Efficacy Evaluation of a VR-2332-Based Modified Live Vaccine Against NADC30-like PRRSV in China
Abstract
1. Introduction
2. Methods
2.1. Viruses, Cells, and Vaccine
2.2. Animals and Experimental Design
2.3. Clinical and Macroscopic Observations
2.4. Sample Collection
2.5. Viral Load
2.6. Analysis of Serum Antibody Response and Cytokines
2.7. Histopathology and Immunohistochemistry Staining
2.8. Statistical Analysis
3. Results
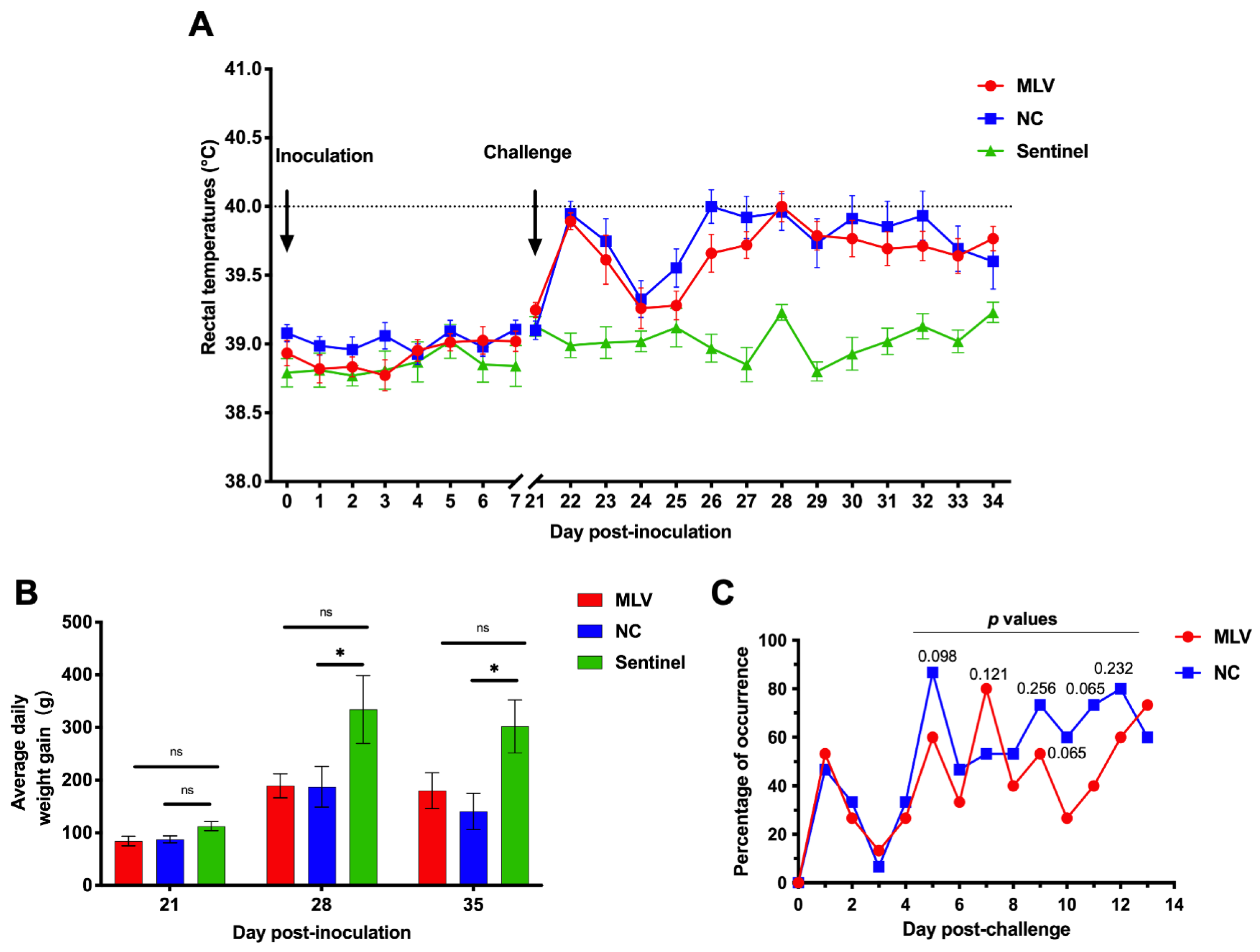
3.1. Clinical Symptoms and Pig Growth Performance
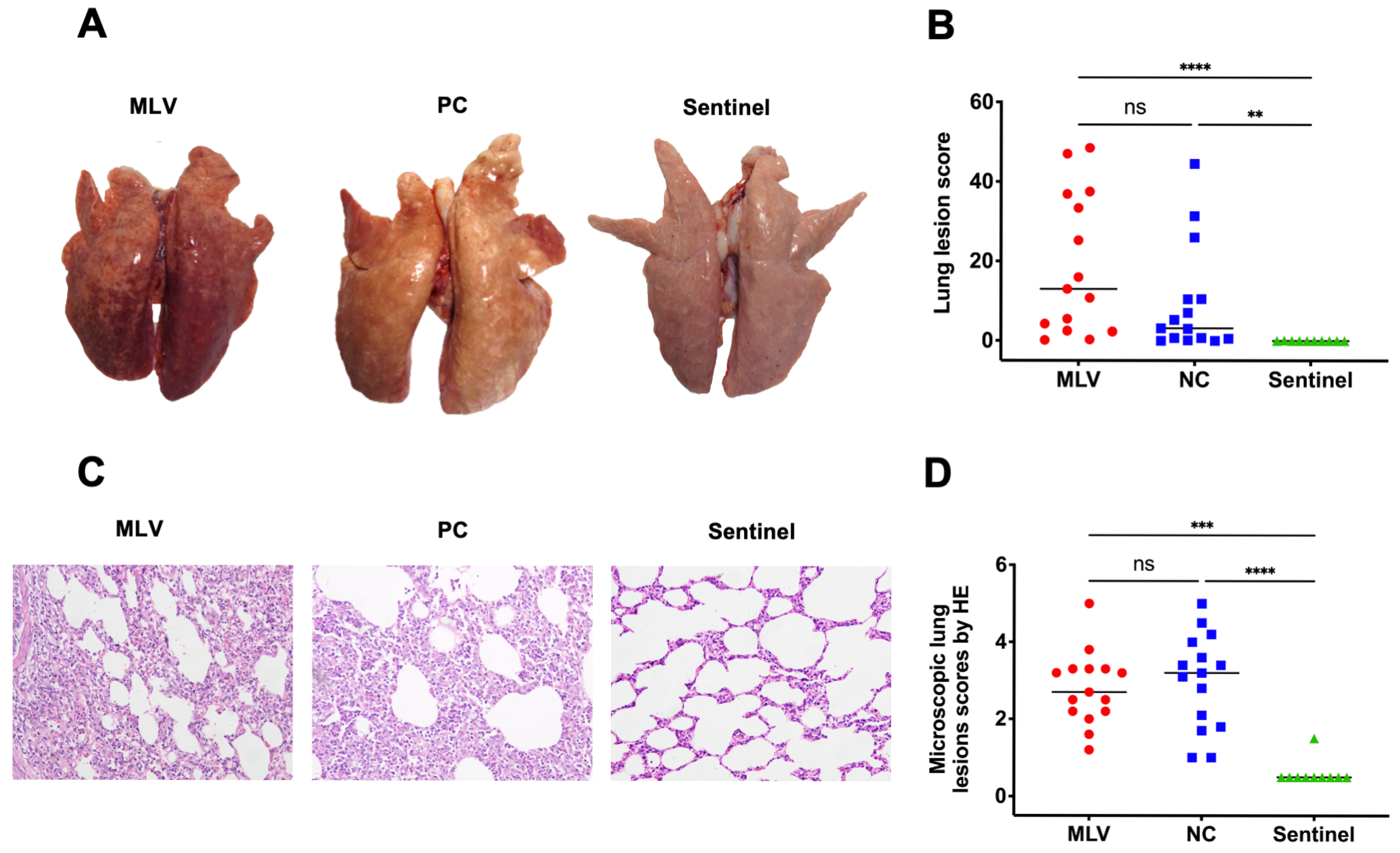
3.2. Pathological and Histopathological Findings
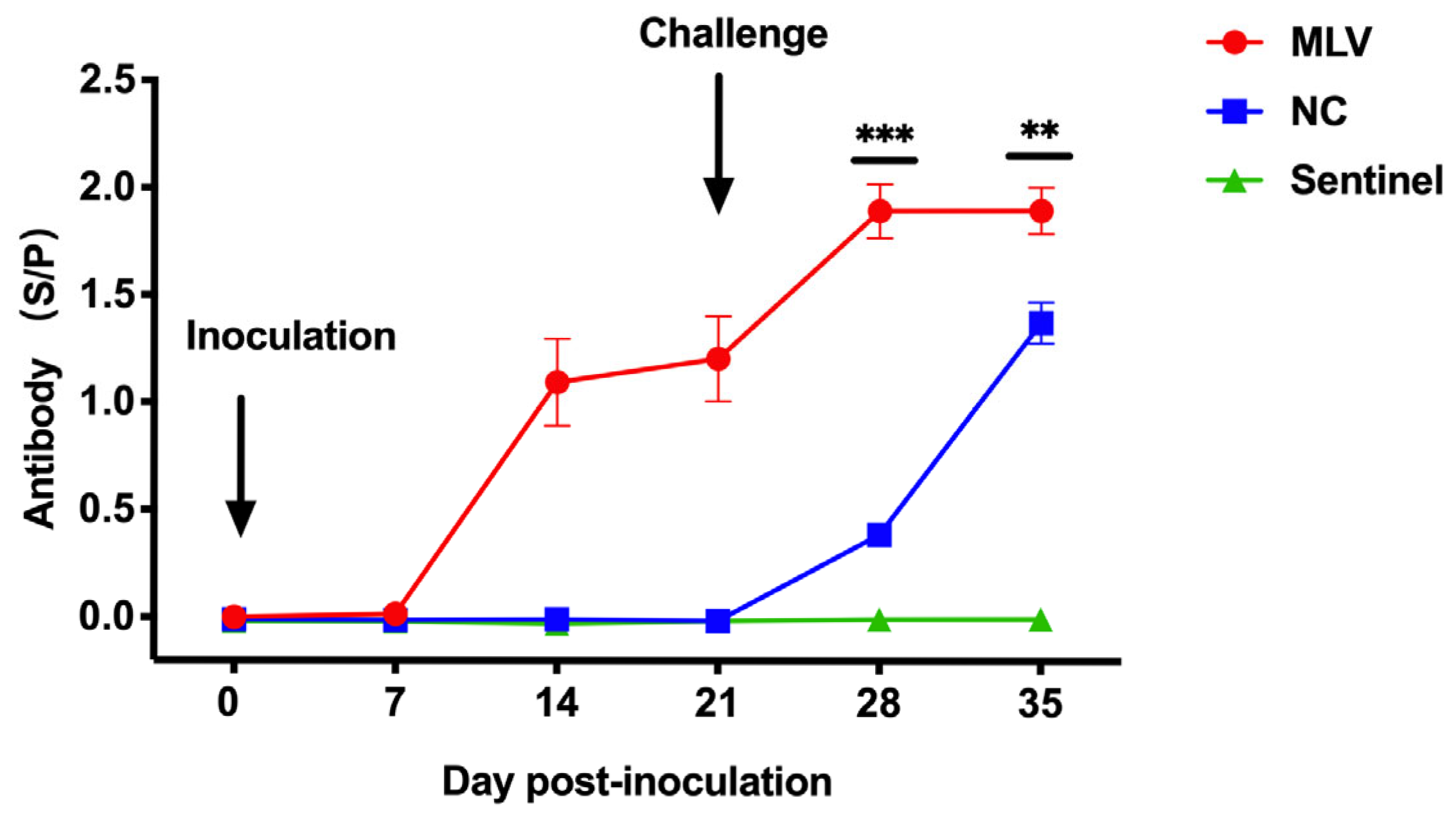
3.3. Antibody Responses
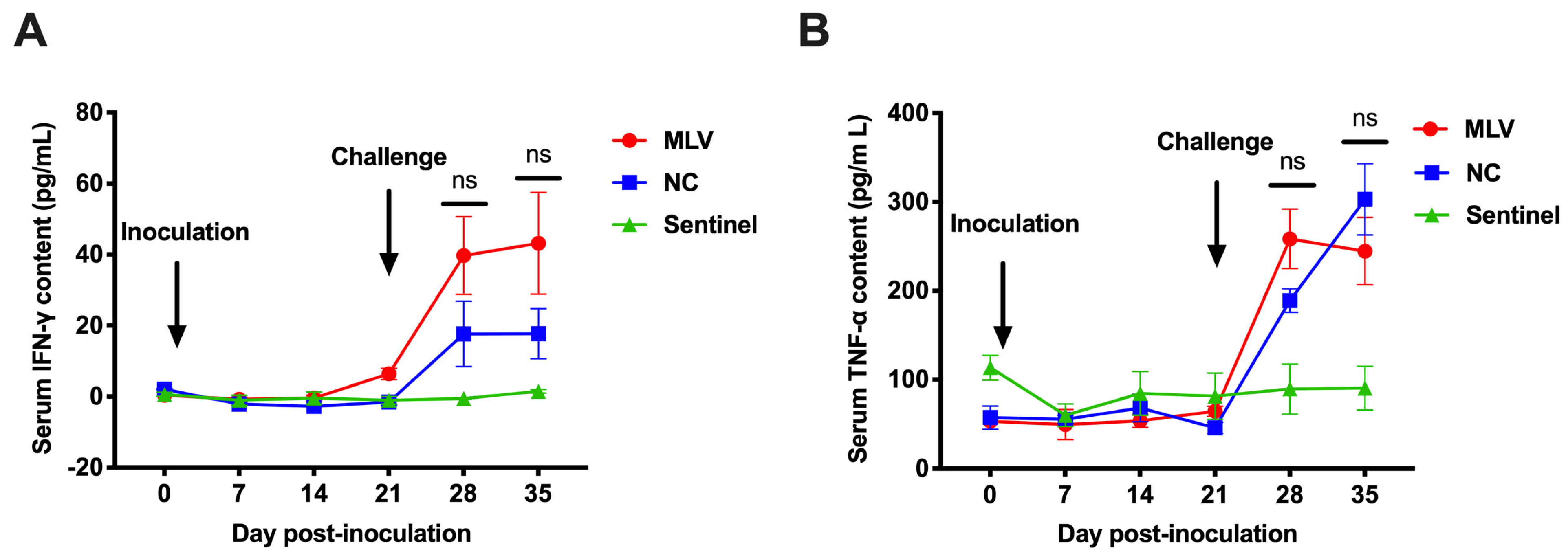
3.4. Cytokines
3.5. Viremia and Tissue Viral Load
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADWG | average daily weight gain |
| Dpc | days post-challenge |
| Dpi | days post-inoculation |
| IN | intranasally |
| IM | intramuscularly |
| IFN-γ | interferon gamma |
| MLV | modified live vaccine |
| RT-PCR | real-time polymerase chain reaction |
| PRRS | porcine reproductive and respiratory syndrome |
| PRRSV | porcine reproductive and respiratory syndrome virus |
| TNF-α | tumor necrosis factor-alpha |
| WOA | weeks of age |
References
- Jeong, J.; Kang, I.; Kim, S.; Park, S.-J.; Park, K.H.; Oh, T.; Yang, S.; Chae, C. A Modified-Live Porcine Reproductive and Respiratory Syndrome Virus (PRRSV)-1 Vaccine Protects Late-Term Pregnancy Gilts against Heterologous PRRSV-1 but Not PRRSV-2 Challenge. Transbound. Emerg. Dis. 2018, 65, 1227–1234. [Google Scholar] [CrossRef] [PubMed]
- Neumann, E.J.; Kliebenstein, J.B.; Johnson, C.D.; Mabry, J.W.; Bush, E.J.; Seitzinger, A.H.; Green, A.L.; Zimmerman, J.J. Assessment of the Economic Impact of Porcine Reproductive and Respiratory Syndrome on Swine Production in the United States. J. Am. Vet. Med. Assoc. 2005, 227, 385–392. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Z.; Li, H.; Yang, S.; Ren, F.; Bian, T.; Sun, L.; Zhou, B.; Zhou, L.; Qu, X. The Economic Impact of Porcine Reproductive and Respiratory Syndrome Outbreak in Four Chinese Farms: Based on Cost and Revenue Analysis. Front. Vet. Sci. 2022, 9, 1024720. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses (ICTV). Taxonomy. Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 9 May 2025).
