Protective Efficacy of Subunit Vaccine Expressing Rv0976c Against Tuberculosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Development Conditions
2.2. Molecular Cloning
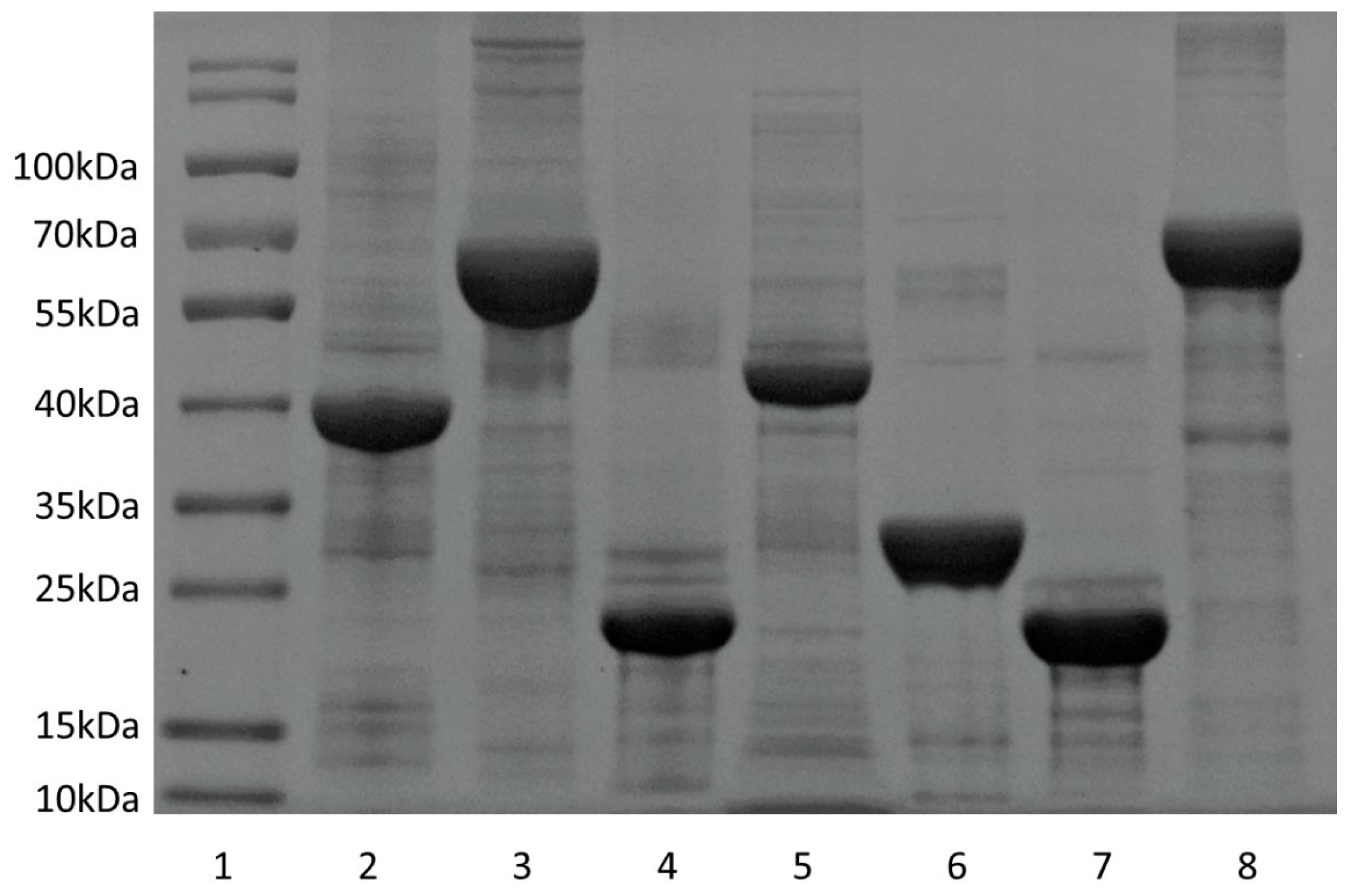
2.3. Expression and Purification of Recombinant Proteins
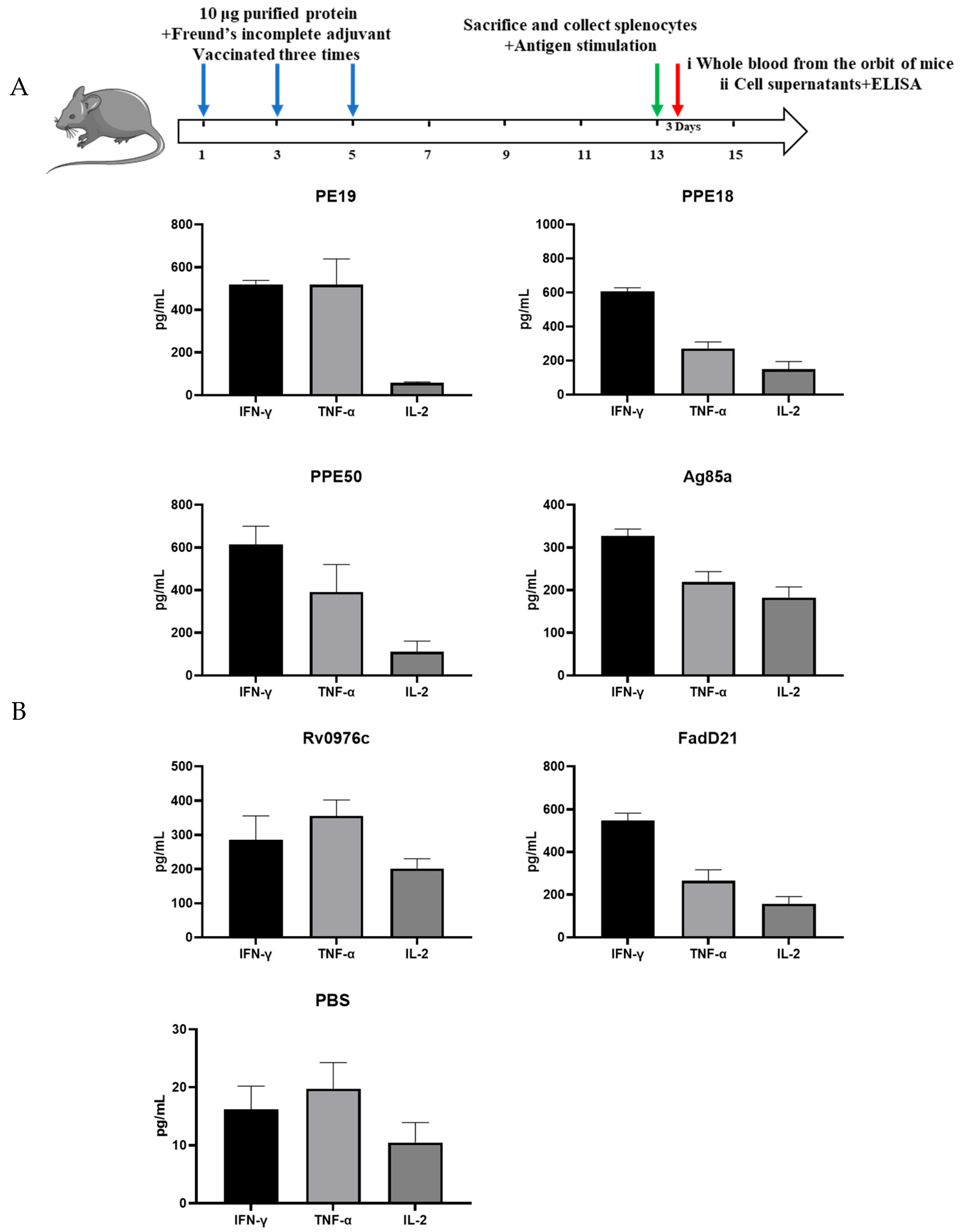
2.4. Immunogenicity Assays
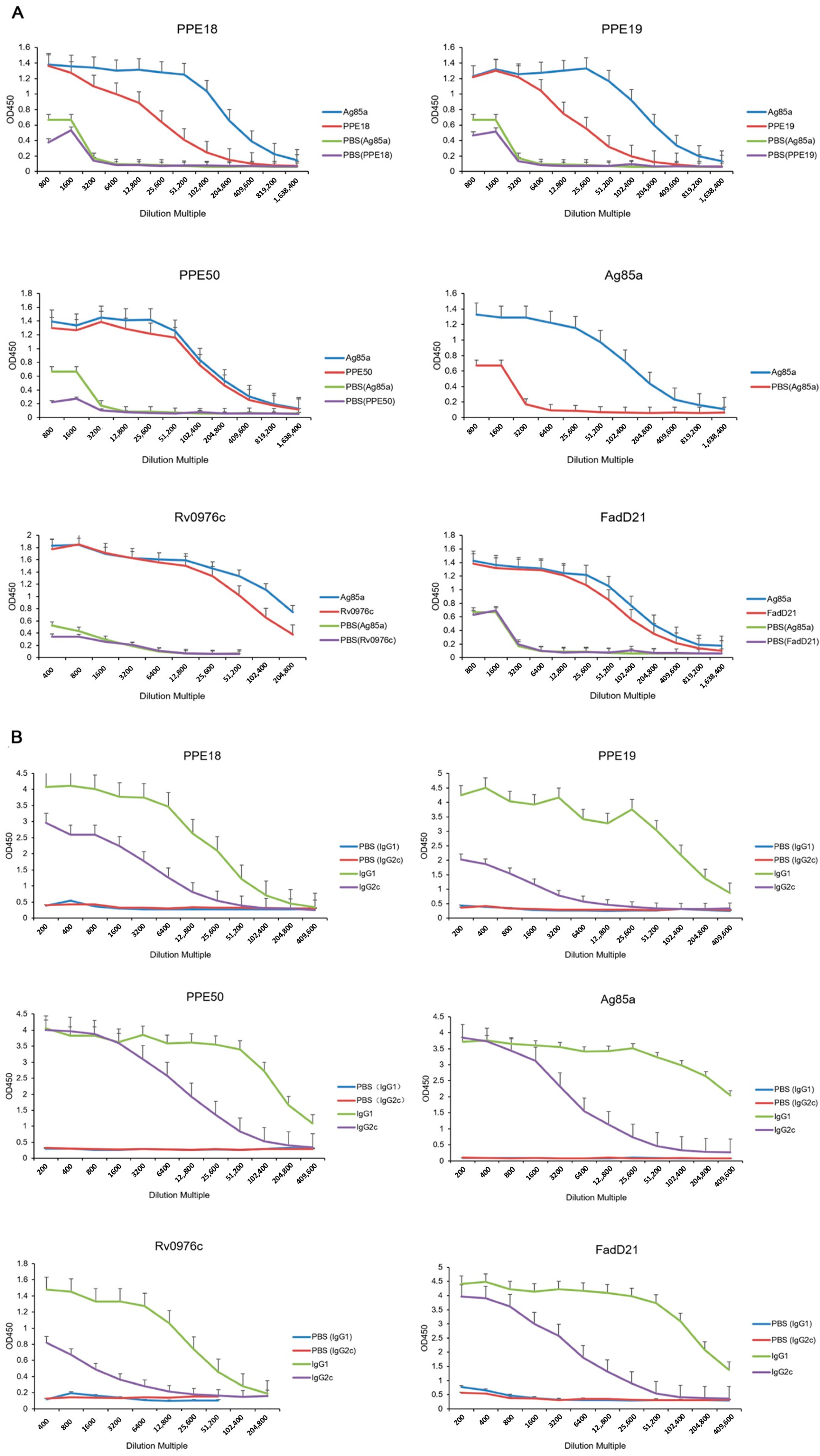
2.5. Evaluation of Serum Antibody Levels of Rv0976c
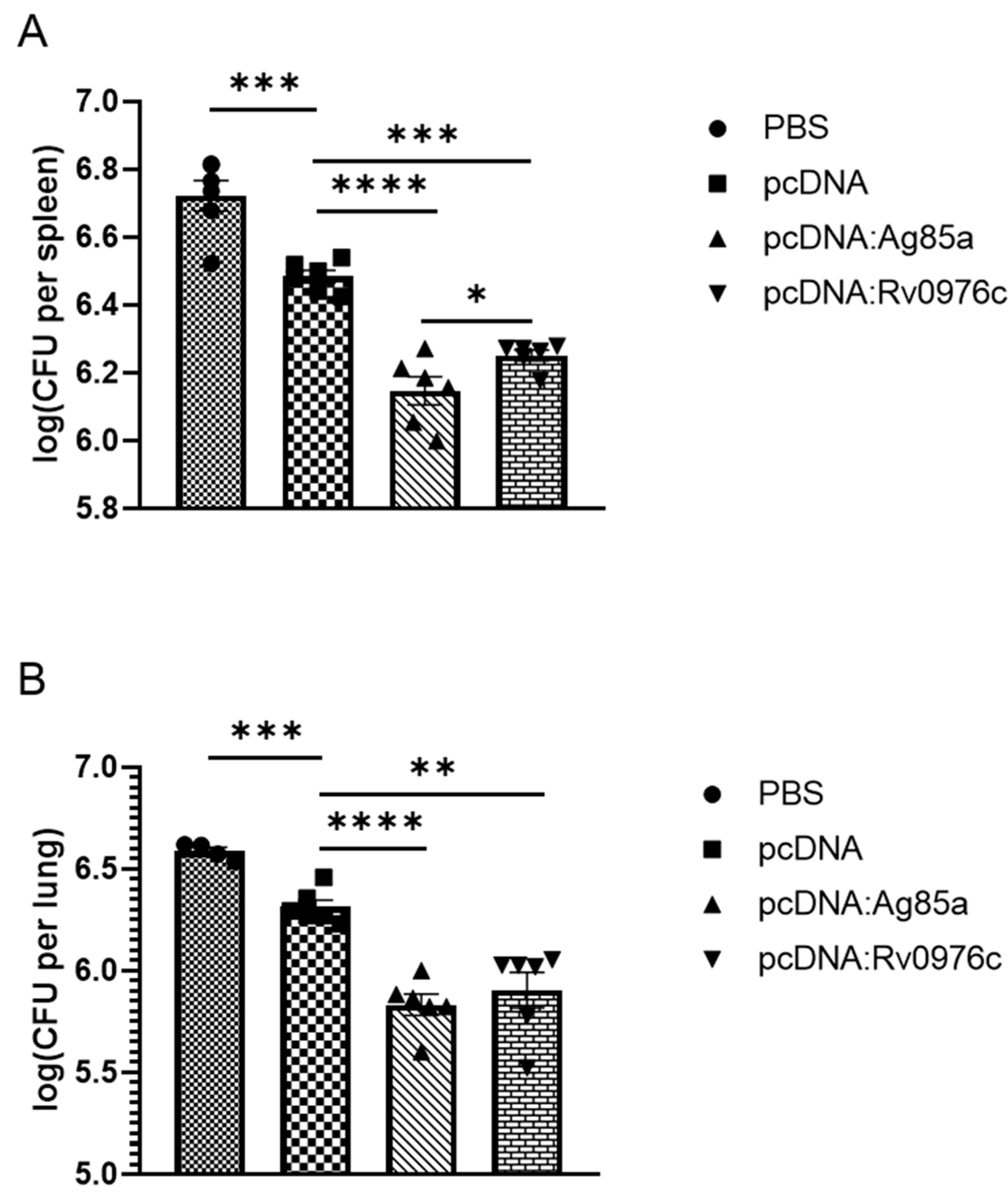
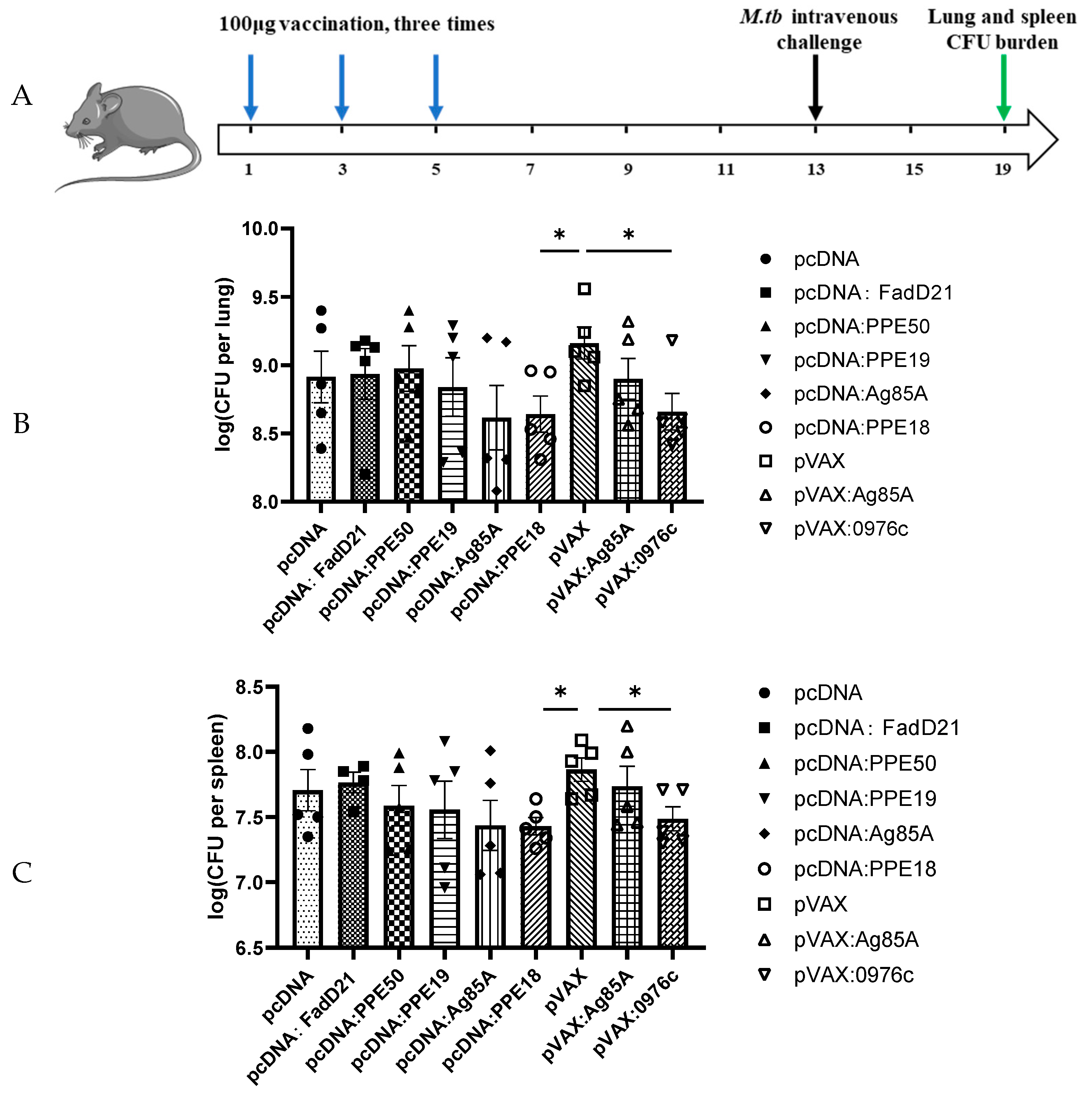
2.6. Animal Protection Assays
3. Results
3.1. Cellular and Humoral Immune Responses Induced by Selected Proteins
3.2. Protective Efficacy of Selected Antigens
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Ahmad, S.; Bhattacharya, D.; Gupta, N.; Rawat, V.; Tousif, S.; Van Kaer, L.; Das, G. Clofazimine enhances the efficacy of BCG revaccination via stem cell-like memory T cells. PLoS Pathog. 2020, 16, e1008356. [Google Scholar] [CrossRef]
- Russell, D.G.; Barry, C.E.; Flynn, J.L. Tuberculosis: What We Don’t Know Can, and Does, Hurt Us. Science 2010, 328, 852–856. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, S.H.E. Fact and fiction in tuberculosis vaccine research: 10 years later. Lancet Infect. Dis. 2011, 11, 633–640. [Google Scholar] [CrossRef]
- Van Rhijn, I.; Moody, D.B. Donor Unrestricted T Cells: A Shared Human T Cell Response. J. Immunol. 2015, 195, 1927–1932. [Google Scholar] [CrossRef]
