | 0 | 21 | 50 | 32 | 13.95 | 7.9 | ++ | - | |
| NS3547-563 | LSQTKQSGENFPYLVAY | 28 | 22 | 0.3 | 1.5 | 0.4 | 1.15 | ++ | - | |
| NS3601-618 | RLGAVQNEVTLTHPITKY | 29 | 12 | 0.08 | 32 | 0.2 | 12.95 | + | - | v |
| NS5A58-75 | HCGAEITGHVKNGTMRIV | 24 | 8 | 5 | 32 | 2.65 | 36.5 | - | - | |
| NS5A98-115 | CTPLPAPNYKFALWRVSA | 20 | 12 | 7 | 32 | 19 | 19 | - | - | |
| NS5A140-157 | CPCQIPSPEFFTELDGVR | 21 | 22 | 8 | 1.5 | 6.3 | 0.5 | - | - | |
| NS5A269-284 | ITRVESENKVVILDSF | 24 | 7 | 4 | 50 | 5.1 | 45.5 | - | - | |
| NS5B1-16 | SMSYSWTGALVTPCAA | 13 | 11 | 2 | 0.03 | 5.3 | 0.2 | + | + | v |
| NS5B46-63 | CQRQKKVTFDRLQVLDSH | 15 | 11 | 15 | 0.05 | 27 | 0.25 | - | - | v |
| NS5B152-169 | GGRKPARLIVFPDLGVRV | 15 | 22 | 32 | 15 | 22 | 12.35 | - | ++ | v |
| NS5B249-266 | ARVAIKSLTERLYVGGPL | 22 | 12 | 0.8 | 3 | 1.35 | 2.8 | - | - | |
| NS5B329-346 | VQEDAASLRAFTEAMTRY | 20 | 12 | 0.4 | 1.5 | 9.7 | 13.95 | ++ | + | |
| NS5B402-419 | HTPVNSWLGNIIMFAPTL | 21 | 8 | 7 | 8 | 12 | 2.7 | - | - | |
| NS5B423-439 | MILMTHFFSVLIARDQL | 13 | 21 | 8 | 0.17 | 9.1 | 0.3 | - | - | v |
1 Fluorescence index of H-2Db: 0.2–0.5 +; 0.5–1.5 ++, >1.5 +++; 2 Fluorescence index of H-2Kb: 0.2–2 +; 2–8 ++; >8 +++.
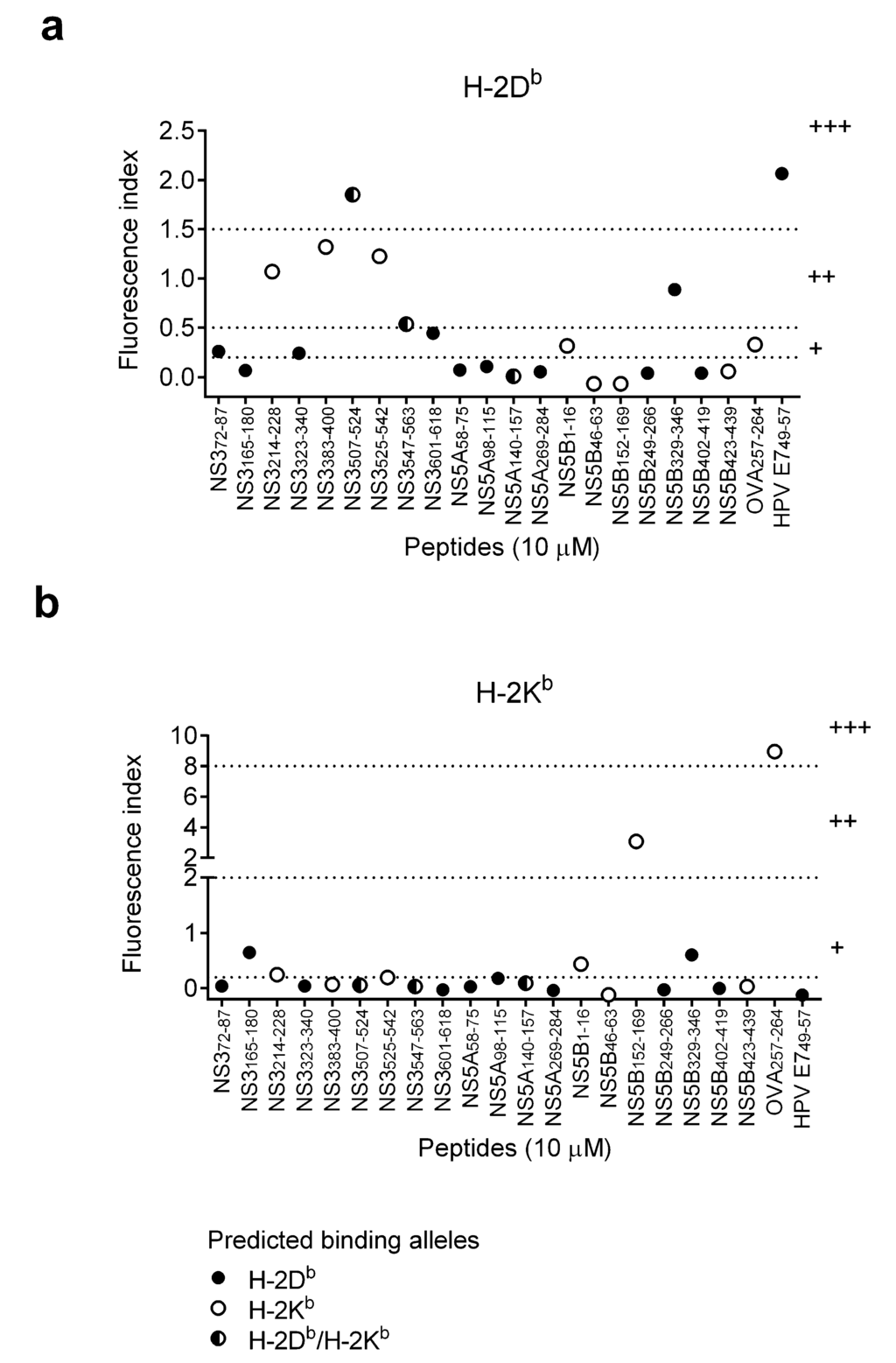
Figure 1.