- Tian, K.; Yu, X.; Zhao, T.; Feng, Y.; Cao, Z.; Wang, C.; Hu, Y.; Chen, X.; Hu, D.; Tian, X.; et al. Emergence of Fatal PRRSV Variants: Unparalleled Outbreaks of Atypical PRRS in China and Molecular Dissection of the Unique Hallmark. PLoS ONE 2007, 2, e526. [Google Scholar] [CrossRef] [PubMed]
- Lunney, J.K.; Fang, Y.; Ladinig, A.; Chen, N.; Li, Y.; Rowland, B.; Renukaradhya, G.J. Porcine Reproductive and Respiratory Syndrome Virus (PRRSV): Pathogenesis and Interaction with the Immune System. Annu. Rev. Anim. Biosci. 2016, 4, 129–154. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Nan, Y.; Xiao, S.; Zhao, Q.; Zhou, E. Antiviral Strategies against PRRSV Infection. Trends Microbiol. 2017, 25, 968–979. [Google Scholar] [CrossRef]
- Keffaber, K.K. Reproductive Failure of Unknown Etiology. Am. Assoc. Swine Pract. Newsl. 1989, 1, 1–10. [Google Scholar]
- Zimmerman, J.J.; Yoon, K.-J.; Pirtle, E.C.; Wills, R.W.; Sanderson, T.J.; McGinley, M.J. Studies of Porcine Reproductive and Respiratory Syndrome (PRRS) Virus Infection in Avian Species. Vet. Microbiol. 1997, 55, 329–336. [Google Scholar] [CrossRef]
- Jian, Y.; Lu, C.; Shi, Y.; Kong, X.; Song, J.; Wang, J. Genetic Evolution Analysis of PRRSV ORF5 Gene in Five Provinces of Northern China in 2024. BMC Vet. Res. 2025, 21, 242. [Google Scholar] [CrossRef]
- Shi, M.; Lam, T.T.; Hon, C.C.; Murtaugh, M.P.; Davies, P.R.; Hui, R.K.; Li, J.; Wong, L.T.; Yip, C.W.; Jiang, J.W.; et al. Phylogeny- based Evolutionary, Demographical, and Geographical Dissection of North American Type 2 Porcine Reproductive and Respiratory Syndrome Viruses. J. Virol. 2010, 84, 8700–8711. [Google Scholar] [CrossRef]
- Liu, J.; Xu, Y.; Lin, Z.; Fan, J.; Dai, A.; Deng, X.; Mao, W.; Huang, X.; Yang, X.; Wei, C. Epidemiology Investigation of PRRSV Discharged by Faecal and Genetic Variation of ORF5. Transbound. Emerg. Dis. 2021, 68, 2334–2344. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Kang, R.; Zhang, Y.; Ding, M.; Xie, B.; Tian, Y.; Wu, X.; Zuo, L.; Yang, X.; Wang, H. Whole Genome Analysis of Two Novel Type 2 Porcine Reproductive and Respiratory Syndrome Viruses with Complex Genome Recombination between Lineage 8, 3, and 1 Strains Identified in Southwestern China. Viruses 2018, 10, 328. