Poxvirus K3 Orthologs Regulate NF-κB-Dependent Inflammatory Responses by Targeting the PKR–eIF2α Axis in Multiple Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells Lines and Viruses
2.2. Plasmids
2.3. Generation of Stable Cell Lines Expressing Human PKR and European Rabbit PKR
2.4. Viral Infection
2.5. Luciferase Assays
2.6. Quantitative (q) RT-PCR
2.7. Western Blot Analysis
2.8. Statistical Analysis
3. Results
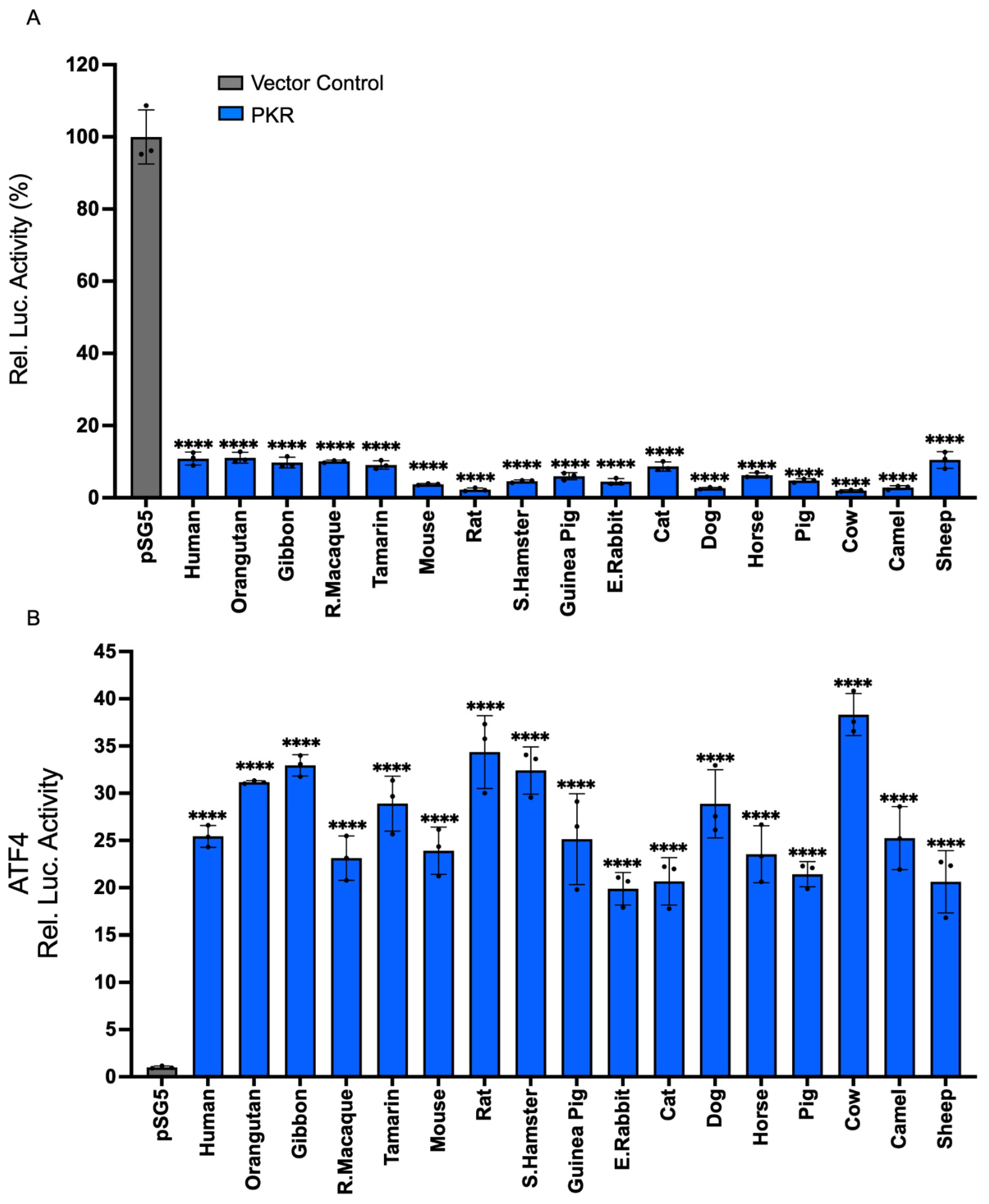
3.1. PKR Orthologs from Various Species Induce NF-κB-Mediated Inflammatory Gene Expression
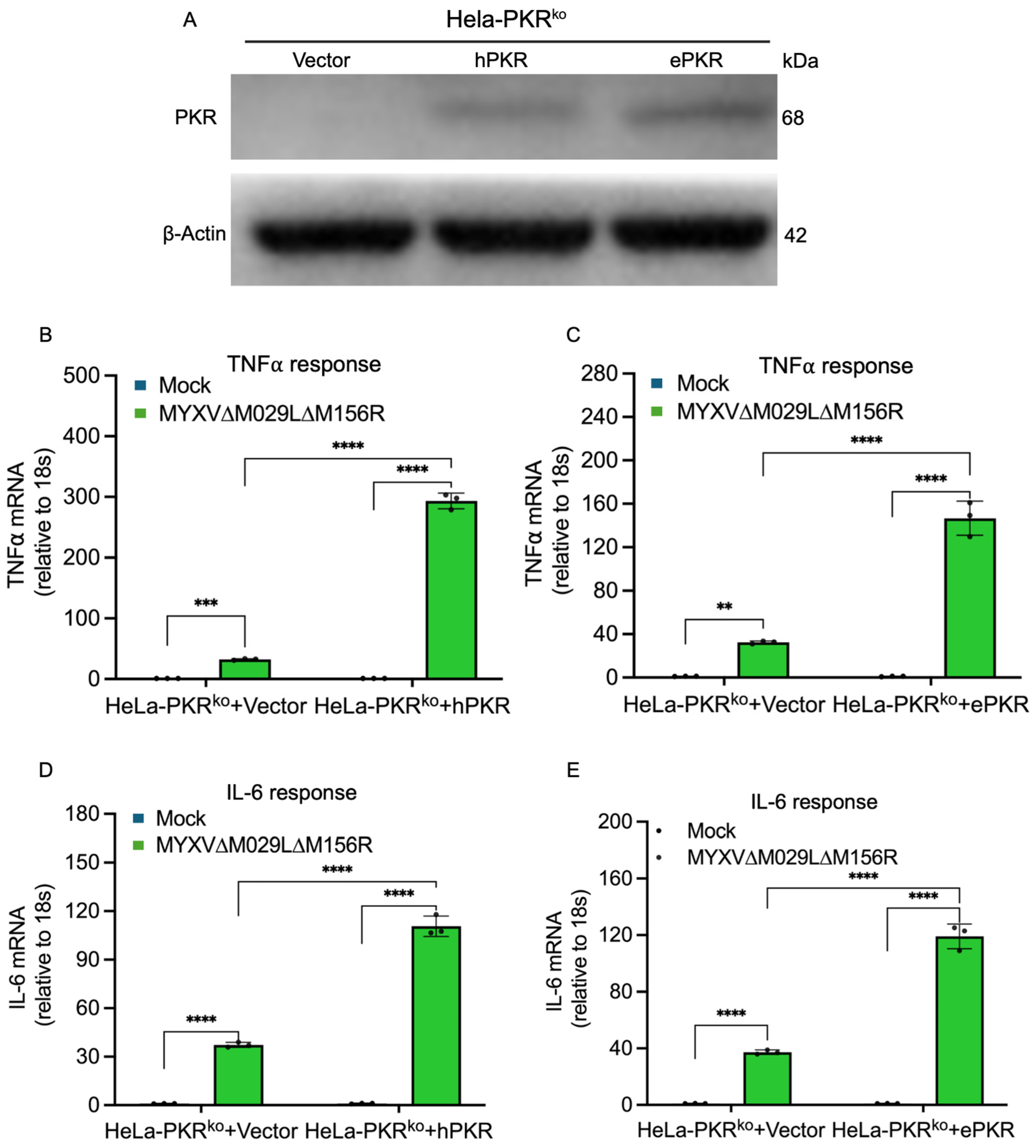
3.2. Functional Comparison of Human and European Rabbit PKR During Infection
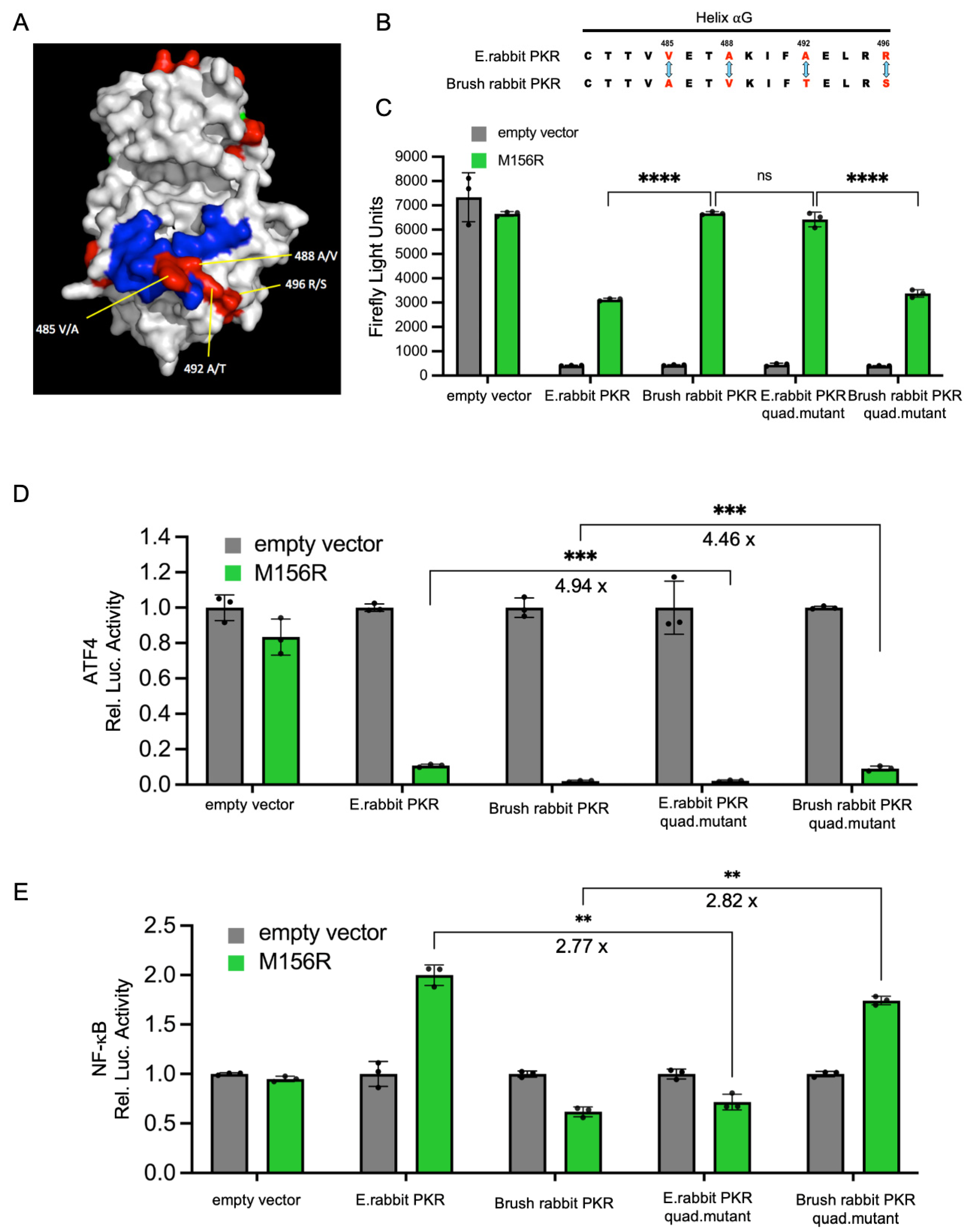
3.3. The Extent of PKR Inhibition Influences Protein Expression and NF-κB Activities
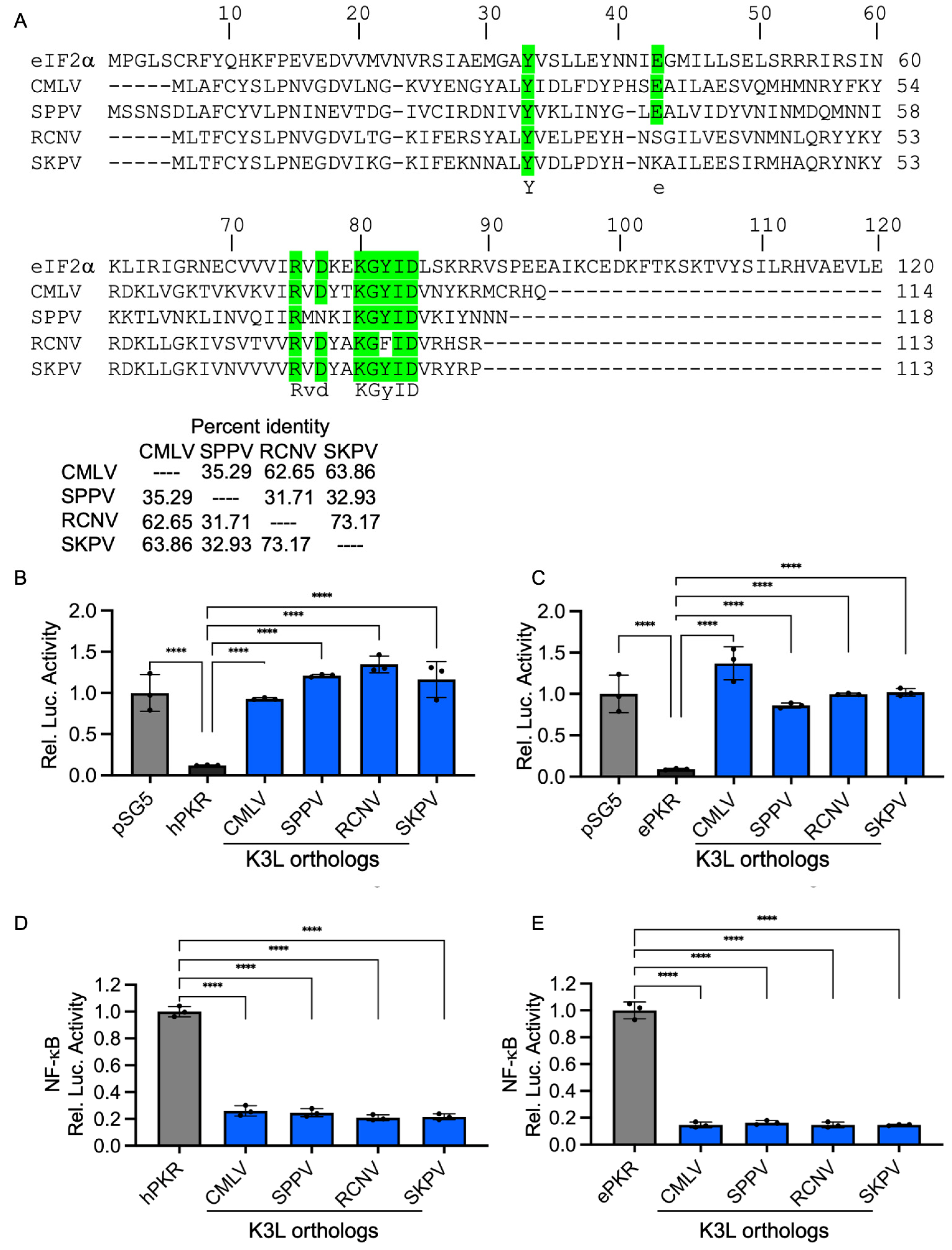
3.4. Inhibition of NF-κB Activity by Complete Inhibition of Human PKR
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IFN | Interferon |
| ISG | Interferon-stimulated gene |
| PKR | Protein kinase R |
| GCN2 | General control nonderepressible 2 kinase |
| PERK | Protein kinase RNA-like ER stress kinase |
| dsRNA | Double-stranded RNA |
| eIF2 | Eukaryotic translation initiation factor 2 |
| ATF4 | Activating transcription factor 4 |
| ISR | Integrated stress response |
| NF-κB | Nuclear factor kappa B |
| VACV | Vaccinia virus |
| MYXV | Myxoma virus |
| mAb | Monoclonal antibody |
| MOI | Multiplicity of infection |
| LBR | Luciferase-based reporter |
| uORF | Upstream open reading frame |
References
- Meurs, E.; Chong, K.; Galabru, J.; Thomas, N.S.B.; Kerr, I.M.; Williams, B.R.G.; Hovanessian, A.G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 1990, 62, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Weber, F.; Wagner, V.; Rasmussen, S.B.; Hartmann, R.; Paludan, S.R. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006, 80, 5059–5064. [Google Scholar] [CrossRef]
- Li, S.; Peters, G.A.; Ding, K.; Zhang, X.; Qin, J.; Sen, G.C. Molecular basis for PKR activation by PACT or dsRNA. Proc. Natl. Acad. Sci. USA 2006, 103, 10005–10010. [Google Scholar] [CrossRef]
- Dey, M.; Cao, C.; Dar, A.C.; Tamura, T.; Ozato, K.; Sicheri, F.; Dever, T.E. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell 2005, 122, 901–913. [Google Scholar] [CrossRef]
