A Novel Flow Cytometry Array for High Throughput Detection of SARS-CoV-2 Antibodies
Abstract
1. Introduction
2. Materials and Methods
- Assay Sensitivity, Specificity, Positive Predictive Value (PPV), and Negative Predictive Value (NPV). We tested 121 patient samples and 299 normal serum samples.
- Assay Performance. Intra- and inter-plate CVs were used to determine the assay precision and reproducibility.
- Class Specificity. Dithiothreitol (DTT) inactivates IgM antibodies, but not IgG antibodies. Here, IgM-, IgG-, and IgA-positive serum samples were either treated or not treated with 5 mM DTT at 37 °C for 15 min prior to running the bead-based assay to determine whether the detection of IgM antibodies was specific.
- Cross-Reactivity. Serum from donors immunized against other infectious agents was employed to determine the cross-reactivity of antibodies to S-RBD.
3. Results
3.1. Establishment of the Bead-Based Assay
3.2. Determination of the Assay Sensitivity and Specificity
3.3. Assay Intra- and Inter-Plate Coefficient of Variation (CV)
3.4. Assay Positive Predictive Value (PPV) and Negative Predictive Value (NPV) Determination
3.5. Comparison of Antibody Measurement Using ELISA
3.6. Class Specificity
3.7. Crossreactivity Investigation with Samples with Non-COVID-19 Infections
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hasöksüz, M.; Kiliç, S.; Saraç, F. Coronaviruses and SARS-CoV-2. Turk. J. Med. Sci. Turk. Klinikleri. 2020, 50, 549–556. [Google Scholar] [CrossRef]
- Sironi, M.; Hasnain, S.E.; Rosenthal, B.; Phan, T.; Luciani, F.; Shaw, M.-A.; Sallum, M.A.; Mirhashemi, M.E.; Morand, S.; González-Candelas, F. SARS-CoV-2 and COVID-19: A genetic, epidemiological, and evolutionary perspective. Infect. Genet. Evolution. 2020, 84, 104384. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020, 11, 1620. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Lee, N.; Hui, D.; Wu, A.; Chan, P.; Cameron, P.; Joynt, G.M.; Ahuja, A.; Yung, M.Y.; Leung, C.B.; To, K.F.; et al. A Major Outbreak of Severe Acute Respiratory Syndrome in Hong Kong. N. Engl. J. Med. 2003, 348, 1986–1994. Available online: www.nejm.org (accessed on 24 March 2022). [CrossRef]
- Al-Omari, A.; Rabaan, A.A.; Salih, S.; Al-Tawfiq, J.A.; Memish, Z.A. MERS coronavirus outbreak: Implications for emerging viral infections. Diagn. Microbiol. Infect. Dis. 2019, 93, 265–285. [Google Scholar] [CrossRef]
- Buitrago-Garcia, D.; Egli-Gany, D.; Counotte, M.J.; Hossmann, S.; Imeri, H.; Ipekci, A.M.; Salanti, G.; Low, N. Occurrence and transmission potential of asymptomatic and presymptomatic SARSCoV-2 infections: A living systematic review and meta-analysis. PLoS Med. Public Libr. Sci. 2020, 17, e1003346. [Google Scholar] [CrossRef]
- Ruan, Q.; Yang, K.; Wang, W.; Jiang, L.; Song, J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020, 46, 846–848. [Google Scholar] [CrossRef]
- Ruan, Q.; Yang, K.; Wang, W.; Jiang, L.; Song, J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020, 46, 846–848, Correction in Intensive Care Med. 2020, 46, 1294–1297. https://doi.org/10.1007/s00134-020-06028-z. [Google Scholar] [CrossRef]
- Izcovich, A.; Ragusa, M.A.; Tortosa, F.; Marzio, M.A.L.; Agnoletti, C.; Bengolea, A.; Ceirano, A.; Espinosa, F.; Saavedra, E.; Sanguine, V.; et al. Prognostic factors for severity and mortality in patients infected with COVID-19: A systematic review. PLoS ONE 2020, 15, e0241955, Erratum in PLoS ONE Public Libr. Sci. 2022, 17, e0241955. https://doi.org/10.1371/journal.pone.0269291. [Google Scholar] [CrossRef]
- Ioannidis, J.P.A.; Axfors, C.; Contopoulos-Ioannidis, D.G. Population-level COVID-19 mortality risk for non-elderly individuals overall and for non-elderly individuals without underlying diseases in pandemic epicenters. Environ. Res. 2020, 188, 109890. [Google Scholar] [CrossRef]