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Wang, Z.; Ha, Z.; Zhang, Y.; Xie, Y.; Zhang, H.; Nan, F.; Zhang, J.; Zhao, G.; Li, Z.; et al. Genetic Characterization of a New NSP2-Deletion Porcine Reproductive and Respiratory Syndrome Virus in China. Microb. Pathog. 2021, 150, 104729. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Xu, T.; Luo, Z.; Deng, L.; Jian, Z.; Lai, S.; Ai, Y.; Zhou, Y.; Ge, L.; Xu, Z.; et al. Prevalence and Genetic Diversity of PRRSV in Sichuan Province of China from 2021 to 2023: Evidence of an Ongoing Epidemic Transition. Virology 2024, 600, 110213. [Google Scholar] [CrossRef]
- Li, C.; Fan, A.; Liu, Z.; Wang, G.; Zhou, L.; Zhang, H.; Huang, L.; Zhang, J.; Zhang, Z.; Zhang, Y. Prevalence, Time of Infection, and Diversity of Porcine Reproductive and Respiratory Syndrome Virus in China. Viruses 2024, 16, 774. [Google Scholar] [CrossRef]
- Xu, H.; Li, C.; Li, W.; Zhao, J.; Gong, B.; Sun, Q.; Tang, Y.; Xiang, L.; Leng, C.; Peng, J.; et al. Novel Characteristics of Chinese NADC34-like PRRSV during 2020–2021. Transbound. Emerg. Dis. 2022, 69, e3215–e3224. [Google Scholar] [CrossRef]
- Murtaugh, M.P.; Stadejek, T.; Abrahante, J.E.; Lam, T.T.Y.; Leung, F.C.-C. The Ever-Expanding Diversity of Porcine Reproductive and Respiratory Syndrome Virus. Virus Res. 2010, 154, 18–30. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Pei, X.; Chen, S.; Jing, Y.; Wu, Y.; Ma, Z.; Li, Z.; Zheng, Z.; Feng, Y.; et al. A Chimeric Strain of Porcine Reproductive and Respiratory Syndrome Virus 2 Derived from HP-PRRSV and NADC30-like PRRSV Confers Cross-Protection against Both Strains. Vet. Res. 2024, 55, 132. [Google Scholar] [CrossRef]
- Zhang, H.; Luo, Q.; He, Y.; Zheng, Y.; Sha, H.; Li, G.; Kong, W.; Liao, J.; Zhao, M. Research Progress on the Development of Porcine Reproductive and Respiratory Syndrome Vaccines. Vet. Sci. 2023, 10, 491. [Google Scholar] [CrossRef]
- Chae, C. Commercial PRRS Modified-Live Virus Vaccines. Vaccines 2021, 9, 185. [Google Scholar] [CrossRef]
- Tian, K. NADC30-Like Porcine Reproductive and Respiratory Syndrome in China. Open Virol. J. 2017, 11, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, F.; Han, X.; Guo, H.; Liu, C.; Hou, L.; Wang, Y.; Zheng, H.; Wang, L.; Wen, Y. Recent Advances in the Study of NADC34-like Porcine Reproductive and Respiratory Syndrome Virus in China. Front. Microbiol. 2022, 13, 950402. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yang, Y.; Xia, Q.; Guan, Z.; Zhang, J.; Li, B.; Qiu, Y.; Liu, K.; Shao, D.; Ma, Z.; et al. Genetic Characterization of Porcine Reproductive and Respiratory Syndrome Virus from Eastern China during 2017–2022. Front. Microbiol. 2022, 13, 971817. [Google Scholar] [CrossRef]