- Jiang, H.-Y.; Wek, S.A.; McGrath, B.C.; Scheuner, D.; Kaufman, R.J.; Cavener, D.R.; Wek, R.C. Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-kappaB in response to diverse cellular stresses. Mol. Cell. Biol. 2003, 23, 5651–5663. [Google Scholar] [CrossRef]
- Deng, J.; Lu, P.D.; Zhang, Y.; Scheuner, D.; Kaufman, R.J.; Sonenberg, N.; Harding, H.P.; Ron, D. Translational repression mediates activation of nuclear factor kappa B by phosphorylated translation initiation factor 2. Mol. Cell. Biol. 2004, 24, 10161–10168. [Google Scholar] [CrossRef]
- Jiang, H.Y.; Wek, R.C. GCN2 phosphorylation of eIF2alpha activates NF-kappaB in response to UV irradiation. Biochem. J. 2005, 385 Pt 2, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Vattem, K.M.; Wek, R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA 2004, 101, 11269–11274. [Google Scholar] [CrossRef]
- Andreev, D.E.; O’Connor, P.B.; Fahey, C.; Kenny, E.M.; Terenin, I.M.; Dmitriev, S.E.; Cormican, P.; Morris, D.W.; Shatsky, I.N.; Baranov, P.V. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife 2015, 4, e03971. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, M.; Cheng, A.; Yang, Q.; Wu, Y.; Jia, R.; Liu, M.; Zhu, D.; Chen, S.; Zhang, S.; et al. The role of host eIF2alpha in viral infection. Virol. J. 2020, 17, 112. [Google Scholar] [CrossRef] [PubMed]
- Gil, J.; Rullas, J.; García, M.A.; Alcamí, J.; Esteban, M. The catalytic activity of dsRNA-dependent protein kinase, PKR, is required for NF-kappaB activation. Oncogene 2001, 20, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, M.C.; Weil, R.; Dam, E.; Hovanessian, A.G.; Meurs, E.F. PKR stimulates NF-kappaB irrespective of its kinase function by interacting with the IkappaB kinase complex. Mol. Cell. Biol. 2000, 20, 4532–4542. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.S.; West, A.P.; Ghosh, S. NF-kappaB and the immune response. Oncogene 2006, 25, 6758–6780. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-kappaB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Zandi, E.; Rothwarf, D.M.; Delhase, M.; Hayakawa, M.; Karin, M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell 1997, 91, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Pahl, H.L. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 1999, 18, 6853–6866. [Google Scholar] [CrossRef]
