TCR-T Cell Recognition of an NY-ESO-1 Epitope Presented by HLA-A2 Supertype: Implications for Cancer Immunotherapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
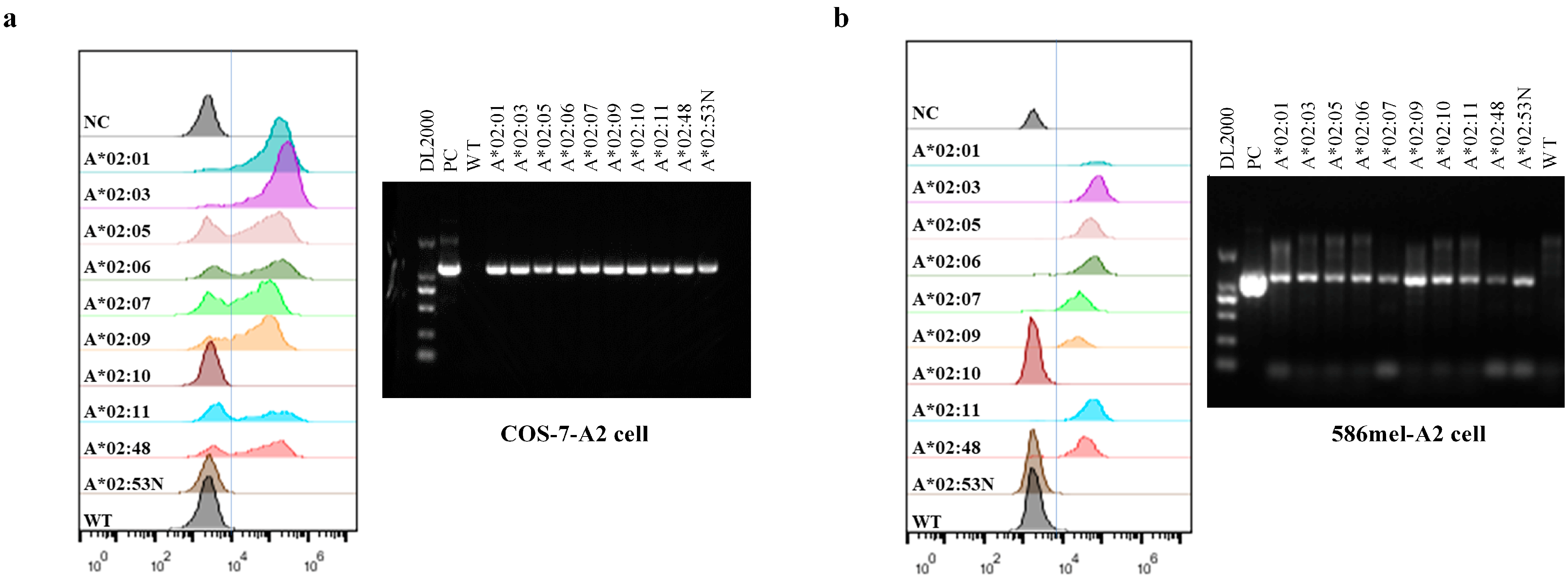
2.2. Generation of COS-7-A2, COS-7-NY-A2, 586 mel-A2, and 5637-NY-A2 Cells
2.3. Incubation of the TCR-T Cells with COS-7-A2, COS-7-NY-A2, 586 mel-A2, and 5637-NY-A2 Cells
2.4. ELISA
2.5. Flow Cytometry (FACS) Analysis
2.6. Statistical Analysis
3. Results
3.1. Population Frequency-Based Selection of HLA Supertype Genes for the Expansion of Population Coverage of a Given TCR
3.2. Target Antigen Peptide Presented by Members of the HLA-A2 Supertype in Cos-7 Cells Could Activate the HLA-A*02:01-Restricted TCR-T Cells
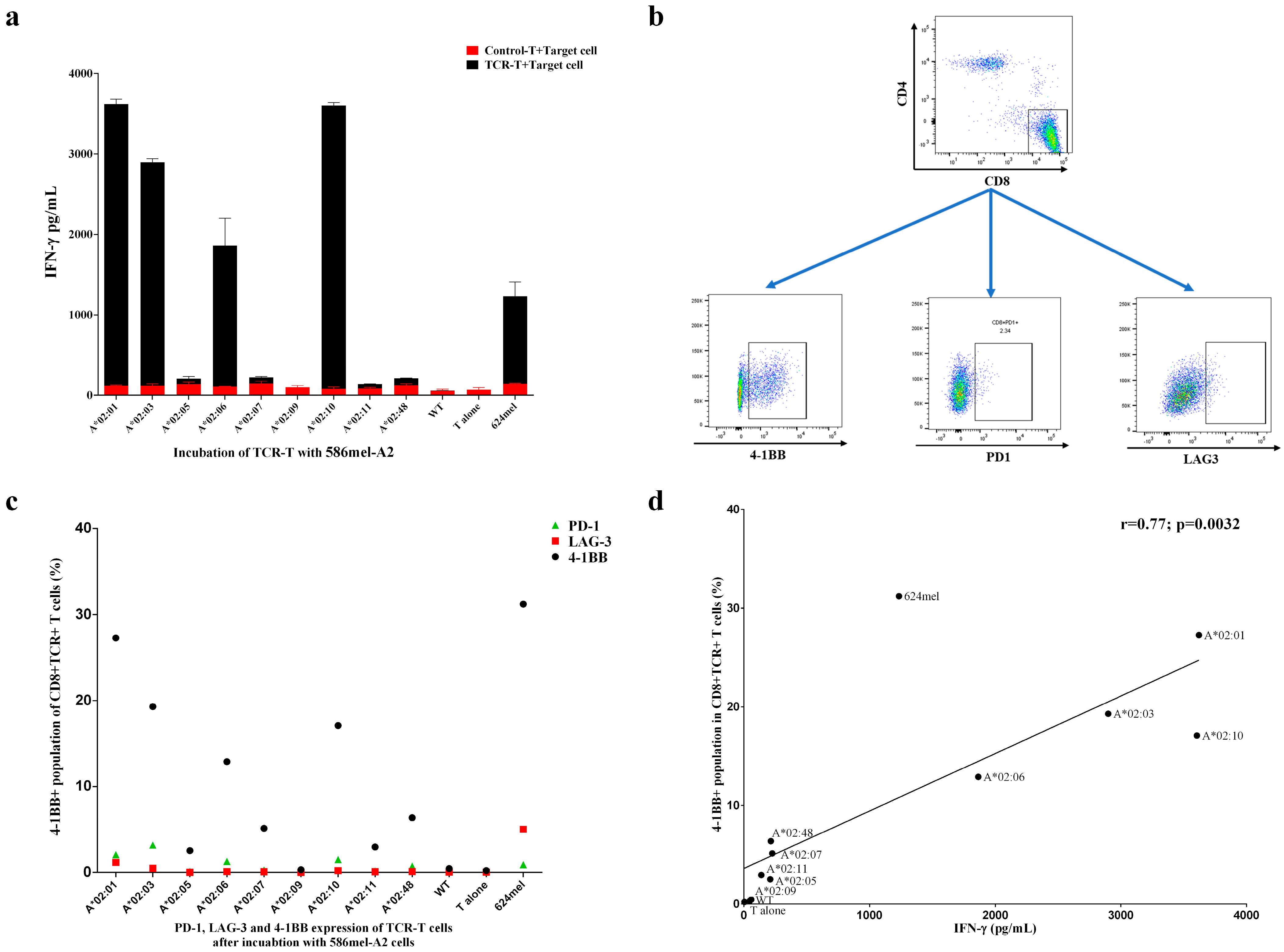
3.3. Evaluation of the Functional Avidity of Target Antigen Peptide Presented by Different HLA-A2 Alleles of the HLA-A2 Supertype
3.4. Target Antigen Peptide Could Be Presented by Transduced HLA-A2 Supertype Members in Tumor Cells and Be Recognized by HLA-A*02:01-Restricted TCR-T Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AFND | Allele Frequency Net Database |
| CAR-T | Chimeric antigen receptor T-cell therapy |
| FDA | US Food and Drug Administration |
| FCM | Flow cytometry |
| GFP | Green Fluorescent Protein |
| HLA | Human leukocyte antigen |
| HPV | Human papillomavirus |
| LCLs | Lymphoblastoid cell lines |
| pMHC | Peptide-major histocompatibility complex |
| TCR-T | T-cell receptor-engineered T-cell therapy |
| WT | Wild type |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Saito, M.; Suzuki, H.; Kono, K.; Takenoshita, S.; Kohno, T. Treatment of lung adenocarcinoma by molecular-targeted therapy and immunotherapy. Surg. Today 2018, 48, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chan, H.L.; Chen, P. Immune Checkpoint Inhibitors: Basics and Challenges. Curr. Med. Chem. 2019, 26, 3009–3025. [Google Scholar] [CrossRef] [PubMed]
- Baulu, E.; Gardet, C.; Chuvin, N.; Depil, S. TCR-engineered T cell therapy in solid tumors: State of the art and perspectives. Sci. Adv. 2023, 9, eadf3700. [Google Scholar] [CrossRef] [PubMed]
- Nagarsheth, N.B.; Norberg, S.M.; Sinkoe, A.L.; Adhikary, S.; Meyer, T.J.; Lack, J.B.; Warner, A.C.; Schweitzer, C.; Doran, S.L.; Korrapati, S.; et al. TCR-engineered T cells targeting E7 for patients with metastatic HPV-associated epithelial cancers. Nat. Med. 2021, 27, 419–425. [Google Scholar] [CrossRef]
