Epitope Profiling of SARS-CoV-2 Spike Antigen Provides a Novel Strategy for Developing ELISAs Specific for Different Spike Protein Variants in Bivalent Vaccine Formulations
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of the SARS-CoV-2 Recombinant Prefusion Spike Antigen
2.2. HuCAL Screening and Generation of Ligands Specific to Ancestral SARS-CoV-2 Spike Antigen
2.3. Pseudovirus-Based Neutralization Assays
2.4. Biolayer Interferometry (BLI) Equipment and General Reagents for BLI Experiments
2.5. Assessment of mAbs for ACE2 Receptor Binding Inhibition Activity Using Biolayer Interferometry (BLI)
2.6. Antibody Binding Studies on Ancestral and Beta Spike Antigens Using BLI
2.7. ELISA for Ancestral SARs-CoV-2 Spike Antigen
2.8. Epitope Mapping by Hydrogen–Deuterium Exchange Mass Spectrometry (HDX-MS)
2.9. Using Biolayer Interferometry (BLI) to Establish Proof of Concept of Blocking ELISA
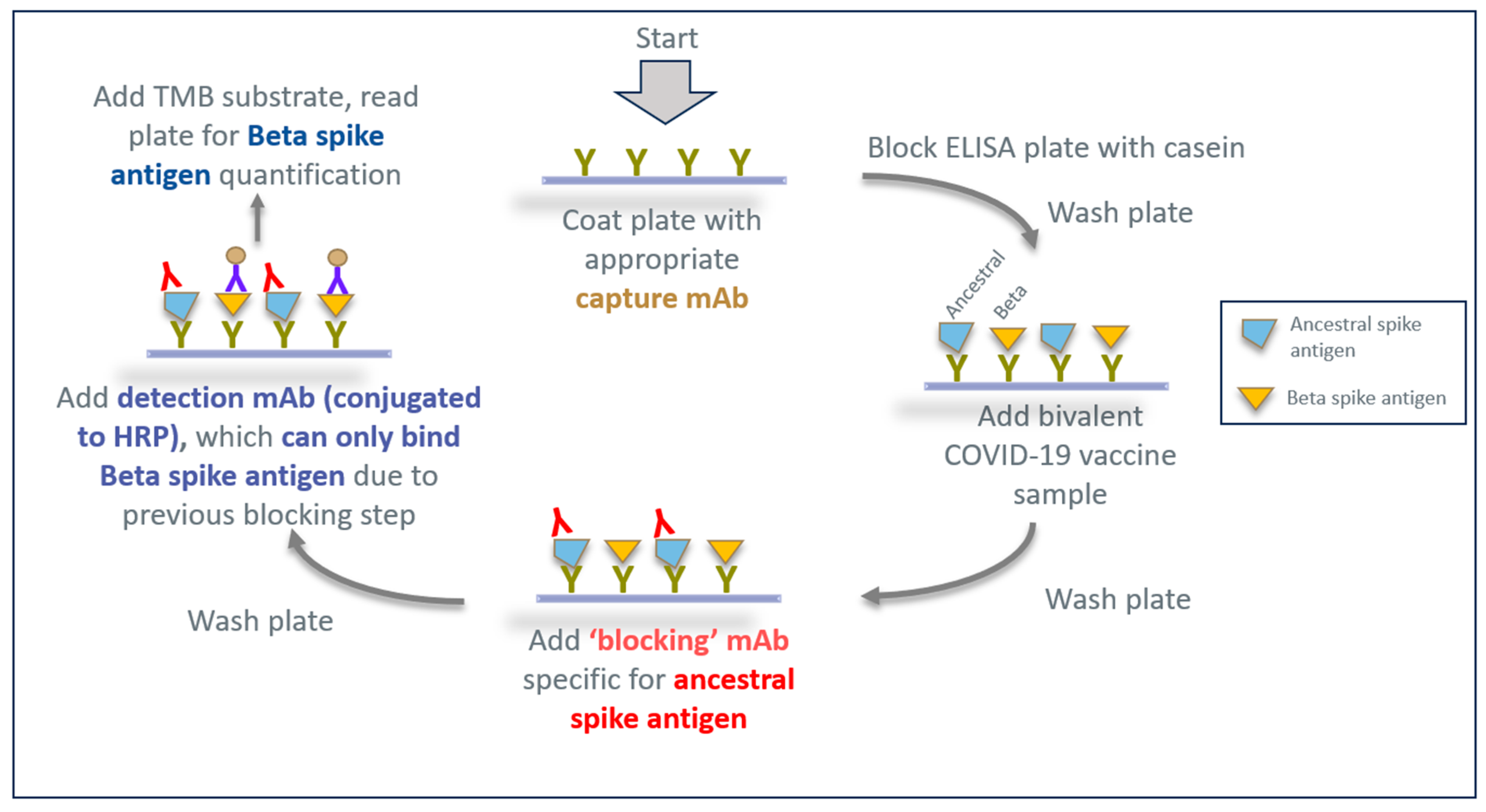
2.10. Epitope-Blocking ELISA for Specific Detection of SARS-CoV-2 Beta Spike Antigen Variant
2.11. Qualification of ELISAs
3. Results
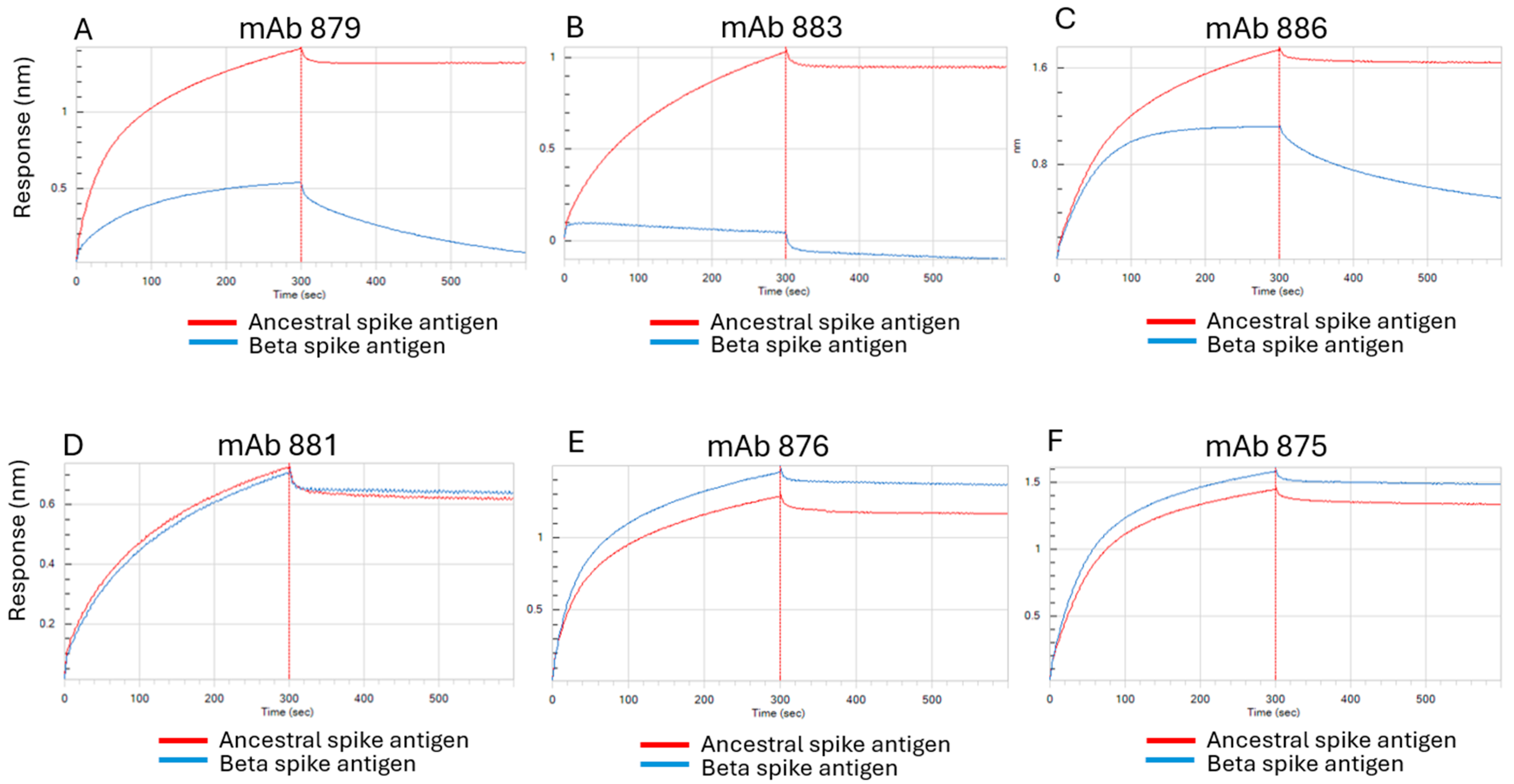
3.1. Characterization of Antibodies Generated Against Ancestral SARS-CoV-2 Spike Antigen
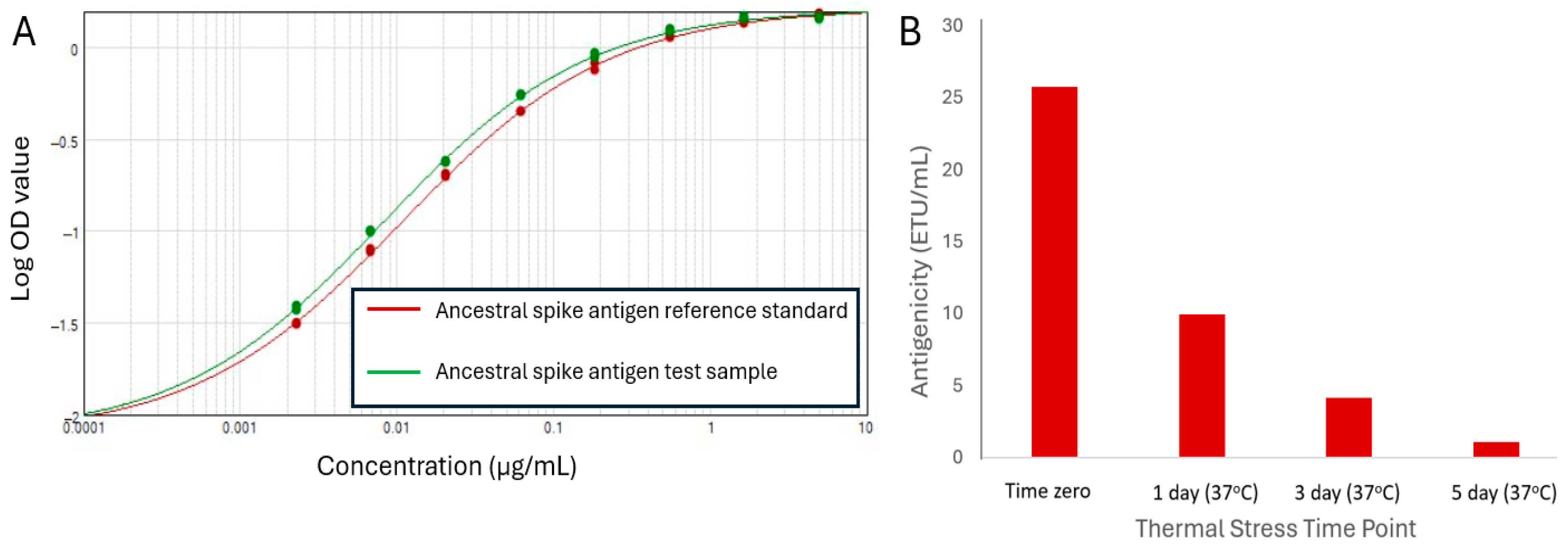
3.2. Antigenicity ELISA Development for Ancestral SARS-CoV-2 Spike Antigen
3.3. Developing an Epitope-Blocking ELISA Strategy to Detect Beta Spike Antigen in Bivalent Vaccine Formulation
3.4. Assessing the Epitope-Blocking ELISA’s Ability to Detect Beta Spike Antigen in Bivalent Vaccine
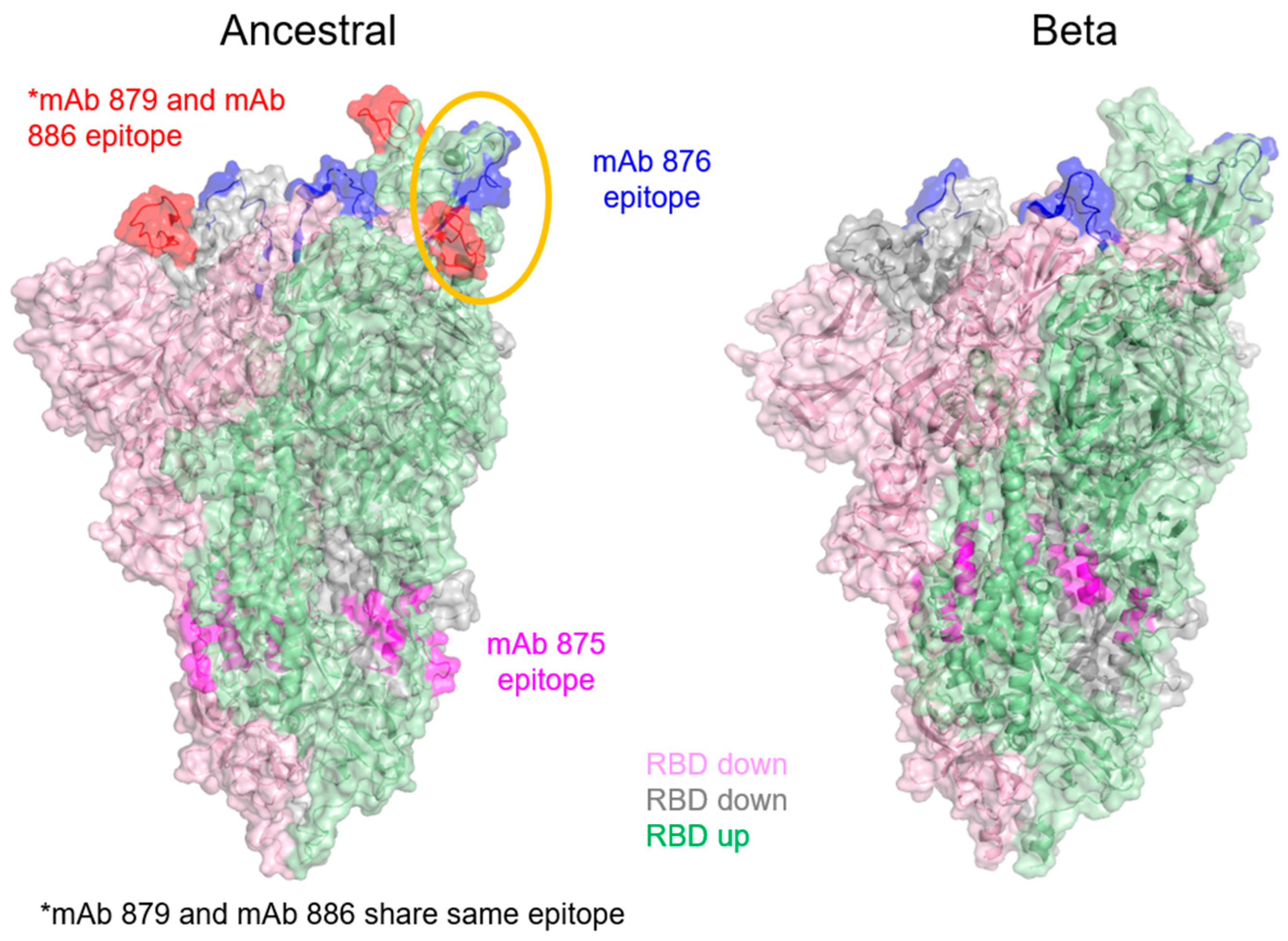
3.5. Epitope Mapping of mAbs Using HDX-MS
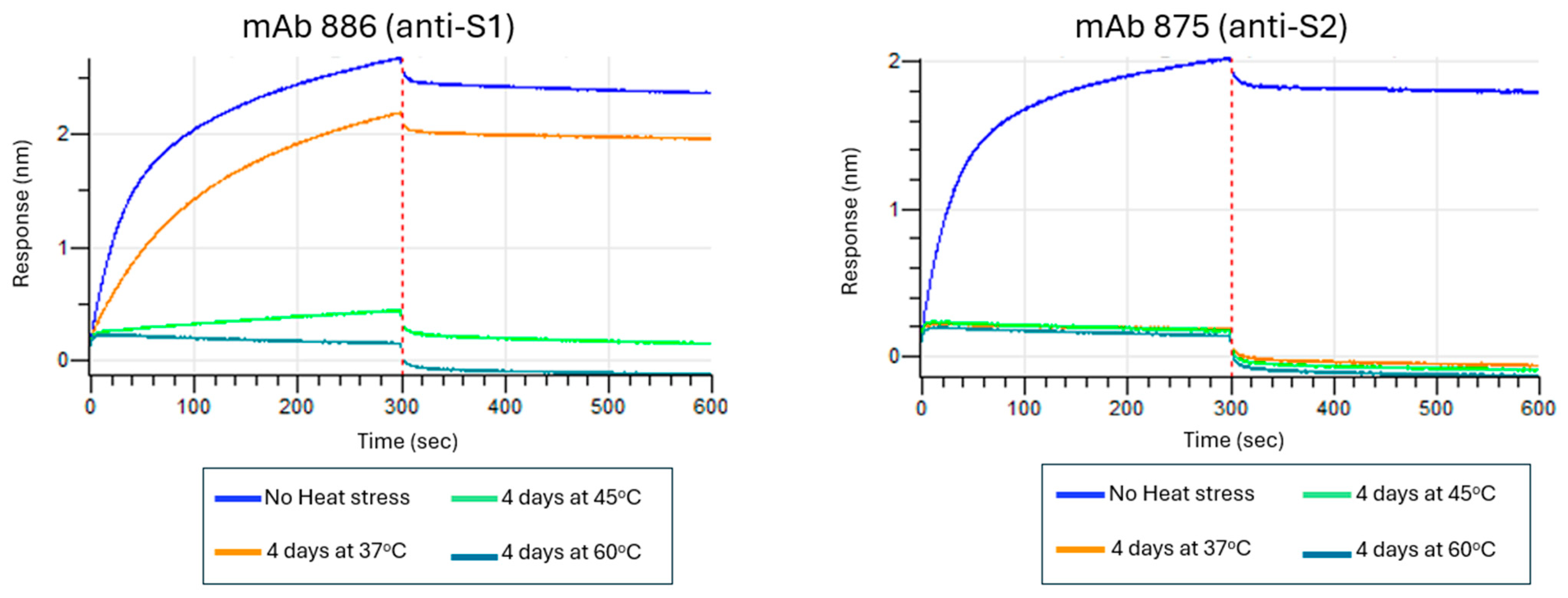
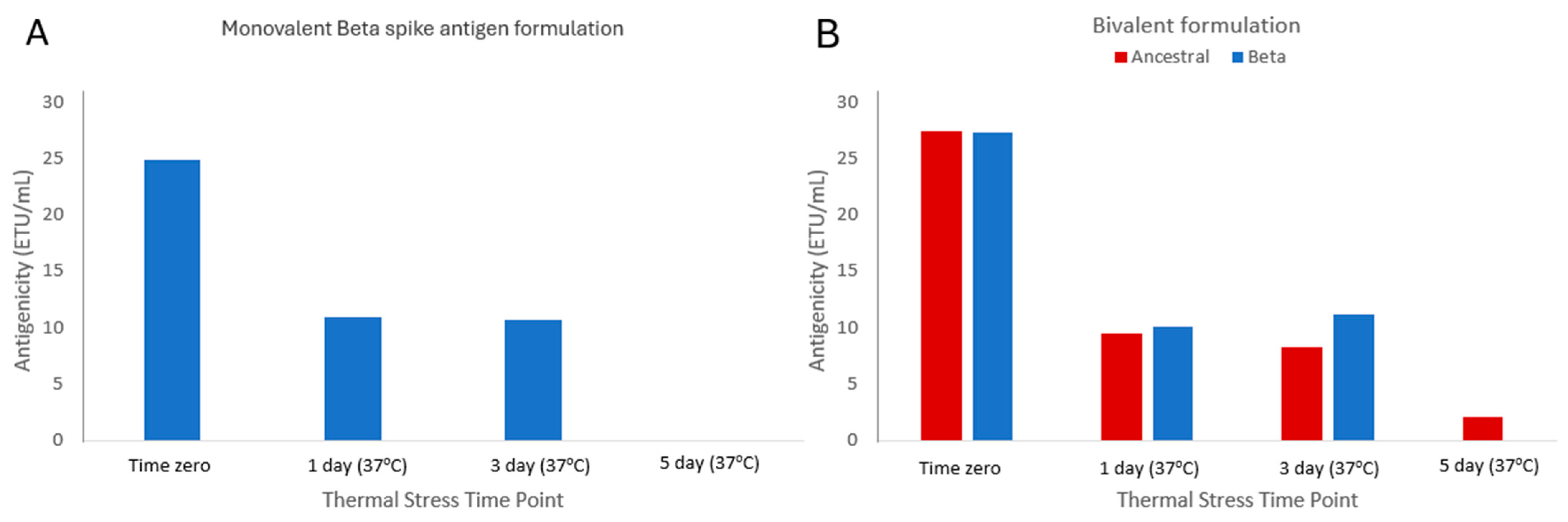