Stabilization of MHC class I molecules with binding of HCV SLPs. To induce MHC class I expression on cell surface, RMA-S cells were cultured at 26 °C for 48 h prior to the incubation with SLPs. Cells were then incubated with 10 μM of SLPs at 26 °C for 4 h, followed by a 1-h cultured at 37 °C. The expression levels of surface MHC class I molecules, (a) H-2Db and (b) H-2Kb, were analyzed using flow cytometry. HPV E749–57 and OVA257–264 short peptides were positive controls for binding to H-2Db and H-2Kb molecules, respectively. Dash lines indicate the cutoff values for H-2Db (0.2–0.5: weak binders (+); 0.5–1.5: intermediate binders (++); >1.5: strong binders (+++)) and H-2Kb (0.2–2: weak binders (+); 2–8: intermediate binders (++); >8: strong binders (+++)).
To test the performance of these selected algorithms, the binding capacity of the HCV synthetic long peptides (SLPs) containing the selected CTL epitopes based on the prediction of the three algorithms (Table 1) were determined on MHC class I expressing cells, RMA-S cells (Figure 1). One concentration (10 μM) of SLPs was used in this MHC stabilization assay to screen the possible H-2Db and H-2Kb binders. HCV SLPs were classified as weak, intermediate and strong binders according to the ability to stabilize MHC class I molecules on the surface of RMA-S cells. Known CTL epitope peptides of ovalbumin (OVA257–264) and human papillomavirus (HPV E749–57) were included as positive controls for H-2Kb and H-2Db binding, respectively. Among 13 predicted HCV H-2Db binders, predicted by at least one algorithm (solid and half-open circles), there was one strong binder (NS3507–524), two intermediate binders (NS3547–563 and NS5B329–346) and three weak binders (NS372–87, NS3323–340, NS3601–618). Three predicted H-2Kb binders (open circles) stabilized H-2Db molecules and were considered intermediate binders (NS3214–228, NS3383–400, NS3525–542) (Figure 1a). In ten SLPs that were predicted to be H-2Kb binders by at least one algorithm (open and half-open circles), one was classified as an intermediate binder (NS5B152–169) and two were weak binders (NS3214–228, NS5B1–16). Two predicted H-2Db binders (solid circle) stabilized H-2Kb molecules and were considered weak binders (NS3165–180 and NS5B329–346) (Figure 1b).
3.2. Binding Affinity of HCV Short Peptides to MHC Class I Molecules
Six out of nine SLPs that contain a CTL epitope predicted by two or more algorithms were able to bind and stabilize MHC class I molecules (Table 1). However, some SLPs that contain CTL epitopes predicted only by SYFPEITHI did not bind to MHC class I molecules. Since SLPs contain extra amino acid flanking the predicted CTL epitopes, binding affinity of SLPs to MHC class I molecules may be reduced [18,19]. Thus, binding capacity of the CTL epitopes was confirmed with short synthetic peptides that contain only the CTL epitope. Short synthetic peptides were selected based on one of the following criteria: (i) CTL epitope has to be predicted by at least 2 algorithms and/or classified as a weak, intermediate or strong binder from the result of the MHC I stabilization assay with SLPs (NS3323–340, NS3507–524, NS3601–618, NS5B1–16, NS5B46–63, NS5B423–439); (ii) CTL epitope has to be predicted by at least 1 algorithm and classified as intermediate or strong binder from the result of MHC I stabilization assay (NS5B152–169). Since the accuracy of prediction greatly increased when peptides were analyzed with two or more algorithms, four extra CTL epitopes, which were predicted by at least two algorithms as strong binders, were selected (NS2139–147, NS3265–273, NS4B38–46 and NS5A280–287) (Table 2). And also for this analysis, the known CTL epitopes of ovalbumin (OVA257–264) and human papillomavirus (HPV E749–57), which are strong H-2Kb and H-2Db binders, respectively, were selected as positive controls.
Binding affinity of selected short synthetic peptides on MHC class I molecules was determined by MHC I stabilization assay with various concentrations of peptide (Figure 2). As expected, stabilization of both H-2Db and H-2Kb molecules increased with increasing concentration of peptide. The kinetic of the stabilization of H-2Db molecules was similar for all selected short peptides. The plateau of stabilization was reached at approximately 30 μM of peptides (Figure 2a). Two kinetic patterns of stabilizations of H-2Kb molecules were observed. For strong binders (at 100 μM, FI > 8; OVA257–264, NS5B2–10, NS3265–273, NS5B157–165, NS2139–147 and NS5B52–60), there was a dose dependent increase of FI and the plateau of stabilization was not detectable even at high peptide concentration (100 μM). For intermediate binders (NS3514–522, NS5B425–433 and NS5A280–287), stabilization of H-2Kb molecules was modest at low peptide concentration (0.3–10 μM). However, the stabilization effect increased exponentially once the concentration was above 30 μM. This observation may explain why we did not see strong H-2Kb binders when SLPs at 10 μM were used in the MHC I stabilization assay (Figure 1). Results of stabilization of MHC class I molecules with short peptides matched the result from all of the MHC class I prediction algorithms used (H-2Db: >90% matched; H-2Kb: 100% matched) (Table 2).
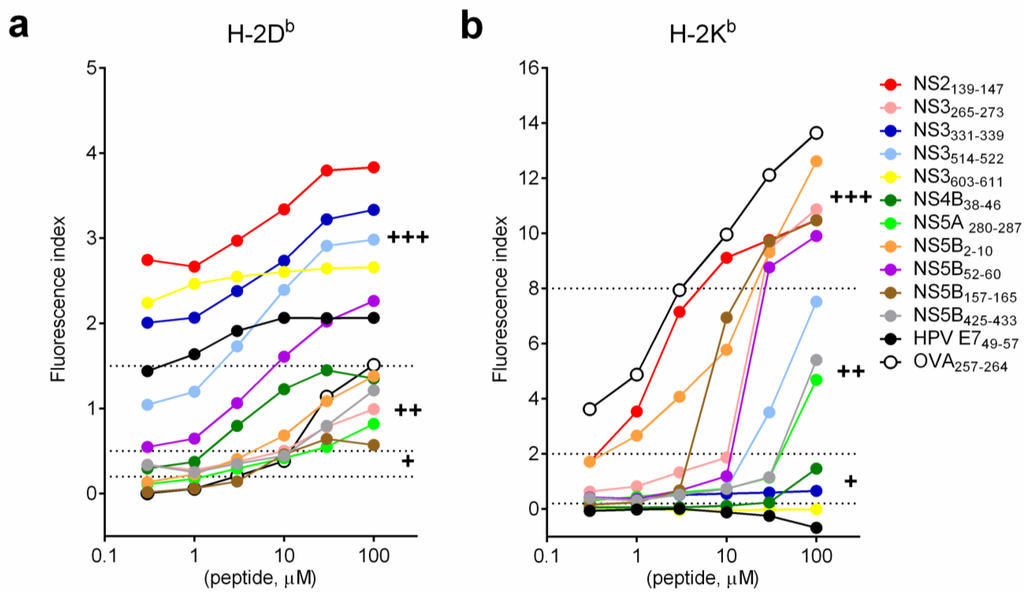
Figure 2.