- Doran, S.L.; Stevanovic, S.; Adhikary, S.; Gartner, J.J.; Jia, L.; Kwong, M.L.M.; Faquin, W.C.; Hewitt, S.M.; Sherry, R.M.; Yang, J.C.; et al. T-Cell Receptor Gene Therapy for Human Papillomavirus-Associated Epithelial Cancers: A First-in-Human, Phase I/II Study. J. Clin. Oncol. 2019, 37, 2759–2768. [Google Scholar] [CrossRef]
- Xia, Y.; Tian, X.; Wang, J.; Qiao, D.; Liu, X.; Xiao, L.; Liang, W.; Ban, D.; Chu, J.; Yu, J.; et al. Treatment of metastatic non-small cell lung cancer with NY-ESO-1 specific TCR engineered-T cells in a phase I clinical trial: A case report. Oncol. Lett. 2018, 16, 6998–7007. [Google Scholar] [CrossRef]
- Robbins, P.F.; Kassim, S.H.; Tran, T.L.; Crystal, J.S.; Morgan, R.A.; Feldman, S.A.; Yang, J.C.; Dudley, M.E.; Wunderlich, J.R.; Sherry, R.M.; et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: Long-term follow-up and correlates with response. Clin. Cancer Res. 2015, 21, 1019–1027. [Google Scholar] [CrossRef]
- US Food & Drug Administration. Approval Letter–TECELRA. Available online: https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/tecelra (accessed on 1 August 2024).
- Wang, R.F.; Wang, H.Y. Immune targets and neoantigens for cancer immunotherapy and precision medicine. Cell Res. 2017, 27, 11–37. [Google Scholar] [CrossRef]
- Gonzalez-Galarza, F.F.; McCabe, A.; Santos, E.; Jones, J.; Takeshita, L.; Ortega-Rivera, N.D.; Cid-Pavon, G.M.D.; Ramsbottom, K.; Ghattaoraya, G.; Alfirevic, A.; et al. Allele frequency net database (AFND) 2020 update: Gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020, 48, D783–D788. [Google Scholar] [CrossRef]
- He, Y.; Li, J.; Mao, W.; Zhang, D.; Liu, M.; Shan, X.; Zhang, B.; Zhu, C.; Shen, J.; Deng, Z.; et al. HLA common and well-documented alleles in China. HLA 2018, 92, 199–205. [Google Scholar] [CrossRef]
- Robinson, J.; Halliwell, J.A.; Hayhurst, J.D.; Flicek, P.; Parham, P.; Marsh, S.G. The IPD and IMGT/HLA database: Allele variant databases. Nucleic Acids Res. 2015, 43, D423–D431. [Google Scholar] [CrossRef]
- Sidney, J.; Peters, B.; Frahm, N.; Brander, C.; Sette, A. HLA class I supertypes: A revised and updated classification. BMC Immunol. 2008, 9, 1. [Google Scholar] [CrossRef]
- Sette, A.; Sidney, J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 1999, 50, 201–212. [Google Scholar] [CrossRef]
- Wang, M.; Claesson, M.H. Classification of human leukocyte antigen (HLA) supertypes. Methods Mol. Biol. 2014, 1184, 309–317. [Google Scholar] [CrossRef]
- Lund, O.; Nielsen, M.; Kesmir, C.; Petersen, A.G.; Lundegaard, C.; Worning, P.; Sylvester-Hvid, C.; Lamberth, K.; Roder, G.; Justesen, S.; et al. Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics 2004, 55, 797–810. [Google Scholar] [CrossRef]
