Accelerating Neoantigen Discovery: A High-Throughput Approach to Immunogenic Target Identification
Abstract
1. Introduction
2. Methods
2.1. Assembly of the Training Datasets
2.2. Characterization of the Training Datasets
2.3. Peptide Encoding
2.4. Algorithm Training
2.5. Bias Testing for HLA and Peptide Length
2.6. Benchmarking
2.7. In Vitro Immunogenicity Validation with ELISpot
2.8. Retrospective Analysis of Personalized Cancer Vaccine Trials
2.9. Biomarker Analysis of a CPI-Treated Cohort
3. Results
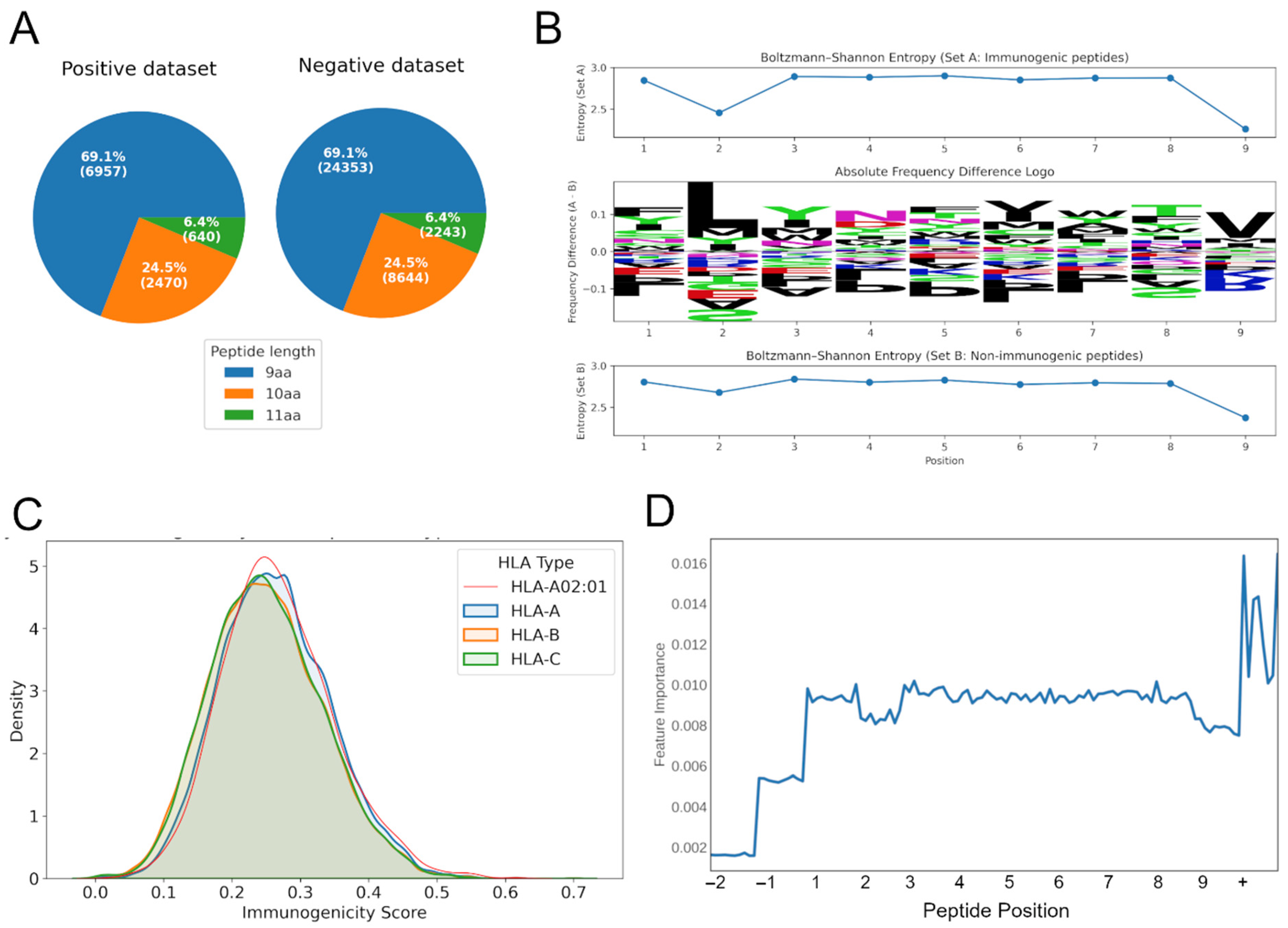
3.1. Building the Training Dataset of the Immunogenicity Model
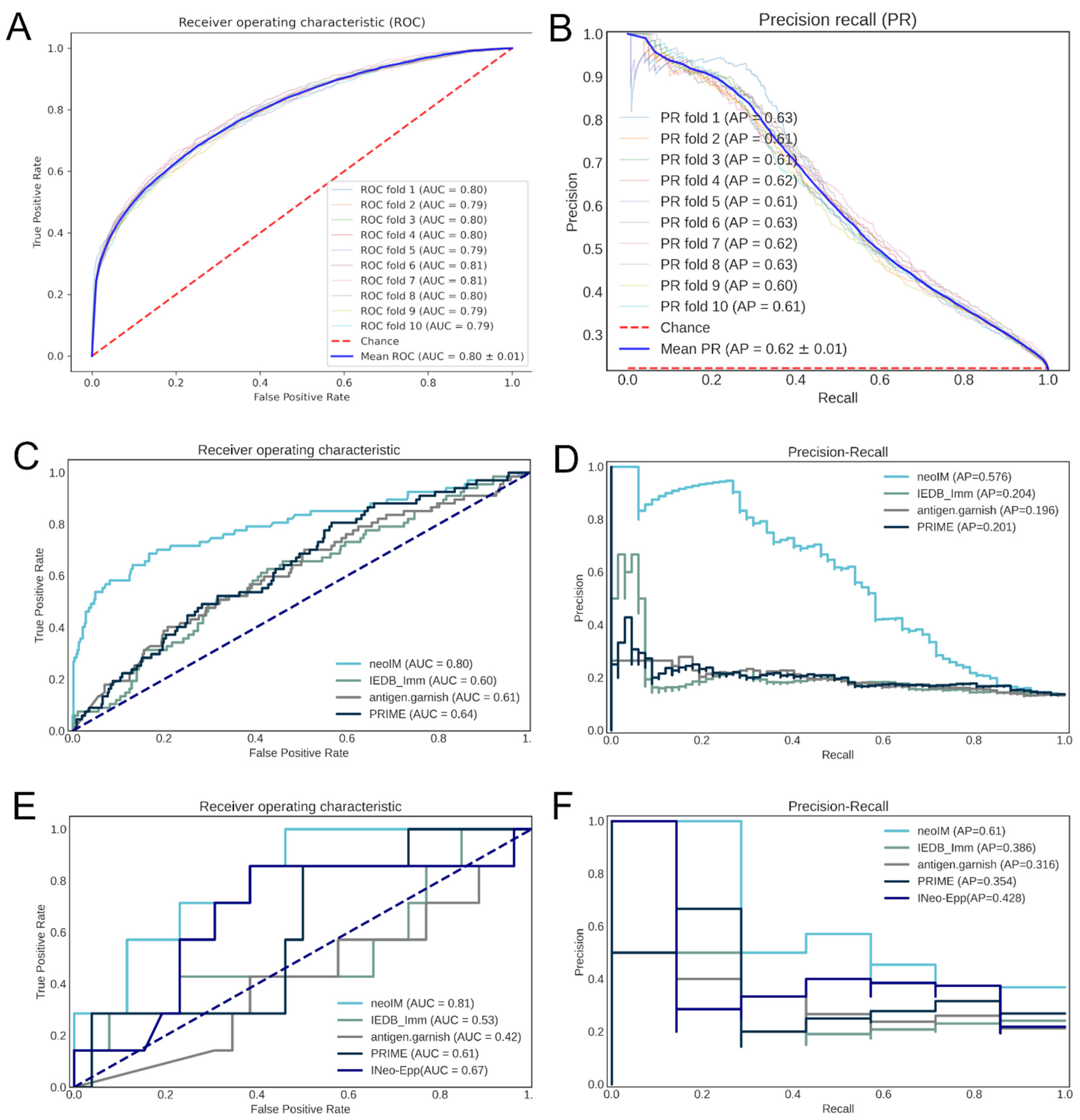
3.2. neoIM Model Training and Performance
3.3. Bias Testing Demonstrates Minor Influence of HLA Allele and Peptide Length
3.4. Benchmarking the neoIM Model
3.5. ELISpot Validation of neoIM Immunogenicity Predictions for Different Neoantigen Types
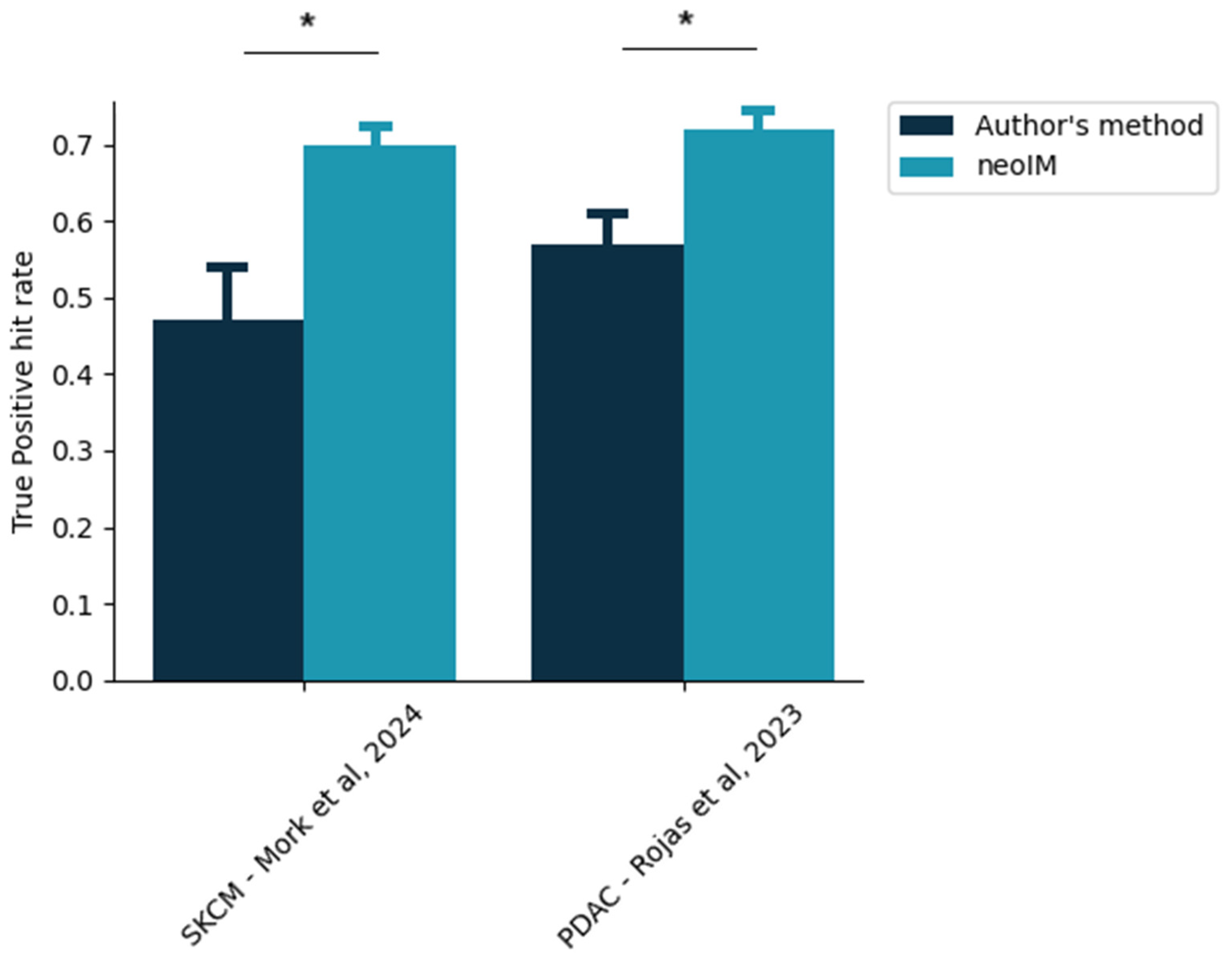
3.6. Retrospective Analysis of Recent Personalized Cancer Vaccine Trials
3.7. neoIM Tumor Immunogenicity as a Predictive Biomarker for CPI Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coulie, P.G.; Van Den Eynde, B.J.; Van Der Bruggen, P.; Boon, T. Tumour Antigens Recognized by T Lymphocytes: At the Core of Cancer Immunotherapy. Nat. Rev. Cancer 2014, 14, 135–146. [Google Scholar] [CrossRef]
- Lang, F.; Schrörs, B.; Löwer, M.; Türeci, Ö.; Sahin, U. Identification of Neoantigens for Individualized Therapeutic Cancer Vaccines. Nat. Rev. Drug Discov. 2022, 21, 261–282. [Google Scholar] [CrossRef]
- Lybaert, L.; Lefever, S.; Fant, B.; Smits, E.; De Geest, B.; Breckpot, K.; Dirix, L.; Feldman, S.A.; van Criekinge, W.; Thielemans, K.; et al. Challenges in Neoantigen-Directed Therapeutics. Cancer Cell 2023, 41, 15–40. [Google Scholar] [CrossRef]