- Martínez-Lobo, F.J.; de Lome, L.C.; Díez-Fuertes, F.; Segalés, J.; García-Artiga, C.; Simarro, I.; Castro, J.M.; Prieto, C. Safety of Porcine Reproductive and Respiratory Syndrome Modified Live Virus (MLV) Vaccine Strains in a Young Pig Infection Model. Vet. Res. 2013, 44, 115. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Wang, J.; Bai, X.; Ji, G.; Yan, H.; Li, Y.; Wang, Y.; Tan, F.; Xiao, Y.; Li, X.; et al. Pathogenicity Comparison between Highly Pathogenic and NADC30-like Porcine Reproductive and Respiratory Syndrome Virus. Arch. Virol. 2016, 161, 2257–2261. [Google Scholar] [CrossRef]
- Bai, X.; Wang, Y.; Xu, X.; Sun, Z.; Xiao, Y.; Ji, G.; Li, Y.; Tan, F.; Li, X.; Tian, K. Commercial Vaccines Provide Limited Protection to NADC30-like PRRSV Infection. Vaccine 2016, 34, 5540–5545. [Google Scholar] [CrossRef] [PubMed]
- Percie du Sert, N.; Hurst, V.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; et al. The ARRIVE Guidelines 2.0: Updated Guidelines for Reporting Animal Research. PLoS Biol. 2020, 18, e3000410. [Google Scholar]
- Sun, F.; Zhou, L.; Bian, T.; Tian, X.; Ren, K.; Lu, C.; Zhang, L.; Li, L.; Cui, S.; Yang, C.; et al. Efficacy Evaluation of Two Commercial Modified-Live Virus Vaccines against a Novel Recombinant Porcine Reproductive and Respiratory Syndrome Virus (PRRSV)-2. Vet. Microbiol. 2018, 216, 176–182. [Google Scholar] [CrossRef]
- Chai, W.; Liu, Z.; Sun, Z.; Su, L.; Zhang, C.; Huang, L. Efficacy of Two Porcine Reproductive and Respiratory Syndrome (PRRS) Modified-Live Virus (MLV) Vaccines against Heterologous NADC30-like PRRS Virus Challenge. Vet. Microbiol. 2020, 248, 108805. [Google Scholar] [CrossRef]
- Choi, H.-Y.; Lee, S.-H.; Ahn, S.-H.; Choi, J.-C.; Jeong, J.-Y.; Lee, B.-J.; Kang, Y.-L.; Hwang, S.-S.; Lee, J.-K.; Lee, S.-W.; et al. A Chimeric Porcine Reproductive and Respiratory Syndrome Virus (PRRSV)-2 Vaccine Is Safe under International Guidelines and Effective Both in Experimental and Field Conditions. Res. Vet. Sci. 2021, 135, 143–152. [Google Scholar] [CrossRef]