- Hoffmann, A.; Levchenko, A.; Scott, M.L.; Baltimore, D. The IkappaB-NF-kappaB signaling module: Temporal control and selective gene activation. Science 2002, 298, 1241–1245. [Google Scholar] [CrossRef]
- Shih, V.F.-S.; Kearns, J.D.; Basak, S.; Savinova, O.V.; Ghosh, G.; Hoffmann, A. Kinetic control of negative feedback regulators of NF-kappaB/RelA determines their pathogen- and cytokine-receptor signaling specificity. Proc. Natl. Acad. Sci. USA 2009, 106, 9619–9624. [Google Scholar] [CrossRef]
- Guo, Q.; Jin, Y.; Chen, X.; Ye, X.; Shen, X.; Lin, M.; Zeng, C.; Zhou, T.; Zhang, J. NF-kappaB in biology and targeted therapy: New insights and translational implications. Signal Transduct. Target. Ther. 2024, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Bruneau, R.C.; Brennan, G.; Rothenburg, S. Battle Royale: Innate Recognition of Poxviruses and Viral Immune Evasion. Biomedicines 2021, 9, 765. [Google Scholar] [CrossRef]
- Reus, J.B.; Rex, E.A.; Gammon, D.B. How to Inhibit Nuclear Factor-Kappa B Signaling: Lessons from Poxviruses. Pathogens 2022, 11, 1061. [Google Scholar] [CrossRef]
- McFadden, G. Poxvirus tropism. Nat. Rev. Microbiol. 2005, 3, 201–213. [Google Scholar] [CrossRef]
- Rothenburg, S.; Yang, Z.; Beard, P.; Sawyer, S.L.; Titanji, B.; Gonsalves, G.; Kindrachuk, J. Monkeypox emergency: Urgent questions and perspectives. Cell 2022, 185, 3279–3281. [Google Scholar] [CrossRef]
- Yu, H.; Resch, W.; Moss, B. Poxvirus structural biology for application to vaccine design. Trends Immunol. 2025, 46, 455–470. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Resch, W.; Cotter, C.A.; Xiao, W.; Karamanolis, T.; Belghith, A.A.; Ignacio, M.A.; Earl, P.L.; Cohen, G.H. Antibody Binding and Neutralizing Targets within the Predicted Structure of the Poxvirus Multiprotein Entry-Fusion Complex. bioRxiv 2025. [Google Scholar] [CrossRef]
- Brennan, G.; Stoian, A.M.M.; Yu, H.; Rahman, M.J.; Banerjee, S.; Stroup, J.N.; Park, C.; Tazi, L.; Rothenburg, S. Molecular Mechanisms of Poxvirus Evolution. mBio 2023, 14, e0152622. [Google Scholar] [CrossRef]