Binding affinity of HCV short peptides to MHC class I molecules. Short synthetic HCV peptides were serial diluted and incubated with RMA-S cells as described in Figure 1. (a) H-2Db; (b) H-2Kb. Dash lines indicate the cutoff values for H-2Db (0.2–0.5: weak binders (+); 0.5–1.5: intermediate binders (++); >1.5: strong binders (+++)) and H-2Kb (0.2–2: weak binders (+); 2–8: intermediate binders (++); >8: strong binders (+++)).
3.3. The Presence of Proteasomal Degradation Sites at the Carboxyterminal Site of the Predicted CTL Epitopes
Under physiological conditions, proteins have to be cleaved by proteasomes into short peptides in order to be loaded onto MHC class I molecules. Proteasomal cleavage sites were predicted with MAPPP, PAProC I and Netchop. MAPPP and Netchop predict the cleavage site of constitutive proteasomes and PAProC I predicts cleavage site of both constitutive proteasomes and immunoproteasomes. From the result of at least two prediction algorithms, proteasomal cleavage sites were present at all strong MHC class I binders (+++) (Table 2).
3.4. The Presence of MHC Class II Epitopes Flanking the Predicted CTL Epitopes
It has been observed that immunodominant CTL epitopes such as Influenza B nucleoprotein are flanked and/or overlap with Th epitopes [20,21,22]. Association of CTL and Th epitopes may indeed enhance the immunogenicity of the CTL epitopes in vivo [23]. To investigate the presence of MHC class II epitopes flanking the strong MHC class I epitopes, we next performed prediction of MHC class II epitopes with IEDB. Most of the identified high affinity MHC class I epitopes were flanked with at least one predicted MHC class II epitope. Also, two MHC class I epitopes (both H-2Db and H-2Kb), overlap with a predicted MHC class II epitope (IEDB rank < 10) (NS3514–522 and NS5B2–10). Flanking of MHC class I epitopes with predicted MHC class II epitopes was also observed in the positive control, OVA257–264 (Table 2).
3.5. Induction of Peptide-Specific Effector CD8+ T Cells in Vivo
Induction of in vivo T-cell response depends on the presentation of peptides and the availability, avidity and affinity of precursor CD8+ T cells [24,25]. Here, we investigated the induction of T-cell response against the predicted CTL epitopes in mice immunized three times with the rSFV particles expressing all HCV nsPs, NS3/4A or NS5A/B' (rSFVeNS2'-5B', rSFVeNS3/4A or rSFVeNS5A/B'). Splenocytes were isolated 1 to 3 weeks after the last immunization and re-stimulated with selected short peptides in order to induce degranulation (surface expression of CD107a/b) and secretion of IFN-γ by peptide-specific CD8+ T cells (Figure 3). rSFV immunizations induced functional effector CD8+ T cells (CD107a/b+IFN-γ+) against NS2139–147, NS3603–611, NS5B2–10 and NS5B157–165. When no peptides were added to the splenocytes, we already observed the presence of endogenous CD107a/b+IFN-γ+CD8+ cells from mice immunized with any rSFV particles but not from mice immunized with PBS. This indicates that rSFV immunizations in general induced functional CD8+ T-cell responses. To check for a specific response against peptide, background (without peptide re-stimulation) subtraction was applied. Frequencies above zero indicate specific response against the re-stimulating peptide. CD8+ T-cell response against NS3603–611 [26] was the highest among all selected peptides and was detected in mice immunized with rSFVeNS2'-5B' or rSFVeNS3/4A. Responses against NS5B2–10 and NS5B157–165 were low and induced in mice immunized with rSFVeNS5A/B' or rSFVeNS2'-5B'. A very low response against NS2139–147 was observed only in mice immunized with rSFVeNS2'-5B', not in mice with other immunizations. Of note, all responding peptides were predicted by at least one algorithm for MHC class I prediction. Proteasomal cleavages sites were presented in the carboxyterminals of all four peptides. Three peptides (NS2139–147, NS3603–611 and NS5B2–10) contained predicted MHC class II epitopes in close proximity to their CTL epitopes with relatively high rankings (Table 2).

Table 2.
Detailed analysis of selected short synthetic HCV peptides. Prediction of proteasomal cleavage sites and MHC class II epitopes flanking the selected CTL epitopes for (a) H-2Db, (b) H-2Kb. The CTL epitope and its flanking amino acids (11 amino acid at 5' end and 11 amino acid at 3' end of the CTL epitope) were analyzed with the MHC class II prediction algorithm (IEDB). Rankings below 10 were considered strong binders. The values represent the ranking range for the analyzed amino acids. Data were sorted according to the binding affinity of short peptides to RMA-S cells (Figure 2), strong binders at the top of the row (a: H-2Db; b: H-2Kb). N.D. not determined. Strong binders are depicted in bold and highlighted in grey.