3.6. Assessing Stability-Indicating Ability of the Epitope-the Blocking ELISA
3.7. Qualification of ELISAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Borkotoky, S.; Dey, D.; Hazarika, Z. Interactions of Angiotensin-Converting Enzyme-2 (ACE2) and SARS-CoV-2 Spike Receptor-Binding Domain (RBD): A Structural Perspective. Mol. Biol. Rep. 2022, 50, 2713–2721. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, X.; Zhou, H.; Zhu, H.; Jiang, S.; Wang, P. Broadly Neutralizing Antibodies to SARS-CoV-2 and Other Human Coronaviruses. Nat. Rev. Immunol. 2022, 23, 189–199. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Hou, Y.J.; Chiba, S.; Halfmann, P.; Here, C.; Kuroda, M.; Dinnon, K.H., III; Leist, S.R.; Schäfer, A.; Nakajima, N.; Takahashi, K.; et al. SARS-Cov-2 D614G Variant Exhibits Efficient Replication Ex Vivo and Transmission in Vivo. Science 2020, 370, 1464–1468. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Koenig, P.-A.; Schmidt, F.I.; Phimister, E.G. Spike D614G—A Candidate Vaccine Antigen Against Covid-19. N. Engl. J. Med. 2021, 384, 2349–2351. [Google Scholar] [CrossRef]
- Moore, J.P.; Offit, P.A. SARS-CoV-2 Vaccines and the Growing Threat of Viral Variants. JAMA 2021, 325, 821–822. [Google Scholar] [CrossRef]
- Riches-Duit, R.; Hassall, L.; Kogelman, A.; Westdijk, J.; Dobly, A.; Francotte, A.; Stickings, P. Characterisation of Diphtheria Monoclonal Antibodies as a First Step Towards the Development of an in Vitro Vaccine Potency Immunoassay. Biologicals 2021, 69, 38–48. [Google Scholar] [CrossRef]