- Ernst, J.D. Mechanisms of M. tuberculosis Immune Evasion as Challenges to TB Vaccine Design. Cell Host Microbe 2018, 24, 34–42. [Google Scholar] [CrossRef]
- Yang, R.; Mele, F.; Worley, L.; Langlais, D.; Rosain, J.; Benhsaien, I.; Elarabi, H.; Croft, C.A.; Doisne, J.M.; Zhang, P.; et al. Human T-bet Governs Innate and Innate-like Adaptive IFN-γ Immunity against Mycobacteria. Cell 2020, 183, 1826–1847. [Google Scholar] [CrossRef]
- Soares, A.P.; Scriba, T.J.; Joseph, S.; Harbacheuski, R.; Murray, R.A.; Gelderbloem, S.J.; Hawkridge, A.; Hussey, G.D.; Maecker, H.; Kaplan, G.; et al. Bacillus Calmette-Guérin vaccination of human newborns induces T cells with complex cytokine and phenotypic profiles. J. Immunol. 2008, 180, 3569–3577. [Google Scholar] [CrossRef]
- Borremans, M.; Dewit, L.; Volckaert, G.; Ooms, J.; Debruyn, J.; Huygen, K.; Vanvooren, J.P.; Stelandre, M.; Verhofstadt, R.; Content, J. Cloning, Sequence Determination, and Expression of a 32-Kilodalton-Protein Gene of Mycobacterium-Tuberculosis. Infect. Immun. 1989, 57, 3123–3130. [Google Scholar] [CrossRef]
- Dietrich, J.; Aagaard, C.; Leah, R.; Olsen, A.W.; Stryhn, A.; Doherty, T.M.; Andersen, P. Exchanging ESAT6 with TB10.4 in an Ag85B fusion molecule-based tuberculosis subunit vaccine: Efficient protection and ESAT6-based sensitive monitoring of vaccine efficacy. J. Immunol. 2005, 174, 6332–6339. [Google Scholar] [CrossRef] [PubMed]