| a | MHC I Stabilization Assay 1 | MHC Class I Prediction | Proteasomal Cleavage | MHC Class II Prediction | |||||||
| SYFPEITHI (>20 strong binding) | NetMHCpan 2.8 (<0.5 strong binding) | IEDB (<0.5 strong binding) | MAPPP (Cleavage Probability 3) | PAProC I (Score 4) | Netchop (Cleavage Probability 3) | IEDB- H-2-IAb (<10 strong binding) | |||||
| Protein/Position | Sequence | H-2Db | H-2Db | H-2Db | H-2Db | 5' of CTL Epitope | Complete CTL Epitope | 3' of CTL Epitope | |||
| NS2139-147 | YVYNHLTPL | +++ | 13 | 0.15 | 0.7 | 1 | 0- | 0.95 | 31.64–58.25 | 1.82–3.90 | 27.51–64.83 |
| NS3331-339 | VSHPNIEEV | +++ | 23 | 0.08 | 0.3 | 0.7502 | 121+++ | 0.97 | 2.21–12.13 | 10.72–48.25 | 31.02–54.21 |
| NS3514-522 | RAYMNTPGL | +++ | 22 | 0.1 | 0.7 | 0 | 78++ | 0.95 | 5.17–73.56 | 5.97–22.88 | 16.57–87.59 |
| NS3603-611 | GAVQNEVTL | +++ | 29 | 0.08 | 0.2 | 0 | 178+++ | 0.88 | 9.35–55.70 | 18.51–32.13 | 16.70–51.34 |
| HPV E749-57 | RAHYNIVTF | +++ | 23 | 0.08 | 0.2 | 1 | N.D. | 0.91 | 33.38–53.38 | 56.31–79.26 | 69.56–86.81 |
| NS5B52-60 | VTFDRLQVL | +++ | 15 | 15 | 27 | 0.5976 | 0- | 0.96 | 10.39–86.63 | 71.97–79.66 | 28.42–85.75 |
| OVA257-264 | SIINFEKL | ++ | 0 | 4 | 0.2 | 1 | N.D. | 0.97 | 51.45–69.97 | 51.88–81.68 | 9.57–82.66 |
| NS4B38-46 | AVQTNWQKL | ++ | 23 | 1 | 1.9 | 0 | 0- | 0.72 | 12.81–32.62 | 23.98–74.51 | 21.59–58.54 |
| NS5B2-10 | MSYSWTGAL | ++ | 13 | 2 | 5.3 | 1 | 0- | 0.87 | N.D. | 0.93–0.95 | 2.62–56.47 |
| NS5B425-433 | LMTHFFSVL | ++ | 13 | 8 | 9.1 | 0.5009 | 40++ | 0.96 | 11.68–61.33 | 36.91–72.20 | 46.08–77.76 |
| NS3265-273 | ITYSTYGKF | ++ | 9 | 32 | 14.3 | 1 | 0- | 0.209 | 10.30–47.29 | 36.17–47.83 | 49.48–84.06 |
| NS5A280-287 | ILDSFDPL | ++ | 0 | 15 | 0.2 | 0.906 | 0- | 0.92 | 51.43–86.16 | 18.50–86.11 | 20.39–86.16 |
| NS5B157-165 | ARLIVFPDL | ++ | 15 | 32 | 22 | 0.6886 | 0- | 0.96 | 44.16–67.37 | 47.47–56.37 | 51.99–81.87 |
| b | MHC I Stabilization Assay 2 | MHC Class I Prediction | Proteasomal Cleavage | MHC Class II Prediction | |||||||
| SYFPEITHI (>20 strong binding) | NetMHCpan 2.8 (<0.5 strong binding) | IEDB (<0.5 strong binding) | MAPPP (Cleavage Probability 3) | PAProC I (Score 4) | Netchop (Cleavage Probability 3) | IEDB- H-2-IAb (<10 strong binding) | |||||
| Protein/Position | Sequence | H-2Kb | H-2Kb | H-2Kb | H-2Kb | 5' of CTL Epitope | Complete CTL Epitope | 3' of CTL Epitope | |||
| OVA257-264 | SIINFEKL | +++ | 25 | 1.5 | 0.35 | 1 | N.D. | 0.97 | 51.45–69.97 | 51.88–81.68 | 9.57–82.66 |
| NS5B2-10 | MSYSWTGAL | +++ | 11 | 0.03 | 0.2 | 1 | 0- | 0.87 | N.D. | 0.93–0.95 | 2.62–56.47 |
| NS3265-273 | ITYSTYGKF | +++ | 11 | 0.4 | 0.3 | 1 | 0- | 0.209 | 10.30–47.29 | 36.17–47.83 | 49.48–84.06 |
| NS5B157-165 | ARLIVFPDL | +++ | 22 | 15 | 12.35 | 0.6886 | 0- | 0.96 | 44.16–67.37 | 47.47–56.37 | 51.99–81.87 |
| NS2139-147 | YVYNHLTPL | +++ | 11 | 0.01 | 0.2 | 1 | 0- | 0.95 | 31.64–58.25 | 1.82–3.90 | 27.51–64.83 |
| NS5B52-60 | VTFDRLQVL | +++ | 11 | 0.05 | 0.25 | 0.5976 | 0- | 0.96 | 10.39–86.63 | 71.97–79.66 | 28.42–85.75 |
| NS3514-522 | RAYMNTPGL | ++ | 11 | 0.25 | 0.7 | 0 | 78++ | 0.95 | 5.17–73.56 | 5.97–22.88 | 16.57 – 87.59 |
| NS5B425-433 | LMTHFFSVL | ++ | 21 | 0.17 | 0.3 | 0.5009 | 40++ | 0.96 | 11.68–61.33 | 36.91–72.20 | 46.08–77.76 |
| NS5A280-287 | ILDSFDPL | ++ | 21 | 6 | 1.45 | 0.906 | 0- | 0.92 | 51.43–86.16 | 18.50–86.11 | 20.39–86.16 |
| NS4B38-46 | AVQTNWQKL | + | 11 | 15 | 8.2 | 0 | 0- | 0.72 | 12.81–32.62 | 23.98–74.51 | 21.59–58.54 |
| NS3331-339 | VSHPNIEEV | + | 9 | 8 | 9.45 | 0.7502 | 121+++ | 0.97 | 2.21–12.13 | 10.72–48.25 | 31.02–54.21 |
| NS3603-611 | GAVQNEVTL | - | 12 | 32 | 12.95 | 0 | 178+++ | 0.88 | 9.35–55.70 | 18.51–32.13 | 16.70–51.34 |
| HPV E749-57 | RAHYNIVTF | - | 8 | 5 | 11.2 | 1 | N.D. | 0.91 | 33.38–53.38 | 56.31–79.26 | 69.56–86.81 |
1 Fluorescence index of H-2Db: 0.2–0.5 +; 0.5–1.5 ++; >1.5 +++; 2 Fluorescence index of H-2Kb: 0.2–2 +; 2–8 ++; >8 +++; 3 Cleavage probability: 1 = 100% cleavage; 4 PAProC I score: high score indicates high chance for cleavage, also depicted by the number of +.