- Verdegaal, E.; Van Der Kooij, M.K.; Visser, M.; Van Der Minne, C.; De Bruin, L.; Meij, P.; Terwisscha Van Scheltinga, A.; Welters, M.J.; Santegoets, S.; De Miranda, N.; et al. Low-Dose Interferon-Alpha Preconditioning and Adoptive Cell Therapy in Patients with Metastatic Melanoma Refractory to Standard (Immune) Therapies: A Phase I/II Study. J. Immunother. Cancer 2020, 8, e000166. [Google Scholar] [CrossRef]
- Wagner, S.; Mullins, C.S.; Linnebacher, M. Colorectal Cancer Vaccines: Tumor-Associated Antigens vs Neoantigens. World J. Gastroenterol. 2018, 24, 5418–5432. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Mutational Landscape Determines Sensitivity to PD-1 Blockade in Non-Small Cell Lung Cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Foppen, M.H.G.; Goldinger, S.M.; et al. Genomic Correlates of Response to CTLA-4 Blockade in Metastatic Melanoma. Science 2015, 350, 207–211. [Google Scholar] [CrossRef]
- Zaidi, N. Can Personalized Neoantigens Raise the T Cell Bar? Cell 2020, 183, 301–302. [Google Scholar] [CrossRef]
- Ott, P.A.; Hu-Lieskovan, S.; Chmielowski, B.; Govindan, R.; Naing, A.; Bhardwaj, N.; Margolin, K.; Awad, M.M.; Hellmann, M.D.; Lin, J.J.; et al. A Phase Ib Trial of Personalized Neoantigen Therapy Plus Anti-PD-1 in Patients with Advanced Melanoma, Non-Small Cell Lung Cancer, or Bladder Cancer. Cell 2020, 183, 347–362.e24. [Google Scholar] [CrossRef]
- Sahin, U.; Derhovanessian, E.; Miller, M.; Kloke, B.P.; Simon, P.; Löwer, M.; Bukur, V.; Tadmor, A.D.; Luxemburger, U.; Schrörs, B.; et al. Personalized RNA Mutanome Vaccines Mobilize Poly-Specific Therapeutic Immunity against Cancer. Nature 2017, 547, 222–226. [Google Scholar] [CrossRef]
- Anagnostou, V.; Smith, K.N.; Forde, P.M.; Niknafs, N.; Bhattacharya, R.; White, J.; Zhang, T.; Adleff, V.; Phallen, J.; Wali, N.; et al. Evolution of Neoantigen Landscape during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Discov. 2017, 7, 264–276. [Google Scholar] [CrossRef]