- Ullah, I.; Bibi, S.; Ul Haq, I.; Safia Ullah, K.; Ge, L.; Shi, X.; Bin, M.; Niu, H.; Tian, J.; Zhu, B. The Systematic Review and Meta-Analysis on the Immunogenicity and Safety of the Tuberculosis Subunit Vaccines M72/AS01E and MVA85A. Front. Immunol. 2020, 11, 1806. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.L.; Dietrich, J.; Tan, E.; Abalos, R.M.; Burgos, J.; Bigbee, C.; Bigbee, M.; Milk, L.; Gideon, H.P.; Rodgers, M.; et al. The multistage vaccine H56 boosts the effects of BCG to protect cynomolgus macaques against active tuberculosis and reactivation of latent Mycobacterium tuberculosis infection. J. Clin. Invest. 2012, 122, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Bertholet, S.; Ireton, G.C.; Ordway, D.J.; Windish, H.P.; Pine, S.O.; Kahn, M.; Phan, T.; Orme, I.M.; Vedvick, T.S.; Baldwin, S.L.; et al. A Defined Tuberculosis Vaccine Candidate Boosts BCG and Protects Against Multidrug-Resistant. Sci. Transl. Med. 2010, 2, 53ra74. [Google Scholar] [CrossRef]
- Reed, S.G.; Coler, R.N.; Dalemans, W.; Tan, E.V.; Dela Cruz, E.C.; Basaraba, R.J.; Orme, I.M.; Skeiky, Y.A.W.; Alderson, M.R.; Cowgill, K.D.; et al. Defined tuberculosis vaccine, Mtb72F/AS02A, evidence of protection in cynomolgus monkeys. Proc. Natl. Acad. Sci. USA 2009, 106, 2301–2306. [Google Scholar] [CrossRef] [PubMed]