- Halbur, P.G.; Paul, P.S.; Frey, M.L.; Landgraf, J.; Eernisse, K.; Meng, X.J.; Lum, M.A.; Andrews, J.J.; Rathje, J.A. Comparison of the Pathogenicity of Two US Porcine Reproductive and Respiratory Syndrome Virus Isolates with That of the Lelystad Virus. Vet. Pathol. 1995, 32, 648–660. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhuang, J.; Wang, J.; Han, L.; Sun, Z.; Xiao, Y.; Ji, G.; Li, Y.; Tan, F.; Li, X.; et al. Outbreak Investigation of NADC30-Like PRRSV in South-East China. Transbound. Emerg. Dis. 2016, 63, 474–479. [Google Scholar] [CrossRef]
- Choi, K.; Park, C.; Jeong, J.; Chae, C. Comparison of Protection Provided by Type 1 and Type 2 Porcine Reproductive and Respiratory Syndrome Field Viruses against Homologous and Heterologous Challenge. Vet. Microbiol. 2016, 191, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Yuan, N.; Yang, Z.; Lv, F.; Dou, L.; Li, X.; Zhao, B.; Dong, S. Molecular Epidemiology and Genetic Evolution of Porcine Reproductive and Respiratory Syndrome Virus in Northern China During 2021–2023. Viruses 2025, 17, 85. [Google Scholar] [CrossRef]
- Tu, T.; Li, Y.; Zhang, G.; Du, C.; Zhou, Y.; Jiang, D.; Luo, Y.; Yao, X.; Yang, Z.; Ren, M.; et al. Isolation, Identification, Recombination Analysis and Pathogenicity Experiment of a PRRSV Recombinant Strain in Sichuan Province, China. Front. Microbiol. 2024, 15, 1362471. [Google Scholar]
- Zhou, L.; Ge, X.; Yang, H. Porcine Reproductive and Respiratory Syndrome Modified Live Virus Vaccine: A “Leaky” Vaccine with Debatable Efficacy and Safety. Vaccines 2021, 9, 362. [Google Scholar] [CrossRef]
- Renukaradhya, G.J.; Meng, X.-J.; Calvert, J.G.; Roof, M.; Lager, K.M. Live Porcine Reproductive and Respiratory Syndrome Virus Vaccines: Current Status and Future Direction. Vaccine 2015, 33, 4069–4080. [Google Scholar] [CrossRef]
- Martelli, P.; Gozio, S.; Ferrari, L.; Rosina, S.; De Angelis, E.; Quintavalla, C.; Bottarelli, E.; Borghetti, P. Efficacy of a Modified Live Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Vaccine in Pigs Naturally Exposed to a Heterologous European (Italian Cluster) Field Strain: Clinical Protection and Cell-Mediated Immunity. Vaccine 2009, 27, 3788–3799. [Google Scholar] [CrossRef]