- Wells, D.K.; van Buuren, M.M.; Dang, K.K.; Hubbard-Lucey, V.M.; Sheehan, K.C.F.; Campbell, K.M.; Lamb, A.; Ward, J.P.; Sidney, J.; Blazquez, A.B.; et al. Key Parameters of Tumor Epitope Immunogenicity Revealed Through a Consortium Approach Improve Neoantigen Prediction. Cell 2020, 183, 818–834.e13. [Google Scholar] [CrossRef]
- Ott, P.; Hu, Z.; Keskin, D.; Shukla, S.A.; Sun, J.; Bozym, D.; Zhang, W.; Luoma, A.; Giobbie-Hurder, A.; Peter, L.; et al. An Immunogenic Personal Neoantigen Vaccine for Melanoma Patients. Nature 2017, 547, 217–221. [Google Scholar] [CrossRef]
- Kiyotani, K.; Chan, H.T.; Nakamura, Y. Immunopharmacogenomics towards Personalized Cancer Immunotherapy Targeting Neoantigens. Cancer Sci. 2018, 109, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, V.N. The Antigenicity of the Tumor Cell—Context Matters. N. Engl. J. Med. 2017, 376, 491–493. [Google Scholar] [CrossRef]
- Ranieri, E. (Ed.) Cytotoxic T-Cells Methods and Protocols Methods in Molecular Biology 1186; Humana: Louisville, KY, USA, 2014. [Google Scholar]
- Hilf, N.; Kuttruff-Coqui, S.; Frenzel, K.; Bukur, V.; Stevanović, S.; Gouttefangeas, C.; Platten, M.; Tabatabai, G.; Dutoit, V.; van der Burg, S.H.; et al. Actively Personalized Vaccination Trial for Newly Diagnosed Glioblastoma. Nature 2019, 565, 240–245. [Google Scholar] [CrossRef]
- Sercarz, E.E.; Lehmann, P.V.; Ametani, A.; Benichou, G.; Miller, A.; Moudgil, K. Dominance and crypticity of t cell antigenic determinants. Annu. Rev. Immunol. 1993, 11, 729–766. [Google Scholar] [CrossRef]
- Rech, A.J.; Balli, D.; Mantero, A.; Ishwaran, H.; Nathanson, K.L.; Stanger, B.Z.; Vonderheide, R.H. Tumor Immunity and Survival as a Function of Alternative Neopeptides in Human Cancer. Cancer Immunol. Res. 2018, 6, 276–287. [Google Scholar] [CrossRef]