- Rahman, M.M.; Liu, J.; Chan, W.M.; Rothenburg, S.; McFadden, G. Myxoma virus protein M029 is a dual function immunomodulator that inhibits PKR and also conscripts RHA/DHX9 to promote expanded host tropism and viral replication. PLoS Pathog. 2013, 9, e1003465. [Google Scholar] [CrossRef]
- Peng, C.; Haller, S.L.; Rahman, M.M.; McFadden, G.; Rothenburg, S. Myxoma virus M156 is a specific inhibitor of rabbit PKR but contains a loss-of-function mutation in Australian virus isolates. Proc. Natl. Acad. Sci. USA 2016, 113, 3855–3860. [Google Scholar] [CrossRef] [PubMed]
- Romano, P.R.; Zhang, F.; Tan, S.-L.; Garcia-Barrio, M.T.; Katze, M.G.; Dever, T.E.; Hinnebusch, A.G. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: Role of complex formation and the E3 N-terminal domain. Mol. Cell. Biol. 1998, 18, 7304–7316. [Google Scholar] [CrossRef]
- Ramelot, T.A.; Cort, J.R.; Yee, A.A.; Liu, F.; Goshe, M.B.; Edwards, A.M.; Smith, R.D.; Arrowsmith, C.H.; Dever, T.E.; Kennedy, M.A. Myxoma virus immunomodulatory protein M156R is a structural mimic of eukaryotic translation initiation factor eIF2alpha. J. Mol. Biol. 2002, 322, 943–954. [Google Scholar] [CrossRef]
- Yu, H.; Peng, C.; Zhang, C.; Stoian, A.M.M.; Tazi, L.; Brennan, G.; Rothenburg, S. Maladaptation after a virus host switch leads to increased activation of the pro-inflammatory NF-kappaB pathway. Proc. Natl. Acad. Sci. USA 2022, 119, e2115354119. [Google Scholar] [CrossRef]
- Yu, H. Poxvirus K3 Orthologs Differentially Modulate NF-kB Activity Through Targeting the PKR-eIF2a Phosphorylation Pathway. Ph.D. Thesis, University of California, Davis, CA, USA, 2021. [Google Scholar]
- Park, C.; Peng, C.; Brennan, G.; Rothenburg, S. Species-specific inhibition of antiviral protein kinase R by capripoxviruses and vaccinia virus. Ann. N. Y. Acad. Sci. 2019, 1438, 18–29. [Google Scholar] [CrossRef]
- Park, C.; Peng, C.; Rahman, M.J.; Haller, S.L.; Tazi, L.; Brennan, G.; Rothenburg, S. Orthopoxvirus K3 orthologs show virus- and host-specific inhibition of the antiviral protein kinase PKR. PLoS Pathog. 2021, 17, e1009183. [Google Scholar] [CrossRef] [PubMed]