- Shen, Y.; Parks, J.M.; Smith, J.C. HLA Class I Supertype Classification Based on Structural Similarity. J. Immunol. 2023, 210, 103–114. [Google Scholar] [CrossRef]
- Wisskirchen, K.; Metzger, K.; Schreiber, S.; Asen, T.; Weigand, L.; Dargel, C.; Witter, K.; Kieback, E.; Sprinzl, M.F.; Uckert, W.; et al. Isolation and functional characterization of hepatitis B virus-specific T-cell receptors as new tools for experimental and clinical use. PLoS ONE 2017, 12, e0182936. [Google Scholar] [CrossRef] [PubMed]
- Fleischhauer, K.; Tanzarella, S.; Wallny, H.J.; Bordignon, C.; Traversari, C. Multiple HLA-A alleles can present an immunodominant peptide of the human melanoma antigen Melan-A/MART-1 to a peptide-specific HLA-A*0201+ cytotoxic T cell line. J. Immunol. 1996, 157, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Fleischhauer, K.; Tanzarella, S.; Russo, V.; Sensi, M.L.; van der Bruggen, P.; Bordignon, C.; Traversari, C. Functional heterogeneity of HLA-A*02 subtypes revealed by presentation of a MAGE-3-encoded peptide to cytotoxic T cell clones. J. Immunol. 1997, 159, 2513–2521. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.F.; Johnston, S.L.; Southwood, S.; Sette, A.; Rosenberg, S.A. Recognition of an antigenic peptide derived from tyrosinase-related protein-2 by CTL in the context of HLA-A31 and -A33. J. Immunol. 1998, 160, 890–897. [Google Scholar] [CrossRef]
- McGranahan, N.; Rosenthal, R.; Hiley, C.T.; Rowan, A.J.; Watkins, T.B.K.; Wilson, G.A.; Birkbak, N.J.; Veeriah, S.; Van Loo, P.; Herrero, J.; et al. Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell 2017, 171, 1259–1271.e11. [Google Scholar] [CrossRef]
- Horlock, C.; Stott, B.; Dyson, J.; Ogg, G.; McPherson, T.; Jones, L.; Sewell, A.K.; Wooldridge, L.; Cole, D.K.; Stebbing, J.; et al. ELISPOT and functional T cell analyses using HLA mono-specific target cells. J. Immunol. Methods 2009, 350, 150–160. [Google Scholar] [CrossRef]
- Jurtz, V.; Paul, S.; Andreatta, M.; Marcatili, P.; Peters, B.; Nielsen, M. NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J. Immunol. 2017, 199, 3360–3368. [Google Scholar] [CrossRef]
- Rammensee, H.; Bachmann, J.; Emmerich, N.P.; Bachor, O.A.; Stevanovic, S. SYFPEITHI: Database for MHC ligands and peptide motifs. Immunogenetics 1999, 50, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.F.; Li, Y.F.; El-Gamil, M.; Zhao, Y.; Wargo, J.A.; Zheng, Z.; Xu, H.; Morgan, R.A.; Feldman, S.A.; Johnson, L.A.; et al. Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. J. Immunol. 2008, 180, 6116–6131. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Barker, D.J.; Georgiou, X.; Cooper, M.A.; Flicek, P.; Marsh, S.G.E. IPD-IMGT/HLA Database. Nucleic Acids Res. 2020, 48, D948–D955. [Google Scholar] [CrossRef]