- Rodo, M.J.; Rozot, V.; Nemes, E.; Dintwe, O.; Hatherill, M.; Little, F.; Scriba, T.J. A comparison of antigen-specific T cell responses induced by six novel tuberculosis vaccine candidates. PLoS Pathog. 2019, 15, e1007643. [Google Scholar] [CrossRef]
- Ru, H.W.; Liu, X.J.; Lin, C.; Yang, J.Y.; Chen, F.Z.; Sun, R.F.; Zhang, L.; Liu, J. The Impact of Genome Region of Difference 4 (RD4) on Mycobacterial Virulence and BCG Efficacy. Front. Cell. Infect. Microbiol. 2017, 7, 239. [Google Scholar] [CrossRef]
- Bajaj, R.A.; Arbing, M.A.; Shin, A.; Cascio, D.; Miallau, L. Crystal structure of the toxin Msmeg_6760, the structural homolog of Rv2035, a novel type II toxin involved in the hypoxic response. Acta Crystallogr. F 2016, 72, 863–869. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Comprehensive comparative-genomic analysis of Type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct 2009, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Bartek, I.L.; Woolhiser, L.K.; Baughn, A.D.; Basaraba, R.J.; Jacobs, W.R.; Lenaerts, A.J.; Voskuil, M.I. Lsr2 Is a Global Transcriptional Regulator Required for Adaptation to Changing Oxygen Levels and Virulence. Mbio 2014, 5, 10-1128. [Google Scholar] [CrossRef]
- Chen, J.M.; German, G.J.; Alexander, D.C.; Ren, H.P.; Tan, T.; Liu, J. Roles of Lsr2 in colony morphology and biofilm formation of Mycobacterium smegmatis. J. Bacteriol. 2006, 188, 633–641. [Google Scholar] [CrossRef]