- Megawati, D.; Stroup, J.N.; Park, C.; Clarkson, T.; Tazi, L.; Brennan, G.; Rothenburg, S. Tanapox Virus and Yaba Monkey Tumor Virus K3 Orthologs Inhibit Primate Protein Kinase R in a Species-Specific Fashion. Viruses 2024, 16, 1095. [Google Scholar] [CrossRef]
- Carpentier, K.S.; Esparo, N.M.; Child, S.J.; Geballe, A.P. A Single Amino Acid Dictates Protein Kinase R Susceptibility to Unrelated Viral Antagonists. PLoS Pathog. 2016, 12, e1005966. [Google Scholar] [CrossRef] [PubMed]
- Krüll, M.; Klucken, A.C.; Wuppermann, F.N.; Fuhrmann, O.; Magerl, C.; Seybold, J.; Hippenstiel, S.; Hegemann, J.H.; Jantos, C.A.; Suttorp, N. Signal transduction pathways activated in endothelial cells following infection with Chlamydia pneumoniae. J. Immunol. 1999, 162, 4834–4841. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Megawati, D.; Zheng, C.; Rothenburg, S. Protein Kinase R (PKR) as a Novel dsRNA Sensor in Antiviral Innate Immunity. Methods Mol. Biol. 2025, 2854, 265–282. [Google Scholar]
- Nejepinska, J.; Malik, R.; Wagner, S.; Svoboda, P. Reporters transiently transfected into mammalian cells are highly sensitive to translational repression induced by dsRNA expression. PLoS ONE 2014, 9, e87517. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Sen, R.; Smale, S.T. Selectivity of the NF-{kappa}B response. Cold Spring Harb. Perspect. Biol. 2010, 2, a000257. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Hayden, M.S.; Ghosh, S. Crosstalk in NF-κB signaling pathways. Nat. Immunol. 2011, 12, 695–708. [Google Scholar] [CrossRef]
- Mitchell, S.; Tsui, R.; Tan, Z.C.; Pack, A.; Hoffmann, A. The NF-κB multidimer system model: A knowledge base to explore diverse biological contexts. Sci. Signal. 2023, 16, eabo2838. [Google Scholar] [CrossRef]
- Razani, B.; Reichardt, A.D.; Cheng, G. Non-canonical NF-κB signaling activation and regulation: Principles and perspectives. Immunol. Rev. 2011, 244, 44–54. [Google Scholar] [CrossRef]
- Kanarek, N.; Ben-Neriah, Y. Regulation of NF-κB by ubiquitination and degradation of the IκBs. Immunol. Rev. 2012, 246, 77–94. [Google Scholar] [CrossRef]
- Sun, S.C. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017, 17, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef]