- Riches-Duit, R.; Hassall, L.; Kogelman, A.; Westdijk, J.; Rajagopal, S.; Davletov, B.; Doran, C.; Dobly, A.; Francotte, A.; Stickings, P. Characterisation of Tetanus Monoclonal Antibodies as a First Step Towards the Development of an in Vitro Vaccine Potency Immunoassay. Biologicals 2021, 71, 31–41. [Google Scholar] [CrossRef]
- Szeto, J.; Beharry, A.; Chen, T.; Zholumbetov, E.; Daigneault, E.; Ming, M.; Lounsbury, I.; Eng, N.; Thangavadivel, N.; Jin, R.; et al. Development of an In Vitro Test Method to Replace an Animal-Based Potency Test for Pertactin Antigen in Multivalent Vaccines. Vaccines 2023, 11, 275. [Google Scholar] [CrossRef]
- Knappik, A.; Ge, L.; Honegger, A.; Pack, P.; Fischer, M.; Wellnhofer, G.; Hoess, A.; Wölle, J.; Plückthun, A.; Virnekäs, B. Fully Synthetic Human Combinatorial Antibody Libraries (HuCAL) Based on Modular Consensus Frameworks and CDRs Randomized with Trinucleotides 1 1Edited by I. A. Wilson. J. Mol. Biol. 2000, 296, 57–86. [Google Scholar] [CrossRef]
- Prassler, J.; Thiel, S.; Pracht, C.; Polzer, A.; Peters, S.; Bauer, M.; Nörenberg, S.; Stark, Y.; Kölln, J.; Popp, A.; et al. HuCAL PLATINUM, a Synthetic Fab Library Optimized for Sequence Diversity and Superior Performance in Mammalian Expression Systems. J. Mol. Biol. 2011, 413, 261–278. [Google Scholar] [CrossRef]
- Tada, T.; Dcosta, B.M.; Samanovic-Golden, M.; Herati, R.S.; Cornelius, A.; Mulligan, M.J.; Landau, N.R. Neutralization of Viruses with European, South African, and United States SARS-CoV-2 Variant Spike Proteins by Convalescent Sera and BNT162b2 mRNA Vaccine-Elicited Antibodies. mBio 2021, 12, 3. [Google Scholar] [CrossRef]
- Shen, X.; Tang, H.; Pajon, R.; Smith, G.; Glenn, G.M.; Shi, W.; Korber, B.; Montefiori, D.C. Neutralization of SARS-Cov-2 Variants, B.1.429 and B.1.351. N. Engl. J. Med. 2021, 384, 2352–2354. [Google Scholar] [CrossRef]