- Chen, J.M.; Ren, H.; Shaw, J.E.; Wang, Y.J.; Li, M.; Leung, A.S.; Tran, V.; Berbenetz, N.M.; Kocíncová, D.; Yip, C.M.; et al. Lsr2 of Mycobacterium tuberculosis is a DNA-bridging protein. Nucleic Acids Res. 2008, 36, 2123–2135. [Google Scholar] [CrossRef]
- Gordon, B.R.G.; Imperial, R.; Wang, L.R.; Navarre, W.W.; Liu, J. Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J. Bacteriol. 2008, 190, 7052–7059. [Google Scholar] [CrossRef]
- Gordon, B.R.; Li, Y.; Wang, L.; Sintsova, A.; van Bakel, H.; Tian, S.; Navarre, W.W.; Xia, B.; Liu, J. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2010, 107, 5154–5159. [Google Scholar] [CrossRef]
- Gordon, B.R.; Li, Y.; Cote, A.; Weirauch, M.T.; Ding, P.; Hughes, T.R.; Navarre, W.W.; Xia, B.; Liu, J. Structural basis for recognition of AT-rich DNA by unrelated xenogeneic silencing proteins. Proc. Natl. Acad. Sci. USA 2011, 108, 10690–10695. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, M.A.; Harth, G.; Dillon, B.J.; Maslesa-Galic, S. Recombinant bacillus calmette guerin (BCG) vaccines expressing the Mycobacterium tuberculosis 30-kDa major secretory protein induce greater protective immunity against tuberculosis than conventional BCG vaccines in a highly susceptible animal model. Proc. Natl. Acad. Sci. USA 2000, 97, 13853–13858. [Google Scholar] [CrossRef]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E.; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 396, 190–198. [Google Scholar] [CrossRef]