- Duan, F.; Duitama, J.; Al Seesi, S.; Ayres, C.M.; Corcelli, S.A.; Pawashe, A.P.; Blanchard, T.; McMahon, D.; Sidney, J.; Sette, A.; et al. Genomic and Bioinformatic Profiling of Mutational Neoepitopes Reveals New Rules to Predict Anticancer Immunogenicity. J. Exp. Med. 2014, 211, 2231–2248. [Google Scholar] [CrossRef]
- Luksza, M.; Riaz, N.; Makarov, V.; Balachandran, V.P.; Hellmann, M.D.; Solovyov, A.; Rizvi, N.A.; Merghoub, T.; Levine, A.J.; Chan, T.A.; et al. A Neoantigen Fitness Model Predicts Tumour Response to Checkpoint Blockade Immunotherapy. Nature 2017, 551, 517–520. [Google Scholar] [CrossRef]
- Rasmussen, M.; Fenoy, E.; Harndahl, M.; Kristensen, A.B.; Nielsen, I.K.; Nielsen, M.; Buus, S. Pan-Specific Prediction of Peptide–MHC Class I Complex Stability, a Correlate of T Cell Immunogenicity. J. Immunol. 2016, 197, 1517–1524. [Google Scholar] [CrossRef]
- Hundal, J.; Carreno, B.M.; Petti, A.A.; Linette, G.P.; Griffith, O.L.; Mardis, E.R.; Griffith, M. PVAC-Seq: A Genome-Guided in Silico Approach to Identifying Tumor Neoantigens. Genome Med. 2016, 8, 11. [Google Scholar] [CrossRef]
- Chowell, D.; Krishna, S.; Becker, P.D.; Cocita, C.; Shu, J.; Tan, X.; Greenberg, P.D.; Klavinskis, L.S.; Blattman, J.N.; Anderson, K.S. TCR Contact Residue Hydrophobicity Is a Hallmark of Immunogenic CD8+ T Cell Epitopes. Proc. Natl. Acad. Sci. USA 2015, 112, E1754–E1762. [Google Scholar] [CrossRef] [PubMed]
- Tareen, A.; Kinney, J.B. Logomaker: Beautiful Sequence Logos in Python. Bioinformatics 2020, 36, 2272–2274. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely Available Python Tools for Computational Molecular Biology and Bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Shao, X.M.; Bhattacharya, R.; Huang, J.; Sivakumar, I.K.A.; Tokheim, C.; Zheng, L.; Hirsch, D.; Kaminow, B.; Omdahl, A.; Bonsack, M.; et al. High-Throughput Prediction of MHC Class I and II Neoantigens with MHCnuggets. Cancer Immunol. Res. 2020, 8, 396–408. [Google Scholar] [CrossRef]