- Dayan, G.H.; Rouphael, N.; Walsh, S.R.; Chen, A.; Grunenberg, N.; Allen, M.; Antony, J.; Asante, K.P.; Bhate, A.S.; Beresnev, T.; et al. Efficacy of a Bivalent (D614 + B.1.351) SARS-CoV-2 Recombinant Protein Vaccine with AS03 Adjuvant in Adults: A Phase 3, Parallel, Randomised, Modified Double-Blind, Placebo-Controlled Trial. Lancet Respir. Med. 2023, 11, 975–990. [Google Scholar] [CrossRef]
- Sridhar, S.; Chicz, R.M.; Warren, W.; Tartaglia, J.; Savarino, S.; Gurunathan, S.; Toussaint, J.-F. The Potential of Beta Variant Containing COVID Booster Vaccines for Chasing Omicron in 2022. Nat. Commun. 2022, 13, 5794. [Google Scholar] [CrossRef]
- Francica, J.R.; Flynn, B.J.; Foulds, K.E.; Noe, A.T.; Werner, A.P.; Moore, I.N.; Gagne, M.; Johnston, T.S.; Tucker, C.; Davis, R.L.; et al. Protective Antibodies Elicited by SARS-CoV-2 Spike Protein Vaccination are Boosted in the Lung After Challenge in Nonhuman Primates. Sci. Transl. Med. 2021, 13, eabi4547. [Google Scholar] [CrossRef]
- Bruch, E.M.; Zhu, S.; Szymkowicz, L.; Blake, T.; Kiss, T.; James, D.A.; Rak, A.; Narayan, K.; Balmer, M.T.; Chicz, R.M. Structural and Biochemical Rationale for Beta Variant Protein Booster Vaccine Broad Cross-Neutralization of SARS-CoV-2. Sci. Rep. 2024, 14, 2038. [Google Scholar] [CrossRef]
- Zhu, S.; Liuni, P.; Chen, T.; Houy, C.; Wilson, D.J.; James, D.A. Epitope screening using Hydrogen/Deuterium Exchange Mass Spectrometry (HDX-MS): An accelerated workflow for evaluation of lead monoclonal antibodies. Biotechnol. J. 2021, 17, 2100358. [Google Scholar] [CrossRef]
- Berbers, G.A.; Marzec, A.H.; Bastmeijer, M.; van Gageldonk, P.G.; Plantinga, A. Blocking ELISA for Detection of Mumps Virus Antibodies in human sera. J. Virol. Methods 1993, 42, 155–168. [Google Scholar] [CrossRef]