- Sassetti, C.M.; Rubin, E.J. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 2003, 100, 12989–12994. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.; Pellegrini, M.; Eisenberg, D. Identifying cognate binding pairs among a large set of paralogs: The case of PE/PPE proteins of Mycobacterium tuberculosis. PLoS Comput. Biol. 2008, 4, e1000174. [Google Scholar] [CrossRef] [PubMed]
- Brosch, R.; Gordon, S.V.; Garnier, T.; Eiglmeier, K.; Frigui, W.; Valenti, P.; Dos Santos, S.; Duthoy, S.; Lacroix, C.; Garcia-Pelayo, C.; et al. Genome plasticity of BCG and impact on vaccine efficacy. Proc. Natl. Acad. Sci. USA 2007, 104, 5596–5601. [Google Scholar] [CrossRef]
- Andersen, P.; Kaufmann, S.H.E. Novel Vaccination Strategies against Tuberculosis. Cold Spring Harb. Perspect. Med. 2014, 4, a018523. [Google Scholar] [CrossRef]
- Beverley, P. TB vaccine failure was predictable. Nature 2013, 503, 469. [Google Scholar] [CrossRef]
- Aagaard, C.; Hoang, T.; Dietrich, J.; Cardona, P.J.; Izzo, A.; Dolganov, G.; Schoolnik, G.K.; Cassidy, J.P.; Billeskov, R.; Andersen, P. A multistage tuberculosis vaccine that confers efficient protection before and after exposure. Nat. Med. 2011, 17, 189–194. [Google Scholar] [CrossRef]
- Commandeur, S.; van Meijgaarden, K.E.; Prins, C.; Pichugin, A.V.; Dijkman, K.; van den Eeden, S.J.; Friggen, A.H.; Franken, K.L.; Dolganov, G.; Kramnik, I.; et al. An unbiased genome-wide Mycobacterium tuberculosis gene expression approach to discover antigens targeted by human T cells expressed during pulmonary infection. J. Immunol. 2013, 190, 1659–1671. [Google Scholar] [CrossRef]