- Lager, K.M.; Schlink, S.N.; Brockmeier, S.L.; Miller, L.C.; Henningson, J.N.; Kappes, M.A.; Kehrli, M.E.; Loving, C.L.; Guo, B.; Swenson, S.L.; et al. Efficacy of Type 2 PRRSV Vaccine against Chinese and Vietnamese HP-PRRSV Challenge in Pigs. Vaccine 2014, 32, 6457–6462. [Google Scholar] [CrossRef]
- Li, Y.; Xu, L.; Jiao, D.; Zheng, Z.; Chen, Z.; Jing, Y.; Li, Z.; Ma, Z.; Feng, Y.; Guo, X.; et al. Genomic Similarity and Antibody-Dependent Enhancement of Immune Serum Potentially Affect the Protective Efficacy of Commercial MLV Vaccines against NADC30-like PRRSV. Virol. Sin. 2023, 38, 813–826. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, X.; Guo, T.; Qiao, S.; Guo, Z.; Li, R.; Jin, Q.; Hu, X.; Xing, G.; Deng, R.; et al. Efficacy of a Live Attenuated Highly Pathogenic PRRSV Vaccine against a NADC30-like Strain Challenge: Implications for ADE of PRRSV. BMC Vet. Res. 2021, 17, 260. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yang, B.; Xu, L.; Jin, H.; Ge, X.; Guo, X.; Han, J.; Yang, H. Efficacy Evaluation of Three Modified-Live Virus Vaccines against a Strain of Porcine Reproductive and Respiratory Syndrome Virus NADC30-Like. Vet. Microbiol. 2017, 207, 108–116. [Google Scholar] [CrossRef]
- Wei, C.; Dai, A.; Fan, J.; Li, Y.; Chen, A.; Zhou, X.; Luo, M.; Yang, X.; Liu, J. Efficacy of Type 2 PRRSV Vaccine against Challenge with the Chinese Lineage 1 (NADC30-like) PRRSVs in Pigs. Sci. Rep. 2019, 9, 10781. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.J.; Wu, L.L.; Zimmerman, J.J.; Hill, H.T.; Platt, K.B. Antibody-Dependent Enhancement (ADE) of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Infection in Pigs. Viral Immunol. 1996, 9, 51–63. [Google Scholar] [CrossRef]
- Cancel-Tirado, S.M.; Evans, R.B.; Yoon, K.J. Monoclonal Antibody Analysis of Porcine Reproductive and Respiratory Syndrome Virus Epitopes Associated with Antibody-Dependent Enhancement and Neutralization of Virus Infection. Vet. Immunol. Immunopathol. 2004, 102, 249–262. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, H.; Li, W.; Feng, X.; Han, F.; Zhang, Y.; Chen, J.; Liu, D.; Xia, P. Activating Fc Gamma Receptors and Viral Receptors Are Required for Antibody-Dependent Enhancement of Porcine Reproductive and Respiratory Syndrome Virus Infection. Vet. Sci. 2022, 9, 470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Feng, X.; Wang, H.; He, S.; Fan, H.; Liu, D. Antibody-Dependent Enhancement of Porcine Reproductive and Respiratory Syndrome Virus Infection Downregulates the Levels of Interferon-Gamma/Lambdas in Porcine Alveolar Macrophages in Vitro. Front. Vet. Sci. 2023, 10, 1150430. [Google Scholar] [CrossRef]