- Bodjo, S.C.; Baziki, J.-D.; Nwankpa, N.; Chitsungo, E.; Koffi, Y.M.; Couacy-Hymann, E.; Diop, M.; Gizaw, D.; Tajelser, I.B.A.; Lelenta, M.; et al. Development and Validation of an Epitope-Blocking ELISA Using an Anti-Haemagglutinin Monoclonal Antibody for Specific Detection of Antibodies in Sheep and Goat Sera Directed Against Peste des Petits Ruminants Virus. Arch. Virol. 2018, 163, 1745–1756. [Google Scholar] [CrossRef]
- Hall, R.; Broom, A.; Hartnett, A.; Howard, M.; Mackenzie, J. Immunodominant Epitopes on the NS1 Protein of MVE and KUN Viruses Serve as Targets for a Blocking ELISA to Detect virus-Specific Antibodies in Sentinel Animal Serum. J. Virol. Methods 1995, 51, 201–210. [Google Scholar] [CrossRef]
- Russell Wms, B.; Burch, R.L. The Principles of Humane Experimental Technique; Universities Federation for Animal Welfare: Wheathampstead, UK, 1959. [Google Scholar]
- De Mattia, F.; Chapsal, J.-M.; Descamps, J.; Halder, M.; Jarrett, N.; Kross, I.; Mortiaux, F.; Ponsar, C.; Redhead, K.; McKelvie, J.; et al. The Consistency Approach for Quality Control of Vaccin—A Strategy to Improve Quality Control and Implement 3Rs. Biologicals 2011, 39, 59–65. [Google Scholar] [CrossRef]
- European Pharmacopoeia. Ph Eur 5.2.14: Substitution of In Vivo Method(S) by In Vitro Method(S) for the Quality Control of Vaccines; European Directorate for the Quality of Medicines & HealthCare of the Council of Europe: Strasbourg, France, 2018. [Google Scholar]
- Park, T.; Hwang, H.; Moon, S.; Kang, S.G.; Song, S.; Kim, Y.H.; Kim, H.; Ko, E.-J.; Yoon, S.-D.; Hwang, H.S. Vaccines against SARS-CoV-2 Variants and Future Pandemics. Expert Rev. Vaccines 2022, 21, 1363–1376. [Google Scholar] [CrossRef]