- Dey, B.; Jain, R.; Gupta, U.D.; Katoch, V.M.; Ramanathan, V.D.; Tyagi, A.K. A booster vaccine expressing a latency-associated antigen augments BCG induced immunity and confers enhanced protection against tuberculosis. PLoS ONE 2011, 6, e23360. [Google Scholar] [CrossRef] [PubMed]
- North, R.J.; LaCourse, R.; Ryan, L. Vaccinated mice remain more susceptible to Mycobacterium tuberculosis infection initiated via the respiratory route than via the intravenous route. Infect. Immun. 1999, 67, 2010–2012. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mansury, D.; Ghazvini, K.; Amel Jamehdar, S.; Badiee, A.; Tafaghodi, M.; Nikpoor, A.R.; Amini, Y.; Jaafari, M.R. Increasing Cellular Immune Response in Liposomal Formulations of DOTAP Encapsulated by Fusion Protein Hspx, PPE44, And Esxv, as a Potential Tuberculosis Vaccine Candidate. Rep. Biochem. Mol. Biol. 2019, 7, 156–166. [Google Scholar]
- Khademi, F.; Sahebkar, A.; Fasihi-Ramandi, M.; Taheri, R.A. Induction of strong immune response against a multicomponent antigen of Mycobacterium tuberculosis in BALB/c mice using PLGA and DOTAP adjuvant. APMIS 2018, 126, 509–514. [Google Scholar] [CrossRef]