- Chang, H.; Gao, X.; Wu, Y.; Wang, F.; Lai, M.; Zheng, J.; Qiu, Y.; He, Y.; Liang, X.; Yuan, K.; et al. Genomic and Pathogenicity Analysis of Two Novel Highly Pathogenic Recombinant NADC30-like PRRSV Strains in China, in 2023. Microbiol. Spectr. 2024, 12, e003682. [Google Scholar] [CrossRef]
- Razzuoli, E.; Armando, F.; De Paolis, L.; Ciurkiewicz, M.; Amadori, M. The Swine IFN System in Viral Infections: Major Advances and Translational Prospects. Pathogens 2022, 11, 175. [Google Scholar] [CrossRef]
- Crisci, E.; Fraile, L.; Montoya, M. Cellular Innate Immunity against PRRSV and Swine Influenza Viruses. Vet. Sci. 2019, 6, 26. [Google Scholar] [CrossRef]
- Charerntantanakul, W.; Pongjaroenkit, S. Co-Administration of Saponin Quil A and PRRSV-1 Modified-Live Virus Vaccine up-Regulates Gene Expression of Type I Interferon-Regulated Gene, Type I and II Interferon, and Inflammatory Cytokines and Reduces Viremia in Response to PRRSV -2 Challenge. Vet. Immunol. Immunopathol. 2018, 205, 24–34. [Google Scholar] [CrossRef]
- Gómez-Laguna, J.; Rodríguez-Gómez, I.M.; Barranco, I.; Pallarés, F.J.; Salguero, F.J.; Carrasco, L. Enhanced Expression of TGFβ Protein in Lymphoid Organs and Lung, but Not in Serum, of Pigs Infected with a European Field Isolate of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV). Vet. Microbiol. 2012, 158, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Kim, S.; Park, C.; Park, K.H.; Kang, I.; Park, S.J.; Chae, C. Commercial Porcine Reproductive and Respiratory Syndrome Virus (PRRSV)-2 Modified Live Virus Vaccine against Heterologous Single and Dual Korean PRRSV-1 and PRRSV-2 Challenge. Vet. Rec. 2018, 182, 485. [Google Scholar] [CrossRef]
- Calzada-Nova, G.; Schnitzlein, W.M.; Husmann, R.J.; Zuckermann, F.A. North American Porcine Reproductive and Respiratory Syndrome Viruses Inhibit Type I Interferon Production by Plasmacytoid Dendritic Cells. J. Virol. 2011, 85, 2703–2713. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Xu, H.; Wang, Y.; Liu, Z.; He, F. Efficacy Evaluation of Two Commercial Vaccines Against a Recombinant PRRSV2 Strain ZJnb16-2 from Lineage 8 and 3 in China. Pathogens 2020, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Miranda, J.; Romero, S.; de Lucas, L.; Saito, F.; Fenech, M.; Díaz, I. Protection Provided by a Commercial Modified-Live Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) 1 Vaccine (PRRSV1-MLV) against a Japanese PRRSV2 Field Strain. J. Vet. Sci. 2023, 24, e54. [Google Scholar] [CrossRef]