- McAllister, C.S.; Taghavi, N.; Samuel, C.E. Protein kinase PKR amplification of interferon β induction occurs through initiation factor eIF-2α-mediated translational control. J. Biol. Chem. 2012, 287, 36384–36392. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Haque, J.; Lacoste, J.; Hiscott, J.; Williams, B.R. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc. Natl. Acad. Sci. USA 1994, 91, 6288–6292. [Google Scholar] [CrossRef]
- Seo, E.J.; Liu, F.; Kawagishi-Kobayashi, M.; Ung, T.L.; Cao, C.; Dar, A.C.; Sicheri, F.; Dever, T.E. Protein kinase PKR mutants resistant to the poxvirus pseudosubstrate K3L protein. Proc. Natl. Acad. Sci. USA 2008, 105, 16894–16899. [Google Scholar] [CrossRef] [PubMed]
- Rothenburg, S.; Seo, E.J.; Gibbs, J.S.; Dever, T.E.; Dittmar, K. Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat. Struct. Mol. Biol. 2009, 16, 63–70. [Google Scholar] [CrossRef]
- Elde, N.C.; Child, S.J.; Geballe, A.P.; Malik, H.S. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature 2009, 457, 485–489. [Google Scholar] [CrossRef]
- Haller, S.L.; Park, C.; Bruneau, R.C.; Megawati, D.; Zhang, C.; Vipat, S.; Peng, C.; Senkevich, T.G.; Brennan, G.; Tazi, L.; et al. Host species-specific activity of the poxvirus PKR inhibitors E3 and K3 mediate host range function. J. Virol. 2024, 98, e0133124. [Google Scholar] [CrossRef]
- Dar, A.C.; Sicheri, F. X-ray crystal structure and functional analysis of vaccinia virus K3L reveals molecular determinants for PKR subversion and substrate recognition. Mol. Cell 2002, 10, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.C.; Dever, T.E.; Sicheri, F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell 2005, 122, 887–900. [Google Scholar] [CrossRef]
- Jacquet, S.; Culbertson, M.; Zhang, C.; El Filali, A.; De La Myre Mory, C.; Pons, J.-B.; Filippi-Codaccioni, O.; Lauterbur, M.E.; Ngoubangoye, B.; Duhayer, J.; et al. Adaptive duplication and genetic diversification of protein kinase R contribute to the specificity of bat-virus interactions. Sci. Adv. 2022, 8, eadd7540. [Google Scholar] [CrossRef]
- Chambers, M.J.; Scobell, S.B.; Sadhu, M.J. Systematic genetic characterization of the human PKR kinase domain highlights its functional malleability to escape a poxvirus substrate mimic. eLife 2024, 13, RP99575. [Google Scholar] [CrossRef]