- Kochenderfer, J.N.; Yu, Z.; Frasheri, D.; Restifo, N.P.; Rosenberg, S.A. Adoptive transfer of syngeneic T cells transduced with a chimeric antigen receptor that recognizes murine CD19 can eradicate lymphoma and normal B cells. Blood 2010, 116, 3875–3886. [Google Scholar] [CrossRef]
- Thomsen, M.; Lundegaard, C.; Buus, S.; Lund, O.; Nielsen, M. MHCcluster, a method for functional clustering of MHC molecules. Immunogenetics 2013, 65, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Sidney, J. HLA supertypes and supermotifs: A functional perspective on HLA polymorphism. Curr. Opin. Immunol. 1998, 10, 478–482. [Google Scholar] [CrossRef]
- Sidney, J.; Grey, H.M.; Kubo, R.T.; Sette, A. Practical, biochemical and evolutionary implications of the discovery of HLA class I supermotifs. Immunol. Today 1996, 17, 261–266. [Google Scholar] [CrossRef]
- Reynisson, B.; Alvarez, B.; Paul, S.; Peters, B.; Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020, 48, W449–W454. [Google Scholar] [CrossRef]
- Bassan, D.; Gozlan, Y.M.; Sharbi-Yunger, A.; Tzehoval, E.; Eisenbach, L. Optimizing T-cell receptor avidity with somatic hypermutation. Int. J. Cancer 2019, 145, 2816–2826. [Google Scholar] [CrossRef]
- Johnson, L.A.; Morgan, R.A.; Dudley, M.E.; Cassard, L.; Yang, J.C.; Hughes, M.S.; Kammula, U.S.; Royal, R.E.; Sherry, R.M.; Wunderlich, J.R.; et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 2009, 114, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Malecek, K.; Johnson, L.A.; Yu, Z.; Vega-Saenz de Miera, E.; Darvishian, F.; McGary, K.; Huang, K.; Boyer, J.; Corse, E.; et al. T-cell receptor affinity and avidity defines antitumor response and autoimmunity in T-cell immunotherapy. Proc. Natl. Acad. Sci. USA 2013, 110, 6973–6978. [Google Scholar] [CrossRef] [PubMed]
- Eiz-Vesper, B.; Schmetzer, H.M. Antigen-Presenting Cells: Potential of Proven und New Players in Immune Therapies. Transfus. Med. Hemother 2020, 47, 429–431. [Google Scholar] [CrossRef] [PubMed]
- Kanaseki, T.; Blanchard, N.; Hammer, G.E.; Gonzalez, F.; Shastri, N. ERAAP synergizes with MHC class I molecules to make the final cut in the antigenic peptide precursors in the endoplasmic reticulum. Immunity 2006, 25, 795–806. [Google Scholar] [CrossRef]
- Serwold, T.; Gaw, S.; Shastri, N. ER aminopeptidases generate a unique pool of peptides for MHC class I molecules. Nat. Immunol. 2001, 2, 644–651. [Google Scholar] [CrossRef]