- Fiers, J.; Cay, A.B.; Maes, D.; Tignon, M. A Comprehensive Review on Porcine Reproductive and Respiratory Syndrome Virus with Emphasis on Immunity. Vaccines 2024, 12, 942. [Google Scholar] [CrossRef]
- Kimman, T.G.; Cornelissen, L.A.; Moormann, R.J.; Rebel, J.M.J.; Stockhofe-Zurwieden, N. Challenges for Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Vaccinology. Vaccine 2009, 27, 3704–3718. [Google Scholar] [CrossRef]
- Lopez, O.J.; Oliveira, M.F.; Garcia, E.A.; Kwon, B.J.; Doster, A.; Osorio, F.A. Protection against Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Infection through Passive Transfer of PRRSV-Neutralizing Antibodies Is Dose Dependent. Clin. Vaccine Immunol. 2007, 14, 269–275. [Google Scholar] [CrossRef]

| Group | Number of Piglets | Vaccination | Route and Dose of Inoculation | Challenge | Route and Dose of Challenge |
|---|---|---|---|---|---|
| MLV | 15 | MLV | 2 mL IM | HNjz15 | 1 mL IN plus 1 mL IM |
| NC | 15 | Sterile PBS | HNjz15 | ||
| Sentinel | 10 | Sterile PBS | Sterile PBS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Tong, X.; Shu, J.; Feng, H.; Quan, Y.; He, Y. Efficacy Evaluation of a VR-2332-Based Modified Live Vaccine Against NADC30-like PRRSV in China. Vaccines 2025, 13, 538. https://doi.org/10.3390/vaccines13050538
Li L, Tong X, Shu J, Feng H, Quan Y, He Y. Efficacy Evaluation of a VR-2332-Based Modified Live Vaccine Against NADC30-like PRRSV in China. Vaccines. 2025; 13(5):538. https://doi.org/10.3390/vaccines13050538
Chicago/Turabian StyleLi, Lixin, Xiaxia Tong, Jianhong Shu, Huapeng Feng, Yanping Quan, and Yulong He. 2025. "Efficacy Evaluation of a VR-2332-Based Modified Live Vaccine Against NADC30-like PRRSV in China" Vaccines 13, no. 5: 538. https://doi.org/10.3390/vaccines13050538
APA StyleLi, L., Tong, X., Shu, J., Feng, H., Quan, Y., & He, Y. (2025). Efficacy Evaluation of a VR-2332-Based Modified Live Vaccine Against NADC30-like PRRSV in China. Vaccines, 13(5), 538. https://doi.org/10.3390/vaccines13050538