- Calis, J.J.A.; Maybeno, M.; Greenbaum, J.A.; Weiskopf, D.; De Silva, A.D.; Sette, A.; Keşmir, C.; Peters, B. Properties of MHC Class I Presented Peptides That Enhance Immunogenicity. PLoS Comput. Biol. 2013, 9, e1003266. [Google Scholar] [CrossRef]
- Richman, L.P.; Vonderheide, R.H.; Rech, A.J. Neoantigen Dissimilarity to the Self-Proteome Predicts Immunogenicity and Response to Immune Checkpoint Blockade Graphical Abstract HHS Public Access ETOC. Cell Syst. 2019, 9, 375–382. [Google Scholar] [CrossRef]
- Schmidt, J.; Smith, A.R.; Magnin, M.; Racle, J.; Devlin, J.R.; Bobisse, S.; Cesbron, J.; Bonnet, V.; Carmona, S.J.; Huber, F.; et al. Prediction of Neo-Epitope Immunogenicity Reveals TCR Recognition Determinants and Provides Insight into Immunoediting. Cell Rep. Med. 2021, 2, 100194. [Google Scholar] [CrossRef]
- Luxenburger, H.; Graß, F.; Baermann, J.; Boettler, T.; Marget, M.; Emmerich, F.; Panning, M.; Thimme, R.; Nitschke, K.; Neumann-Haefelin, C. Differential Virus-Specific CD8+ T-Cell Epitope Repertoire in Hepatitis C Virus Genotype 1 versus 4. J. Viral Hepat. 2018, 25, 779–790. [Google Scholar] [CrossRef]
- Weiskopf, D.; Yauch, L.E.; Angelo, M.A.; John, D.V.; Greenbaum, J.A.; Sidney, J.; Kolla, R.V.; De Silva, A.D.; de Silva, A.M.; Grey, H.; et al. Insights into HLA-Restricted T Cell Responses in a Novel Mouse Model of Dengue Virus Infection Point toward New Implications for Vaccine Design. J. Immunol. 2011, 187, 4268–4279. [Google Scholar] [CrossRef]
- Wang, G.; Wan, H.; Jian, X.; Ouyang, J.; Li, Y. INeo-Epp: T-Cell HLA Class I Immunogenic or Neoantigenic Epitope Prediction via Random Forest Algorithm Based on Sequence Related Amino Acid Features. Biomed. Res. Int. 2019, 2020, 5798356. [Google Scholar]