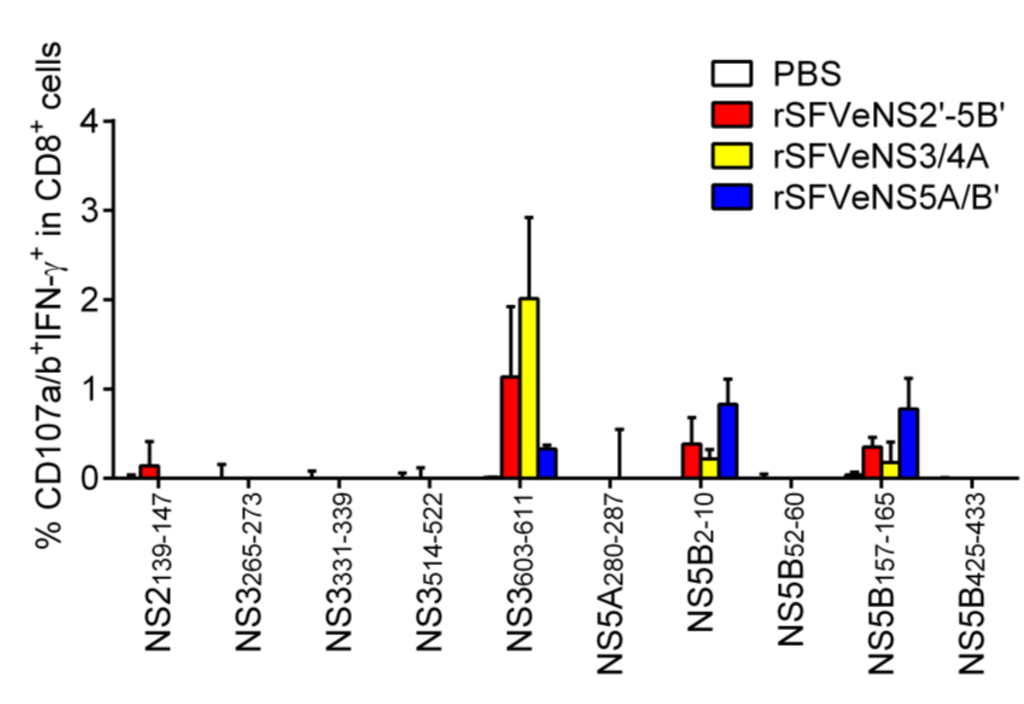
Figure 3.
Induction of peptide-specific effector CD8+ T cells in vivo. Mice were intramuscularly immunized thrice with rSFV expressing HCV nsPs (rSFVeNS2'-5B', rSFVeNS3/4A or rSFVeNS5A/B') or PBS control with a one-week interval. Mice were sacrificed 1 to 3 weeks after the boost immunization. Splenocytes were isolated and cultured with 10 μg/mL of peptides for 5 h before surface and intracellular staining. Background (splenocytes incubated with an equivalent concentration of DMSO) subtraction was applied and values above background are shown. Data represent combined results from three independent experiments, showing the mean +SEM (3–5 mice per group).
4. Discussion
Identification of protective HCV CTL epitopes is essential in the development of vaccines for patients with chronic HCV infection. Treatment can be personalized based on the circulating HCV genotype(s). Ideally one should identify HCV epitopes expressed by the patients’ infected cells and induce T-cell responses against the identified epitopes with vaccine immunizations. Identification of HCV epitopes from cell lines expressing HLA-A2 and HCV proteins has been performed with electro-spray ionization quadrupole time-of-flight mass spectrometry/tandem mass spectrometry (ESI Q-TOF-MS/MS) and the data were consistent with the results predicted from mathematical algorithms [27]. This method could also be used to identify naturally processed and presented HCV peptides from HCV-infected hepatocytes. However, widespread use of this analysis is limited, as liver biopsies are required for the analysis and taking biopsies is not standard procedure for these patients. Moreover, the percentage of HCV-infected hepatocytes in biopsies may be too low for MS analysis [28,29]. An alternative method is to identify CTL epitopes in silico based on the HLA phenotype and the viral sequence identified from blood of HCV-infected patients.
Here we show that the prediction accuracy for the identification of potential CTL epitopes is improved by combining several algorithms for MHC class I epitope and proteasomal cleavage site prediction, and possibly even MHC class II epitope prediction. We narrowed down all HCV nsPs to 22 CTL epitopes using prediction algorithms, of which 10 were proven to bind to H-2b molecules on RMA-S cells. We next showed that immunization with rSFV expressing all nsPs induced a strong epitope-specific T-cell response against one out of ten H-2b binders and a weak response against three epitopes.
Computational analyses used in this study are machine-learning algorithms, i.e., the programs were trained with large databases for reliable results. Therefore, most algorithms favor the detection of immunodominant epitopes. Predictions were made with allele-specific motif methods based on amino acid sequences [30,31]. For example, peptide positions 2 and 9 are classical anchor residues most important for binding to the HLA-A*0201 allele and are typically occupied by leucine, valine and isoleucine [32]. With allele-specific motif methods, peptides containing appropriate amino acids at anchoring positions are predicted to be MHC class I binders, but this type of prediction does not necessarily correlate with the physiological function of the epitopes.
Antigen processing involves a complex regulation of antigen degradation, interaction with chaperones, binding to the peptide-loading complex, loading onto MHC class I molecules and transport to the cell surface [33]. For both presentation of intracellular antigens and cross-presentation of extracellular antigens, antigen degradation by constitutive and/or immune proteasome is required. Upon exposure of IFN-γ in lymphoid cells, the constitutive proteasomes are replaced by immunoproteasomes which have altered preferences for cleavage sites [34]. The immunoproteasomes favor the processing of immunodominant peptides [35]. Normal human liver cells also express immunoproteasomes, as well as constitutive and intermediate proteasomes that enable the broadening of the repertoire of antigenic peptides [36]. To define the physiological function of predicted CTL epitopes, we analyzed the proteasomal cleavage sites recognized by constitutive proteasomes (MAPPP, PAProC I and Netchop) or immunoproteasomes (PAProC I) present at the carboxyterminals of the predicted CTL epitopes. Combining MHC I prediction with proteasomal cleavage prediction indeed increases the accuracy of MHC I peptide prediction [37]. In this study, all predicted strong H-2Db and H-2Kb binders contained proteasomal cleavage sites predicted by at least two algorithms.