- Stanford, M.M.; Werden, S.J.; McFadden, G. Myxoma virus in the European rabbit: Interactions between the virus and its susceptible host. Vet. Res. 2007, 38, 299–318. [Google Scholar] [CrossRef]
- Best, S.M.; Collins, S.V.; Kerr, P.J. Coevolution of host and virus: Cellular localization of virus in myxoma virus infection of resistant and susceptible European rabbits. Virology 2000, 277, 76–91. [Google Scholar] [CrossRef] [PubMed]
- Rothenburg, S.; Brennan, G. Species-Specific Host-Virus Interactions: Implications for Viral Host Range and Virulence. Trends Microbiol. 2020, 28, 46–56. [Google Scholar] [CrossRef] [PubMed]


| Percent identity | ||||||||||||||||||||
| Divergence | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |||
| 1 | 92.2 | 91.5 | 80.9 | 85.3 | 59.8 | 60.4 | 61.8 | 55.9 | 68.0 | 65.5 | 64.4 | 64.0 | 63.5 | 64.3 | 66.6 | 62.3 | 1 | Human | ||
| 2 | 8.3 | 91.8 | 81.6 | 84.0 | 59.8 | 59.6 | 61.8 | 55.3 | 67.2 | 65.3 | 63.9 | 63.5 | 62.7 | 63.6 | 65.7 | 61.5 | 2 | Orangutan | ||
| 3 | 9.1 | 8.7 | 81.2 | 83.8 | 59.4 | 59.4 | 60.8 | 55.5 | 68.1 | 64.9 | 63.9 | 64.2 | 62.9 | 63.9 | 67.0 | 62.5 | 3 | White-cheeked Gibbon | ||
| 4 | 22.1 | 21.2 | 21.6 | 78.5 | 57.3 | 57.3 | 58.0 | 53.3 | 62.8 | 62.8 | 61.1 | 60.8 | 60.6 | 60.1 | 62.2 | 58.8 | 4 | Rhesus macaque | ||
| 5 | 16.4 | 18.0 | 18.2 | 25.4 | 59.2 | 59.2 | 61.2 | 56.3 | 67.4 | 64.4 | 65.2 | 65.3 | 62.5 | 64.1 | 65.8 | 61.9 | 5 | Tamarin | ||
| 6 | 57.0 | 57.0 | 57.8 | 62.2 | 58.2 | 77.4 | 70.5 | 51.2 | 57.7 | 54.0 | 55.1 | 54.8 | 58.2 | 56.3 | 58.4 | 55.4 | 6 | Mouse | ||
| 7 | 55.8 | 57.4 | 57.8 | 62.2 | 58.2 | 26.9 | 69.2 | 55.6 | 56.9 | 55.1 | 55.6 | 56.7 | 57.3 | 56.1 | 58.4 | 53.8 | 7 | Rat | ||
| 8 | 53.0 | 53.0 | 54.9 | 60.6 | 54.1 | 37.4 | 39.7 | 50.7 | 57.7 | 55.6 | 55.3 | 55.1 | 56.3 | 55.2 | 56.6 | 53.8 | 8 | S. hamster | ||
| 9 | 65.4 | 66.7 | 66.3 | 71.5 | 64.5 | 76.7 | 66.1 | 78.0 | 56.8 | 52.7 | 52.5 | 52.2 | 55.6 | 55.1 | 55.7 | 52.4 | 9 | Guinea pig | ||
| 10 | 41.7 | 43.0 | 41.4 | 51.0 | 42.7 | 61.3 | 63.1 | 61.5 | 63.5 | 64.3 | 65.2 | 63.6 | 63.7 | 63.6 | 64.5 | 60.4 | 10 | European rabbit | ||
| 11 | 46.0 | 46.4 | 47.1 | 51.0 | 48.1 | 69.8 | 67.1 | 66.2 | 72.9 | 48.2 | 76.9 | 63.3 | 64.6 | 64.1 | 68.4 | 62.2 | 11 | Cat | ||