- Markwalter, C.; Jang, I.; Burton, R.; Domingo, G.; Wright, D. Biolayer Interferometry Predicts ELISA Performance of Monoclonal Antibody Pairs for Plasmodium Falciparum Histidine-Rich Protein 2. Anal. Biochem. 2017, 534, 10–13. [Google Scholar] [CrossRef]
- Tulsian, N.K.; Palur, R.V.; Qian, X.; Gu, Y.; Shunmuganathan, B.D.; Samsudin, F.; Wong, Y.H.; Lin, J.; Purushotorman, K.; Kozma, M.M.; et al. Defining Neutralization and Allostery by Antibodies Against COVID-19 Variants. Nat. Commun. 2023, 14, 6967. [Google Scholar] [CrossRef]
- Chen, B.; Farzan, M.; Choe, H. SARS-CoV-2 Spike Protein: Structure, Viral Entry and Variants. Nat. Rev. Microbiol. 2025, 23, 455. [Google Scholar] [CrossRef]
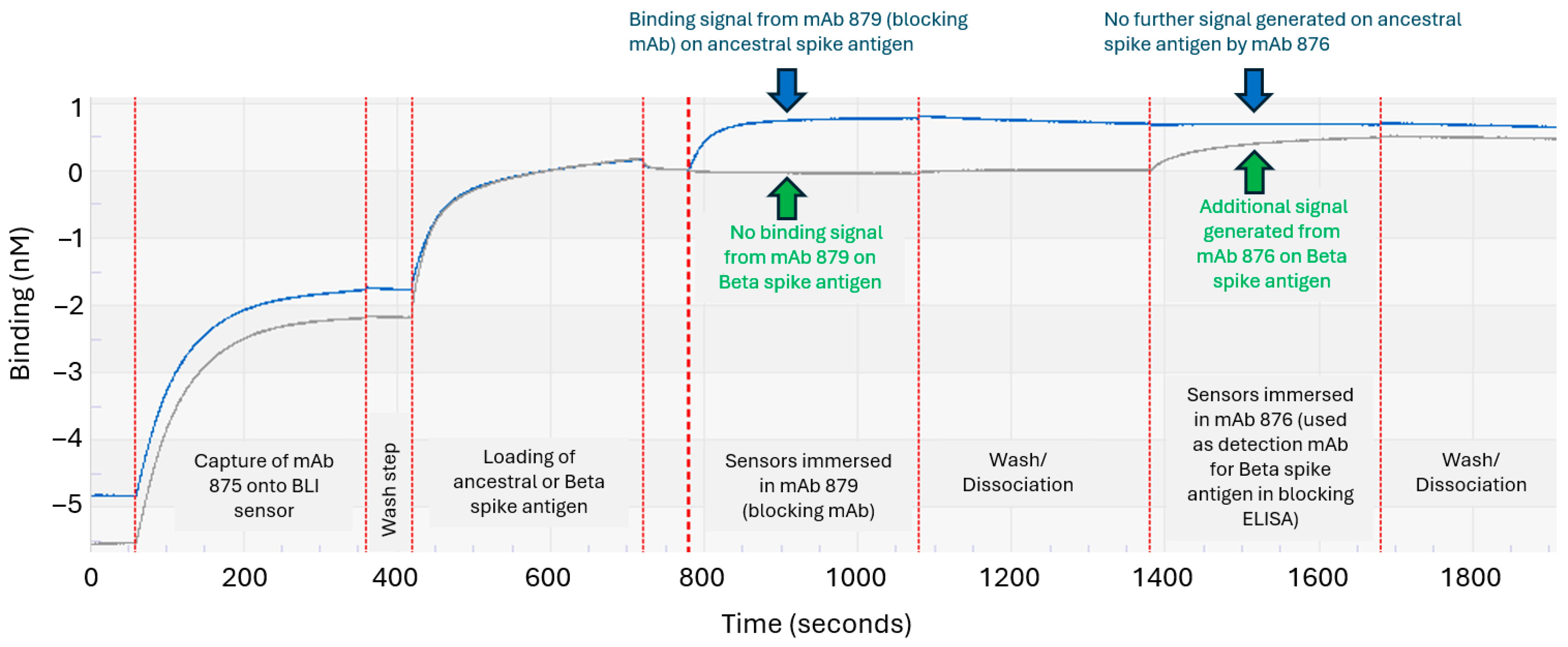
| Ancestral Spike Antigen Domain | mAb Clone Number | Equilibrium Dissociation Constant; KD (M) | Association kon (1/Ms) | Dissociation koff (1/s) | Neutralizing Titer (ng/mL) 1 | Reduction/Inhibition of Ancestral Spike Antigen Binding to ACE2 Receptor 2 |
|---|---|---|---|---|---|---|
| S1 | 876 | 3.01 × 10−10 | 2.60 × 105 | 7.83 × 10−5 | 76 | + |
| S1 | 879 | 6.52 × 10−10 | 3.91 × 105 | 2.55 × 10−4 | 93 | +++ |
| S1 | 886 | 5.39 × 10−10 | 2.00 × 105 | 1.08 × 10−4 | 27 | +++ |
| S1 | 962 | 1.51 × 10−9 | 3.22 × 105 | 4.87 × 10−4 | 6 | +++ |
| S2 | 875 | 9.29 × 10−10 | 4.19 × 105 | 3.89 × 10−4 | Not assessed | Not assessed |
| Bivalent Sample (Ancestral/Beta) (μg/mL) | Beta Spike Antigen Antigenicity Using Epitope-Blocking ELISA (ETU/mL) 1 | % Recovery of Beta Spike Antigen with Epitope-Blocking ELISA 2 | Antigenicity Results Using mAb 875 (Capture) and mAb 876 (Detection) Without any mAb 879 Blocking Step 1, 3 | Ancestral Spike Antigen Antigenicity Using Originally Developed ELISA 1 | % Recovery of Ancestral Spike Antigen Using Originally Developed ELISA 2 |