Activation of CD8+ T cells relies on both recognition of MHC I-peptide complexes on APCs and co-stimulatory activation signals provided by adjacent cells such as APCs and/or CD4+ helper T cells. Activated CD4+ helper T cells orchestrate the activation of CD8+ T cells through secretion of IL-2 and/or activation of dendritic cells for up-regulation of MHC I molecules and antigen presentation [23]. Interestingly, some known immunodominant CD8+ T cell epitopes such as OVA257–264 contain predicted high-affinity Th epitopes that overlap or are in close proximity to one another [23,38]. This may allow presentation of both MHC I and MHC II peptides by the same dendritic cells, resulting in activation of both CD8+ and CD4+ T cells simultaneously [39]. Alternatively, activated helper T cells acquire MHC II-peptide complexes from APCs as well as co-stimulating molecules, leading to activation of Th1 responses and central memory CD8+ T-cells responses [40,41,42]. It is proposed that the uptake of MHC II peptides may be further processed into MHC I peptides which can be loaded onto recycled MHC I molecules and presented to CD8+ T cells [23,43,44]. This will lead to higher activation of CD8+ T cells as helper T cells provide both antigen stimulation and co-stimulation [45]. Peptides recognized by splenocytes from rSFV-immunized mice (NS2139–147 and NS5B2–10) overlapped with strong predicted Th epitopes (Table 2). The immunodominant epitope (NS3603–611) contains intermediate predicted Th epitopes at the 5' ends of the CTL epitope. Remarkably, MHC class II epitope prediction by IEDB may not identify all possible Th epitopes, since, for example, it has been shown that E744–62, binds to MHC class II molecules in vitro but was not predicted as a strong binder [46]. As MHC class II binding sites are abundantly present, however, further studies are needed to prove or reject the hypothesis that there is a correlation between the immunogenicity of class I epitopes and overlap with high-affinity MHC class II epitopes. Besides the results of prediction algorithm, the presence of Th epitopes should be determined using in vitro experiments such as an MHC class II binding assay. For the design of vaccines expressing specific CTL epitopes, this could be considered to include Th epitopes that overlap or are in close proximity with CTL epitopes to enhance immune responses. In a separate study, we demonstrated that inclusion of strong universal helper-epitopes in an SFV replicon vaccine expressing human papillomavirus (HPV) antigens strongly enhanced the frequency of HPV specific CTLs [47].
In this study, we identified 1 out of 22 predicted CTL epitopes inducing strong CD8+ T-cell responses (NS3603–611) and 3 out of 22 inducing minor CD8+ T-cell responses (NS2139–147, NS5B2–10 and NS5B157–165). Subdominant CTL epitopes could not be identified with rSFVeNS2'-5B', rSFVeNS3/4A or rSFVeNS5A/B' immunizations. Inclusion of more epitopes may increase competition between T cells of different specificity, resulting in a reduced response to subdominant epitopes [48]. Furthermore, expression of full proteins may reduce the expression level of one particular peptide compared to expression of only one peptide. This may lead to insufficient antigen presentation, resulting in unstable contact between CD8+ T cells and APC and thus inability to fully activate T cells [49]. Although the biological function of responses against subdominant epitopes is unknown, an alternative approach to checking the accuracy of in silico predictions is to immunize with synthetic long peptides expressing only one particular CTL epitope and its flanking sequences. In this way, the immune response will be directed to that specific epitope.
5. Conclusions
To conclude, we validated in silico approaches to epitope prediction in vivo. We showed that responses against predicted dominant CTL epitopes were induced in mice immunized with HCV-nsPs expressing rSFV. In vivo relevance of subdominant epitopes, which were predicted by mathematical algorithms, may have to be confirmed with peptide immunizations. Combining epitope prediction algorithms with proteasomal cleavage site algorithms increases the accuracy of CTL epitope identification. The predictive value of strong Th epitopes overlapping with, or in close proximity to, CTL epitopes remains to be established. Thus, potential CTL with or without Th epitopes as candidates for therapeutic human vaccine development can be identified and narrowed down. For clinical practice, the establishment of an ex vivo stimulation assay with patients’ PBMCs is warranted to select the rationally designed candidate vaccine that is most immunogenic for that specific patient.
Acknowledgments
We thank Cornelis J.M. Melief (Leiden University Medical Center, the Netherlands) for helpful discussion. We thank Can Kesmir (Utrecht University, the Netherlands) for fruitful discussion on the selection and interpretation of mathematical algorithms. We thank Annemarie Boerma and Joke Regts (University Medical Center Groningen, The Netherlands) for their technical support. This study is funded by an Ubbo Emmius grant.
Author Contributions
Peng Peng Ip, Hans W. Nijman and Toos Daemen conceived and designed the experiments; Peng Peng Ip performed the experiments; Peng Peng Ip, Hans W. Nijman and Toos Daemen analyzed the data; Peng Peng Ip, Hans W. Nijman and Toos Daemen wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ogata, N.; Alter, H.J.; Miller, R.H.; Purcell, R.H. Nucleotide sequence and mutation rate of the h strain of Hepatitis C virus. Proc. Natl. Acad. Sci. USA 1991, 88, 3392–3396. [Google Scholar] [CrossRef] [PubMed]