| 12 | 47.9 | 49.0 | 49.0 | 54.3 | 46.5 | 67.1 | 66.2 | 66.8 | 73.4 | 46.6 | 27.7 | 65.5 | 64.4 | 60.9 | 65.4 | 60.2 | 12 | Dog | ||
| 13 | 48.7 | 49.8 | 48.4 | 55.0 | 46.3 | 67.8 | 63.7 | 67.2 | 74.1 | 49.5 | 50.1 | 46.1 | 63.8 | 59.9 | 64.5 | 59.4 | 13 | Horse | ||
| 14 | 49.7 | 51.2 | 50.8 | 55.3 | 51.5 | 60.4 | 62.2 | 64.4 | 66.0 | 49.3 | 47.7 | 47.9 | 49.1 | 73.6 | 74.9 | 71.4 | 14 | Pig | ||
| 15 | 48.2 | 49.6 | 48.9 | 56.4 | 48.5 | 64.6 | 65.0 | 67.0 | 67.2 | 49.5 | 48.6 | 54.7 | 56.7 | 32.6 | 75.9 | 88.2 | 15 | Cow | ||
| 16 | 44.0 | 45.7 | 43.4 | 52.2 | 45.4 | 59.9 | 60.0 | 63.8 | 65.9 | 47.8 | 40.9 | 46.2 | 47.8 | 30.7 | 29.1 | 74.6 | 16 | Camel | ||
| 17 | 52.0 | 53.5 | 51.6 | 59.1 | 52.7 | 66.5 | 70.4 | 70.1 | 73.6 | 55.7 | 52.2 | 56.2 | 57.9 | 36.0 | 12.9 | 31.0 | 17 | Sheep | ||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | ||||
| Human | Orangutan | White-cheeked Gibbon | Rhesus macaque | Tamarin | Mouse | Rat | S. hamster | Guinea pig | European rabbit | Cat | Dog | Horse | Pig | Cow | Camel | Sheep | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.; Fernandez, M.E.L.; Peng, C.; Megawati, D.; Brennan, G.; Tazi, L.; Rothenburg, S. Poxvirus K3 Orthologs Regulate NF-κB-Dependent Inflammatory Responses by Targeting the PKR–eIF2α Axis in Multiple Species. Vaccines 2025, 13, 800. https://doi.org/10.3390/vaccines13080800
Yu H, Fernandez MEL, Peng C, Megawati D, Brennan G, Tazi L, Rothenburg S. Poxvirus K3 Orthologs Regulate NF-κB-Dependent Inflammatory Responses by Targeting the PKR–eIF2α Axis in Multiple Species. Vaccines. 2025; 13(8):800. https://doi.org/10.3390/vaccines13080800
Chicago/Turabian StyleYu, Huibin, Mary Eloise L. Fernandez, Chen Peng, Dewi Megawati, Greg Brennan, Loubna Tazi, and Stefan Rothenburg. 2025. "Poxvirus K3 Orthologs Regulate NF-κB-Dependent Inflammatory Responses by Targeting the PKR–eIF2α Axis in Multiple Species" Vaccines 13, no. 8: 800. https://doi.org/10.3390/vaccines13080800
APA StyleYu, H., Fernandez, M. E. L., Peng, C., Megawati, D., Brennan, G., Tazi, L., & Rothenburg, S. (2025). Poxvirus K3 Orthologs Regulate NF-κB-Dependent Inflammatory Responses by Targeting the PKR–eIF2α Axis in Multiple Species. Vaccines, 13(8), 800. https://doi.org/10.3390/vaccines13080800








