The authors would like to make the following corrections to this published paper [1].
In the original publication, there was a mistake in Figure 1H,I as these images were accidentally duplicated. There was also a mistake in the cell line name in Figure 1F. The corrected Figure 1 is attached below.
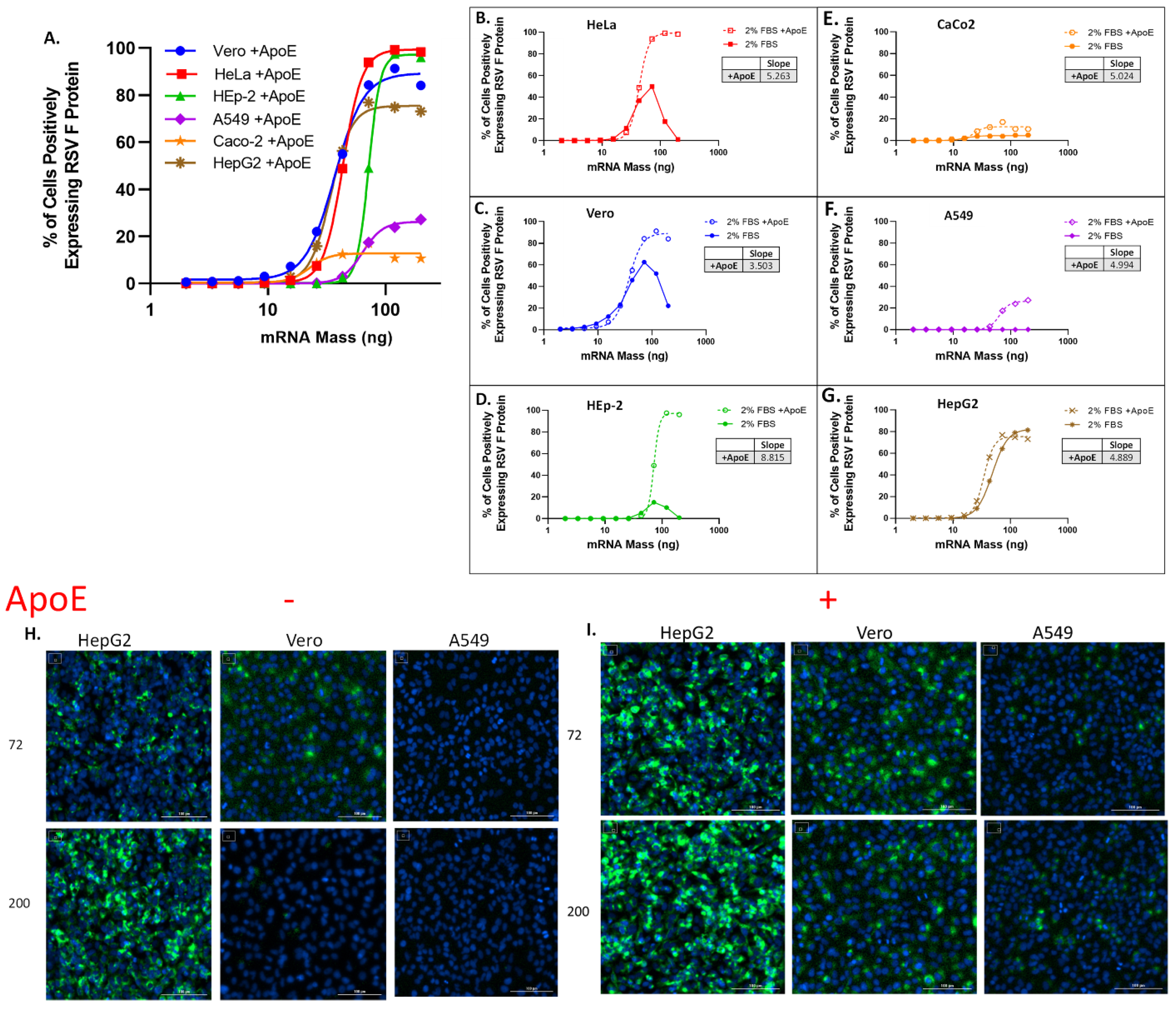
Figure 1.
LNP transfection curves across varying cell lines indicate variable levels of transfection, which improve with the addition of ApoE during transfection. (A) RSV-F protein expression efficiency in Vero, HeLA, HEp-2, A549, CaCo-2 and HepG2 cells was measured by counting percentage of cells positive for RSV-F protein. All cells were transfected for 16 ± 2 h with titration of LNPs starting with 200 ng dose of mRNA in media with 2% FBS. Data were fit using variable slope-four parameter logistics regression (4-PL) model in GraphPad Prism software (version: 6.0). (B–G) RSV-F protein expression efficiency following transfection with LNPs in media + 2%FBS, and media + 2%FBS supplemented with ApoE at 4 ug/mL. Hill-slope values [Y = Bottom + (Top − Bottom)/(1 + 10^((LogEC50-X) × Hill-slope)] from 4-PL regression are given for curves obtained with FBS supplemented with ApoE. (B) HeLa cells. (C) Vero cells. (D) Hep-2 cells. (E) CaCo-2 cells. (F) A549 cells. (G) HepG2 cells. (H) Representative immunofluorescence images of HepG2, Vero and A549 cells without ApoE and (I) with ApoE shown at 72 (top row) and 200 ng/mL (bottom row) of mRNA dose representing the bottom and top of the dose response curve, respectively.
The authors state that the scientific conclusions are unaffected. This correction was approved by the Academic Editor. The original publication has also been updated.
Reference
- Patel, N.; Davis, Z.; Hofmann, C.; Vlasak, J.; Loughney, J.W.; DePhillips, P.; Mukherjee, M. Development and Characterization of an In Vitro Cell-Based Assay to Predict Potency of mRNA–LNP-Based Vaccines. Vaccines 2023, 11, 1224. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).