|---|---|---|---|---|---|
| 5.0/5.0 | 5.12 | 102% | 5.96 | 4.55 | 91% |
| 2.5/2.5 | 2.52 | 101% | 3.1 | 2.13 | 85% |
| 1.25/1.25 | 1.21 | 97% | 1.47 | 1.02 | 82% |
| 0.625/0.625 | 0.6 | 96% | 0.71 | 0.53 | 85% |
| 2.5/0 | 0 | No recovery, as expected | 0.63 | 2.3 | 92% |
| 0/2.5 | 2.48 | 99% | 2.34 | 0 | No recovery, as expected |
| 5.0/2.5 | 2.45 | 98% | 3.54 | 4.95 | 99% |
| mAb Clone | mAb Role | Epitope (aa) on Ancestral Spike Antigen | Epitope (aa) on Beta Spike Antigen |
|---|---|---|---|
| Anti-S1 | |||
| 876 | Detection antibody for Beta spike antigen in blocking ELISA | 420–439 (VIAWNSNNLDSKVGGNYNYL) | 425–436 (LDSKVGGNYNYL) |
| 879 | Blocking antibody for ancestral spike antigen | 459–475 (IYQAGSTPCNGVEGFNC) | No mAb binding (no notable HDX-MS difference observed; see Supplementary Figure S8) |
| 886 | Detection antibody for ancestral spike antigen | 459–475 (IYQAGSTPCNGVEGFNC) | No data available |
| Anti-S2 | |||
| 875 | Capture mAb | 771–781 (QVKQIYKTPPI) 856–872 (IAQYTSALLAGTITSG) | 754–766 (IAVEQDKNTQEVF) 853–861 (MIAQYTSAL) |
| Sample (Level of Beta Spike Antigen in Bivalent Formulation) | Antigenicity (ETU/mL) 1 | Average Antigenicity (ETU/mL) | %CV | Concentration of Beta Spike Antigen Used in Formulation (µg/mL) | Average % Recovery |
|---|---|---|---|---|---|
| Level 5 (200%) | 41.4 | 40.2 | 2.5 | 40 | 101 |
| 39.7 | |||||
| 39.6 | |||||
| Level 4 (100%) | 21.3 | 20.5 | 6.1 | 20 | 103 |
| 21.2 | |||||
| 19.1 | |||||