- Skeiky, Y.A.; Sadoff, J.C. Advances in tuberculosis vaccine strategies. Nat. Rev. Microbiol. 2006, 4, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Talaat, A.M.; Lyons, R.; Howard, S.T.; Johnston, S.A. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. USA 2004, 101, 4602–4607. [Google Scholar] [CrossRef]
- Gautam, U.S.; Mehra, S.; Kaushal, D. In-Vivo Gene Signatures of Mycobacterium tuberculosis in C3HeB/FeJ Mice. PLoS ONE 2015, 10, e0135208. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Z.; Chen, D.; Chen, F.; Xu, W.; Pan, Z.; Xiang, Z.; Gao, X.; Li, Y.; Zhong, F.; Liu, J.; et al. Protective Efficacy of Subunit Vaccine Expressing Rv0976c Against Tuberculosis. Vaccines 2025, 13, 872. https://doi.org/10.3390/vaccines13080872
Zhou Z, Chen D, Chen F, Xu W, Pan Z, Xiang Z, Gao X, Li Y, Zhong F, Liu J, et al. Protective Efficacy of Subunit Vaccine Expressing Rv0976c Against Tuberculosis. Vaccines. 2025; 13(8):872. https://doi.org/10.3390/vaccines13080872
Chicago/Turabian StyleZhou, Ziwei, Dan Chen, Fuzeng Chen, Wenxi Xu, Zhifen Pan, Zhihao Xiang, Xiaoxiao Gao, Yeyu Li, Fagang Zhong, Jun Liu, and et al. 2025. "Protective Efficacy of Subunit Vaccine Expressing Rv0976c Against Tuberculosis" Vaccines 13, no. 8: 872. https://doi.org/10.3390/vaccines13080872
APA StyleZhou, Z., Chen, D., Chen, F., Xu, W., Pan, Z., Xiang, Z., Gao, X., Li, Y., Zhong, F., Liu, J., & Zhang, L. (2025). Protective Efficacy of Subunit Vaccine Expressing Rv0976c Against Tuberculosis. Vaccines, 13(8), 872. https://doi.org/10.3390/vaccines13080872