- Erickson, A.L.; Kimura, Y.; Igarashi, S.; Eichelberger, J.; Houghton, M.; Sidney, J.; McKinney, D.; Sette, A.; Hughes, A.L.; Walker, C.M. The outcome of Hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity 2001, 15, 883–895. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, I.M.; McHutchison, J.G.; Dusheiko, G.; di Bisceglie, A.M.; Reddy, K.R.; Bzowej, N.H.; Marcellin, P.; Muir, A.J.; Ferenci, P.; Flisiak, R.; et al. Telaprevir for previously untreated chronic Hepatitis C virus infection. N. Engl. J. Med. 2011, 364, 2405–2416. [Google Scholar] [CrossRef] [PubMed]
- Poordad, F.; McCone, J., Jr.; Bacon, B.R.; Bruno, S.; Manns, M.P.; Sulkowski, M.S.; Jacobson, I.M.; Reddy, K.R.; Goodman, Z.D.; Boparai, N.; et al. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 2011, 364, 1195–1206. [Google Scholar] [CrossRef] [PubMed]
- Rehermann, B. Hepatitis C virus versus innate and adaptive immune responses: A tale of coevolution and coexistence. J. Clin. Investig. 2009, 119, 1745–1754. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.H.; Cox, A.; Hoover, D.R.; Wang, X.H.; Mao, Q.; Ray, S.; Strathdee, S.A.; Vlahov, D.; Thomas, D.L. Protection against persistence of Hepatitis C. Lancet 2002, 359, 1478–1483. [Google Scholar] [CrossRef] [PubMed]
- Shoukry, N.H.; Grakoui, A.; Houghton, M.; Chien, D.Y.; Ghrayeb, J.; Reimann, K.A.; Walker, C.M. Memory CD8+ T cells are required for protection from persistent Hepatitis C virus infection. J. Exp. Med. 2003, 197, 1645–1655. [Google Scholar] [CrossRef] [PubMed]
- McKiernan, S.M.; Hagan, R.; Curry, M.; McDonald, G.S.; Kelly, A.; Nolan, N.; Walsh, A.; Hegarty, J.; Lawlor, E.; Kelleher, D. Distinct MHC class I and II alleles are associated with Hepatitis C viral clearance, originating from a single source. Hepatology 2004, 40, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Neumann-Haefelin, C.; McKiernan, S.; Ward, S.; Viazov, S.; Spangenberg, H.C.; Killinger, T.; Baumert, T.F.; Nazarova, N.; Sheridan, I.; Pybus, O.; et al. Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology 2006, 43, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Fitzmaurice, K.; Petrovic, D.; Ramamurthy, N.; Simmons, R.; Merani, S.; Gaudieri, S.; Sims, S.; Dempsey, E.; Freitas, E.; Lea, S.; et al. Molecular footprints reveal the impact of the protective HLA-A*03 allele in Hepatitis C virus infection. Gut 2011, 60, 1563–1571. [Google Scholar] [CrossRef] [PubMed]
- Ip, P.P.; Boerma, A.; Regts, J.; Meijerhof, T.; Wilschut, J.; Nijman, H.W.; Daemen, T. Alphavirus-based vaccines encoding nonstructural proteins of Hepatitis C virus induce robust and protective T-cell responses. Mol. Ther. 2014, 22, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Kolykhalov, A.A.; Agapov, E.V.; Blight, K.J.; Mihalik, K.; Feinstone, S.M.; Rice, C.M. Transmission of Hepatitis C by intrahepatic inoculation with transcribed RNA. Science 1997, 277, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Rammensee, H.; Bachmann, J.; Emmerich, N.P.; Bachor, O.A.; Stevanovic, S. Syfpeithi: Database for MHC ligands and peptide motifs. Immunogenetics 1999, 50, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Hoof, I.; Peters, B.; Sidney, J.; Pedersen, L.E.; Sette, A.; Lund, O.; Buus, S.; Nielsen, M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 2009, 61, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.; Sidney, J.; Bourne, P.; Bui, H.H.; Buus, S.; Doh, G.; Fleri, W.; Kronenberg, M.; Kubo, R.; Lund, O.; et al. The immune epitope database and analysis resource: From vision to blueprint. PLOS Biol. 2005, 3, e91. [Google Scholar] [CrossRef] [PubMed]
- Shirai, M.; Akatsuka, T.; Pendleton, C.D.; Houghten, R.; Wychowski, C.; Mihalik, K.; Feinstone, S.; Berzofsky, J.A. Induction of cytotoxic T cells to a cross-reactive epitope in the Hepatitis C virus nonstructural RNA polymerase-like protein. J. Virol. 1992, 66, 4098–4106. [Google Scholar] [PubMed]
- Lauer, G.M.; Barnes, E.; Lucas, M.; Timm, J.; Ouchi, K.; Kim, A.Y.; Day, C.L.; Robbins, G.K.; Casson, D.R.; Reiser, M.; et al. High resolution analysis of cellular immune responses in resolved and persistent Hepatitis C virus infection. Gastroenterology 2004, 127, 924–936. [Google Scholar] [CrossRef] [PubMed]
- Christinck, E.R.; Luscher, M.A.; Barber, B.H.; Williams, D.B. Peptide binding to class I MHC on living cells and quantitation of complexes required for CTL lysis. Nature 1991, 352, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.C.; Jardetzky, T.S.; Garrett, T.P.; Lane, W.S.; Strominger, J.L.; Wiley, D.C. Different length peptides bind to HLA-AW68 similarly at their ends but bulge out in the middle. Nature 1992, 360, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Crowe, S.R.; Miller, S.C.; Brown, D.M.; Adams, P.S.; Dutton, R.W.; Harmsen, A.G.; Lund, F.E.; Randall, T.D.; Swain, S.L.; Woodland, D.L. Uneven distribution of MHC class II epitopes within the influenza virus. Vaccine 2006, 24, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Dow, C.; Oseroff, C.; Peters, B.; Nance-Sotelo, C.; Sidney, J.; Buchmeier, M.; Sette, A.; Mothe, B.R. Lymphocytic choriomeningitis virus infection yields overlapping CD4+ and CD8+ T-cell responses. J. Virol. 2008, 82, 11734–11741. [Google Scholar] [CrossRef] [PubMed]
- Mothe, B.R.; Stewart, B.S.; Oseroff, C.; Bui, H.H.; Stogiera, S.; Garcia, Z.; Dow, C.; Rodriguez-Carreno, M.P.; Kotturi, M.; Pasquetto, V.; et al. Chronic lymphocytic choriomeningitis virus infection actively down-regulates CD4+ T cell responses directed against a broad range of epitopes. J. Immunol. 2007, 179, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, R.; Celis, E. Multiple roles for CD4+ T cells in anti-tumor immune responses. Immunol. Rev. 2008, 222, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, M.K.; Moon, J.J. The role of naive T cell precursor frequency and recruitment in dictating immune response magnitude. J. Immunol. 2012, 188, 4135–4140. [Google Scholar] [CrossRef] [PubMed]
- Kotturi, M.F.; Scott, I.; Wolfe, T.; Peters, B.; Sidney, J.; Cheroutre, H.; von Herrath, M.G.; Buchmeier, M.J.; Grey, H.; Sette, A. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J. Immunol. 2008, 181, 2124–2133. [Google Scholar] [CrossRef] [PubMed]