- Hill, A.; Jugovic, P.; York, I.; Russ, G.; Bennink, J.; Yewdell, J.; Ploegh, H.; Johnson, D. Herpes simplex virus turns off the TAP to evade host immunity. Nature 1995, 375, 411–415. [Google Scholar] [CrossRef]
- Wang, Q.; Douglass, J.; Hwang, M.S.; Hsiue, E.H.; Mog, B.J.; Zhang, M.; Papadopoulos, N.; Kinzler, K.W.; Zhou, S.; Vogelstein, B. Direct Detection and Quantification of Neoantigens. Cancer Immunol. Res. 2019, 7, 1748–1754. [Google Scholar] [CrossRef]
- Osawa, R.; Tsunoda, T.; Yoshimura, S.; Watanabe, T.; Miyazawa, M.; Tani, M.; Takeda, K.; Nakagawa, H.; Nakamura, Y.; Yamaue, H. Identification of HLA-A24-restricted novel T Cell epitope peptides derived from P-cadherin and kinesin family member 20A. J. Biomed. Biotechnol. 2012, 2012, 848042. [Google Scholar] [CrossRef] [PubMed]
- Marincola, F.M.; Shamamian, P.; Alexander, R.B.; Gnarra, J.R.; Turetskaya, R.L.; Nedospasov, S.A.; Simonis, T.B.; Taubenberger, J.K.; Yannelli, J.; Mixon, A.; et al. Loss of HLA haplotype and B locus down-regulation in melanoma cell lines. J. Immunol. 1994, 153, 1225–1237. [Google Scholar] [CrossRef]
- Marincola, F.M.; Shamamian, P.; Simonis, T.B.; Abati, A.; Hackett, J.; O’Dea, T.; Fetsch, P.; Yannelli, J.; Restifo, N.P.; Mule, J.J.; et al. Locus-specific analysis of human leukocyte antigen class I expression in melanoma cell lines. J. Immunother. Emphas. Tumor Immunol. 1994, 16, 13–23. [Google Scholar] [CrossRef]
- Xu, X.; Wang, X.; Fu, B.; Meng, L.; Lang, B. Differentially expressed genes and microRNAs in bladder carcinoma cell line 5637 and T24 detected by RNA sequencing. Int. J. Clin. Exp. Pathol. 2015, 8, 12678–12687. [Google Scholar]
- Leibowitz, M.S.; Andrade Filho, P.A.; Ferrone, S.; Ferris, R.L. Deficiency of activated STAT1 in head and neck cancer cells mediates TAP1-dependent escape from cytotoxic T lymphocytes. Cancer Immunol. Immunother. 2011, 60, 525–535. [Google Scholar] [CrossRef]
- O’Donnell, T.J.; Rubinsteyn, A.; Bonsack, M.; Riemer, A.B.; Laserson, U.; Hammerbacher, J. MHCflurry: Open-Source Class I MHC Binding Affinity Prediction. Cell Syst. 2018, 7, 129–132.e4. [Google Scholar] [CrossRef] [PubMed]
- Assarsson, E.; Sidney, J.; Oseroff, C.; Pasquetto, V.; Bui, H.H.; Frahm, N.; Brander, C.; Peters, B.; Grey, H.; Sette, A. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J. Immunol. 2007, 178, 7890–7901. [Google Scholar] [CrossRef] [PubMed]
- Busch, D.H.; Pamer, E.G. MHC class I/peptide stability: Implications for immunodominance, in vitro proliferation, and diversity of responding CTL. J. Immunol. 1998, 160, 4441–4448. [Google Scholar] [CrossRef]