- Rojas, L.A.; Sethna, Z.; Soares, K.C.; Olcese, C.; Pang, N.; Patterson, E.; Lihm, J.; Ceglia, N.; Guasp, P.; Chu, A.; et al. Personalized RNA Neoantigen Vaccines Stimulate T Cells in Pancreatic Cancer. Nature 2023, 618, 144–150. [Google Scholar] [CrossRef]
- Mørk, S.K.; Skadborg, S.K.; Albieri, B.; Draghi, A.; Bol, K.; Kadivar, M.; Westergaard, M.C.W.; Stoltenborg Granhøj, J.; Borch, A.; Petersen, N.V.; et al. Dose Escalation Study of a Personalized Peptide-Based Neoantigen Vaccine (EVX-01) in Patients with Metastatic Melanoma. J. Immunother. Cancer 2024, 12, e008817. [Google Scholar] [CrossRef]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef]
- Rajasagi, M.; Shukla, S.A.; Fritsch, E.F.; Keskin, D.B.; DeLuca, D.; Carmona, E.; Zhang, W.; Sougnez, C.; Cibulskis, K.; Sidney, J.; et al. Systematic Identification of Personal Tumor-Specific Neoantigens in Chronic Lymphocytic Leukemia. Blood 2014, 124, 453–462. [Google Scholar] [CrossRef]
- Li, G.; Iyer, B.; Prasath, V.B.S.; Ni, Y.; Salomonis, N. DeepImmuno: Deep Learning-Empowered Prediction and Generation of Immunogenic Peptides for T-Cell Immunity. Brief. Bioinform. 2021, 22, bbab160. [Google Scholar] [CrossRef]

| Year of Publication | PMID | Source | Non-Immunogenic Peptides | Immunogenic Peptides |
|---|---|---|---|---|
| 2011 | 21918184 | 477 | 42 | |
| 2018 | 29397015 | 100 | 26 |
| HLA Dependency | Positive Data | Negative Data | Predictive Parameters | ROC AUC—Viral (Neoantigen) Dataset | AP—Viral (Neoantigen) Dataset | |
|---|---|---|---|---|---|---|
| neoIM | Input peptides should be presented. | Positive T-cell assay | Non-self MS-eluted ligands | Amino acid physicochemical properties. | 0.80 (0.81) | 0.58 (0.61) |
| IEDB_imm | Input peptides should be presented. | Positive T-cell assay | Negative T-cell assay | Enrichment of an amino acid in immunogenic peptides. | 0.60 (0.53) | 0.20 (0.39) |