| Level 3 (50%) | 10.6 | 10.0 | 5.6 | 10 | 100 |
| 9.9 | |||||
| 9.5 | |||||
| Level 2 (25%) | 4.9 | 4.8 | 5.5 | 5 | 96 |
| 5.0 | |||||
| 4.5 | |||||
| Level 1 (12.5%) | 2.7 | 2.5 | 8.4 | 2.5 | 99 |
| 2.3 | |||||
| 2.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ettorre, L.; Williams, T.; Houy, C.; Zhu, S.; Kishko, M.; Azizi, A.; James, A.D.; Gajewska, B.; Szeto, J. Epitope Profiling of SARS-CoV-2 Spike Antigen Provides a Novel Strategy for Developing ELISAs Specific for Different Spike Protein Variants in Bivalent Vaccine Formulations. Vaccines 2025, 13, 794. https://doi.org/10.3390/vaccines13080794
Ettorre L, Williams T, Houy C, Zhu S, Kishko M, Azizi A, James AD, Gajewska B, Szeto J. Epitope Profiling of SARS-CoV-2 Spike Antigen Provides a Novel Strategy for Developing ELISAs Specific for Different Spike Protein Variants in Bivalent Vaccine Formulations. Vaccines. 2025; 13(8):794. https://doi.org/10.3390/vaccines13080794
Chicago/Turabian StyleEttorre, Luciano, Trevor Williams, Camille Houy, Shaolong Zhu, Michael Kishko, Ali Azizi, Andrew D. James, Beata Gajewska, and Jason Szeto. 2025. "Epitope Profiling of SARS-CoV-2 Spike Antigen Provides a Novel Strategy for Developing ELISAs Specific for Different Spike Protein Variants in Bivalent Vaccine Formulations" Vaccines 13, no. 8: 794. https://doi.org/10.3390/vaccines13080794
APA StyleEttorre, L., Williams, T., Houy, C., Zhu, S., Kishko, M., Azizi, A., James, A. D., Gajewska, B., & Szeto, J. (2025). Epitope Profiling of SARS-CoV-2 Spike Antigen Provides a Novel Strategy for Developing ELISAs Specific for Different Spike Protein Variants in Bivalent Vaccine Formulations. Vaccines, 13(8), 794. https://doi.org/10.3390/vaccines13080794