- Lazarski, C.A.; Chaves, F.A.; Jenks, S.A.; Wu, S.; Richards, K.A.; Weaver, J.M.; Sant, A.J. The kinetic stability of MHC class II:peptide complexes is a key parameter that dictates immunodominance. Immunity 2005, 23, 29–40. [Google Scholar] [CrossRef]
- van der Burg, S.H.; Visseren, M.J.; Brandt, R.M.; Kast, W.M.; Melief, C.J. Immunogenicity of peptides bound to MHC class I molecules depends on the MHC-peptide complex stability. J. Immunol. 1996, 156, 3308–3314. [Google Scholar] [CrossRef]
- Harndahl, M.; Rasmussen, M.; Roder, G.; Dalgaard Pedersen, I.; Sorensen, M.; Nielsen, M.; Buus, S. Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity. Eur. J. Immunol. 2012, 42, 1405–1416. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, K.W.; Rasmussen, M.; Buus, S.; Nielsen, M. NetMHCstab–predicting stability of peptide-MHC-I complexes; impacts for cytotoxic T lymphocyte epitope discovery. Immunology 2014, 141, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M.; Fenoy, E.; Harndahl, M.; Kristensen, A.B.; Nielsen, I.K.; Nielsen, M.; Buus, S. Pan-Specific Prediction of Peptide-MHC Class I Complex Stability, a Correlate of T Cell Immunogenicity. J. Immunol. 2016, 197, 1517–1524. [Google Scholar] [CrossRef] [PubMed]
- Fasoulis, R.; Rigo, M.M.; Antunes, D.A.; Paliouras, G.; Kavraki, L.E. Transfer learning improves pMHC kinetic stability and immunogenicity predictions. Immunoinformatics 2024, 13, 100030. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Q.; Liu, F.; Ma, Y.; Li, Y.; Lin, T.; Chen, X.; Zhang, J.; Sun, H.; Wang, Z.; Xia, X.; et al. TCR-T Cell Recognition of an NY-ESO-1 Epitope Presented by HLA-A2 Supertype: Implications for Cancer Immunotherapy. Vaccines 2025, 13, 898. https://doi.org/10.3390/vaccines13090898
Lin Q, Liu F, Ma Y, Li Y, Lin T, Chen X, Zhang J, Sun H, Wang Z, Xia X, et al. TCR-T Cell Recognition of an NY-ESO-1 Epitope Presented by HLA-A2 Supertype: Implications for Cancer Immunotherapy. Vaccines. 2025; 13(9):898. https://doi.org/10.3390/vaccines13090898
Chicago/Turabian StyleLin, Qingqing, Fenglan Liu, Yipeng Ma, Yanwei Li, Tong Lin, Xiaochun Chen, Jinling Zhang, Heng Sun, Zhi Wang, Xiaojun Xia, and et al. 2025. "TCR-T Cell Recognition of an NY-ESO-1 Epitope Presented by HLA-A2 Supertype: Implications for Cancer Immunotherapy" Vaccines 13, no. 9: 898. https://doi.org/10.3390/vaccines13090898
APA StyleLin, Q., Liu, F., Ma, Y., Li, Y., Lin, T., Chen, X., Zhang, J., Sun, H., Wang, Z., Xia, X., Tian, G., Jin, S., & Wang, M. (2025). TCR-T Cell Recognition of an NY-ESO-1 Epitope Presented by HLA-A2 Supertype: Implications for Cancer Immunotherapy. Vaccines, 13(9), 898. https://doi.org/10.3390/vaccines13090898