| antigen.garnish Click or tap here to enter text. | Input peptides should be presented. | Positive T-cell assay | Self-proteome | Similarity (BLAST) to IEDB epitopes or non-mutated proteome. | 0.61 (0.58) | 0.20 (0.32) |
| PRIME Click or tap here to enter text. | Final score dependent on single HLA subtype. | Positive T-cell assay | Negative T-cell assay + random peptides | MHC affinity, amino acid frequencies at TCR-contact positions. | 0.64 (0.61) | 0.20 (0.35) |
| INeo-Epp Click or tap here to enter text. | Final score dependent on single HLA subtype. | Positive T-cell assay | Negative T-cell assay | Amino acid physicochemical property, EL rank (%), peptide entropy. | NA (0.67) | NA (0.43) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pfitzer, L.; Boons, G.; Lybaert, L.; van Criekinge, W.; Bogaert, C.; Fant, B. Accelerating Neoantigen Discovery: A High-Throughput Approach to Immunogenic Target Identification. Vaccines 2025, 13, 865. https://doi.org/10.3390/vaccines13080865
Pfitzer L, Boons G, Lybaert L, van Criekinge W, Bogaert C, Fant B. Accelerating Neoantigen Discovery: A High-Throughput Approach to Immunogenic Target Identification. Vaccines. 2025; 13(8):865. https://doi.org/10.3390/vaccines13080865
Chicago/Turabian StylePfitzer, Lena, Gitta Boons, Lien Lybaert, Wim van Criekinge, Cedric Bogaert, and Bruno Fant. 2025. "Accelerating Neoantigen Discovery: A High-Throughput Approach to Immunogenic Target Identification" Vaccines 13, no. 8: 865. https://doi.org/10.3390/vaccines13080865
APA StylePfitzer, L., Boons, G., Lybaert, L., van Criekinge, W., Bogaert, C., & Fant, B. (2025). Accelerating Neoantigen Discovery: A High-Throughput Approach to Immunogenic Target Identification. Vaccines, 13(8), 865. https://doi.org/10.3390/vaccines13080865