- Ahlen, G.; Nystrom, J.; Pult, I.; Frelin, L.; Hultgren, C.; Sallberg, M. In vivo clearance of Hepatitis C virus nonstructural 3/4A-expressing hepatocytes by DNA vaccine-primed cytotoxic T lymphocytes. J. Infect. Dis. 2005, 192, 2112–2116. [Google Scholar] [CrossRef] [PubMed]
- Wolk, B.; Trautwein, C.; Buchele, B.; Kersting, N.; Blum, H.E.; Rammensee, H.G.; Cerny, A.; Stevanovic, S.; Moradpour, D.; Brass, V. Identification of naturally processed Hepatitis C virus-derived major histocompatibility complex class I ligands. PLOS ONE 2012, 7, e29286. [Google Scholar] [CrossRef] [PubMed]
- De Lucas, S.; Bartolome, J.; Rodriguez-Inigo, E.; Casqueiro, M.; Millan, A.; Ruiz-Moreno, M.; Oliva, H.; Carreno, V. Distribution of Hepatitis C virus infection in liver biopsies from children and adults with chronic Hepatitis C. J. Med. Virol. 2001, 64, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.M.; Diamond, D.L.; Chan, E.Y.; Gritsenko, M.A.; Qian, W.; Stastna, M.; Baas, T.; Camp, D.G., 2nd.; Carithers, R.L., Jr.; Smith, R.D.; et al. Proteome analysis of liver cells expressing a full-length Hepatitis C virus (HCV) replicon and biopsy specimens of posttransplantation liver from HCV-infected patients. J. Virol. 2005, 79, 7558–7569. [Google Scholar] [CrossRef] [PubMed]
- Rotzschke, O.; Falk, K.; Stevanovic, S.; Jung, G.; Walden, P.; Rammensee, H.G. Exact prediction of a natural T cell epitope. Eur. J. Immunol. 1991, 21, 2891–2894. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Buus, S.; Appella, E.; Smith, J.A.; Chesnut, R.; Miles, C.; Colon, S.M.; Grey, H.M. Prediction of major histocompatibility complex binding regions of protein antigens by sequence pattern analysis. Proc. Natl. Acad. Sci. USA 1989, 86, 3296–3300. [Google Scholar] [CrossRef] [PubMed]
- Falk, K.; Rotzschke, O.; Stevanovic, S.; Jung, G.; Rammensee, H.G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 1991, 351, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Groettrup, M.; Kirk, C.J.; Basler, M. Proteasomes in immune cells: More than peptide producers? Nat. Rev. Immunol. 2010, 10, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Rock, K.L.; Goldberg, A.L. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 1999, 17, 739–779. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Norbury, C.C.; Cho, Y.; Yewdell, J.W.; Bennink, J.R. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8+ T cells at the levels of T cell repertoire and presentation of viral antigens. J. Exp. Med. 2001, 193, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Guillaume, B.; Chapiro, J.; Stroobant, V.; Colau, D.; van Holle, B.; Parvizi, G.; Bousquet-Dubouch, M.P.; Theate, I.; Parmentier, N.; van den Eynde, B.J. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc. Natl. Acad. Sci. USA 2010, 107, 18599–18604. [Google Scholar] [CrossRef] [PubMed]
- Kesmir, C.; Nussbaum, A.K.; Schild, H.; Detours, V.; Brunak, S. Prediction of proteasome cleavage motifs by neural networks. Protein Eng. 2002, 15, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Piontkivska, H. Frequent associations between CTL and T-helper epitopes in HIV-1 genomes and implications for multi-epitope vaccine designs. BMC Microbiol. 2010. [Google Scholar] [CrossRef]
- Rice, J.; Ottensmeier, C.H.; Stevenson, F.K. DNA vaccines: Precision tools for activating effective immunity against cancer. Na. Rev. Cancer 2008, 8, 108–120. [Google Scholar] [CrossRef]
- Tsang, J.Y.; Chai, J.G.; Lechler, R. Antigen presentation by mouse CD4+ T cells involving acquired MCH class II: Peptide complexes: Another mechanism to limit clonal expansion? Blood 2003, 101, 2704–2710. [Google Scholar] [CrossRef] [PubMed]
- Umeshappa, C.S.; Huang, H.; Xie, Y.; Wei, Y.; Mulligan, S.J.; Deng, Y.; Xiang, J. CD4+ Th-APC with acquired peptide/MHC class I and II complexes stimulate type 1 helper CD4+ and central memory CD8+ T cell responses. J. Immunol. 2009, 182, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Wetzel, S.A.; McKeithan, T.W.; Parker, D.C. Peptide-specific intercellular transfer of MHC class II to CD4+ T cells directly from the immunological synapse upon cellular dissociation. J. Immunol. 2005, 174, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Gromme, M.; Uytdehaag, F.G.; Janssen, H.; Calafat, J.; van Binnendijk, R.S.; Kenter, M.J.; Tulp, A.; Verwoerd, D.; Neefjes, J. Recycling MHC class I molecules and endosomal peptide loading. Proc. Natl. Acad. Sci. USA 1999, 96, 10326–10331. [Google Scholar] [CrossRef] [PubMed]
- Reimann, J.; Schirmbeck, R. Alternative pathways for processing exogenous and endogenous antigens that can generate peptides for MHC class I-restricted presentation. Immunol. Rev. 1999, 172, 131–152. [Google Scholar] [CrossRef] [PubMed]
- Lanzavecchia, A. From antigen presentation to T-cell activation. Res. Immunol. 1998, 149, 626. [Google Scholar] [CrossRef] [PubMed]
- Ossevoort, M.A.; de Bruijn, M.L.; van Veen, K.J.; Kast, W.M.; Melief, C.J. Peptide specificity of alloreactive CD4 positive T lymphocytes directed against a major histocompatibility complex class I disparity. Transplantation 1996, 62, 1485–1491. [Google Scholar] [CrossRef] [PubMed]
- Ip, P.P.; Boerma, A.; Walczak, M.; Oosterhuis, K.; Haanen, J.B.; Schumacher, T.N.; Nijman, H.W.; Daemen, T. Antigen design enhances the immunogenicity of Semliki Forest virus-based therapeutic human papillomavirus vaccines. Gene Ther. 2015. [Google Scholar] [CrossRef]
- Kedl, R.M.; Kappler, J.W.; Marrack, P. Epitope dominance, competition and T cell affinity maturation. Curr. Opin. Immunol. 2003, 15, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Henrickson, S.E.; Perro, M.; Loughhead, S.M.; Senman, B.; Stutte, S.; Quigley, M.; Alexe, G.; Iannacone, M.; Flynn, M.P.; Omid, S.; et al. Antigen availability determines CD8+ T cell-dendritic cell interaction kinetics and memory fate decisions. Immunity 2013, 39, 496–507. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).