- Rothan, H.A.; Byrareddy, S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. Acad. Press 2020, 109, 102433. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269, Correction in Nat. Nat. Res. 2020, 580, E7. https://doi.org/10.1038/s41586-020-2202-3. [Google Scholar] [CrossRef]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef]
- Moser, C.; Li, J.Z.; Eron, J.J.; Aga, E.; Daar, E.S.; Wohl, D.A.; Coombs, R.W.; Javan, A.C.; Ignacio, R.A.B.; Jagannathan, P.; et al. Predictors of SARS-CoV-2 RNA From Nasopharyngeal Swabs and Concordance With Other Compartments in Nonhospitalized Adults With Mild to Moderate COVID-19. Open Forum Infect. Dis. 2022, 9, ofac618. [Google Scholar] [CrossRef]
- Fox, T.; Geppert, J.; Dinnes, J.; Scandrett, K.; Bigio, J.; Sulis, G.; Hettiarachchi, D.; Mathangasinghe, Y.; Weeratunga, P.; Wickramasinghe, D.; et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst. Rev. 2022, 11, Cd013652. [Google Scholar] [CrossRef]
- Asamoah-Boaheng, M.; Goldfarb, D.M.; Barakauskas, V.; Kirkham, T.L.; Demers, P.A.; Karim, M.E.; Lavoie, P.M.; Marquez, A.C.; Jassem, A.N.; Jenneson, S.; et al. Evaluation of the Performance of a Multiplexed Serological Assay in the Detection of SARS-CoV-2 Infections in a Predominantly Vaccinated Population. Microbiol. Spectr. 2022, 10, e0145421. [Google Scholar] [CrossRef]
- Shengule, S.; Alai, S.; Bhandare, S.; Patil, S.; Gautam, M.; Mangaonkar, B.; Gupta, S.; Shaligram, U.; Gairola, S. Validation and Suitability Assessment of Multiplex Mesoscale Discovery Immunogenicity Assay for Establishing Serological Signatures Using Vaccinated, Non-Vaccinated and Breakthrough SARS-CoV-2 Infected Cases. Vaccines 2024, 12, 433. [Google Scholar] [CrossRef]
- Rottmayer, K.; Schwarze, M.; Jassoy, C.; Hoffmann, R.; Loeffler-Wirth, H.; Lehmann, C. Potential of a Bead-Based Multiplex Assay for SARS-CoV-2 Antibody Detection. Biology 2024, 13, 273. [Google Scholar] [CrossRef]
- Bray, R.A.; Lee, J.H.; Brescia, P.; Kumar, D.; Nong, T.B.; Shih, R.; Woodle, E.S.; Maltzman, J.S.; Gebel, H.M. Development and Validation of a Multiplex, Bead-based Assay to Detect Antibodies Directed Against SARS-CoV-2 Proteins. Transplantation 2021, 105, 79–89. [Google Scholar] [CrossRef]
- Heaney, C.D.; Pisanic, N.; Randad, P.R.; Kruczynski, K.; Howard, T.; Zhu, X.; Littlefield, K.; Patel, E.U.; Shrestha, R.; Laeyendecker, O.; et al. Comparative performance of multiplex salivary and commercially available serologic assays to detect SARS-CoV-2 IgG and neutralization titers. J. Clin. Virol. 2021, 145, 104997. [Google Scholar] [CrossRef]
- Yu, S.; An, J.; Liao, X.; Wang, H.; Ma, F.; Li, D.; Li, A.; Liu, W.; Zhang, S.; Liao, M.; et al. Distinct kinetics of immunoglobulin isotypes reveal early diagnosis and disease severity of COVID-19: A 6-month follow-up. Clin. Transl. Med. 2021, 11, e342. [Google Scholar] [CrossRef]
- Rubio, R.; Macià, D.; Barrios, D.; Vidal, M.; Jiménez, A.; Molinos-Albert, L.M.; Díaz, N.; Canyelles, M.; Lara-Escandell, M.; Planchais, C.; et al. High-resolution kinetics and cellular determinants of SARS-CoV-2 antibody response over two years after COVID-19 vaccination. Microbes Infect. 2025, 27, 105423. [Google Scholar] [CrossRef]
- Galipeau, Y.; Greig, M.; Liu, G.; Driedger, M.; Langlois, M.A. Humoral Responses and Serological Assays in SARS-CoV-2 Infections. Front. Immunol. 2020, 11, 610688. [Google Scholar] [CrossRef]
- Chen, M.; Qin, R.; Jiang, M.; Yang, Z.; Wen, W.; Li, J. Clinical applications of detecting IgG, IgM or IgA antibody for the diagnosis of COVID-19: A meta-analysis and systematic review. Int. J. Infect. Dis. 2021, 104, 415–422. [Google Scholar] [CrossRef]
- Abbasi, J. Combining Rapid PCR and Antibody Tests Improved COVID-19 Diagnosis. JAMA 2020, 324, 1386. [Google Scholar] [CrossRef]
- Filomena, A.; Pessler, F.; Akmatov, M.K.; Krause, G.; Duffy, D.; Gärtner, B.; Gerhard, M.; Albert, M.L.; Joos, T.O.; Schneiderhan-Marra, N. Development of a bead-based multiplex assay for the analysis of the serological response against the six pathogens HAV, HBV, HCV, CMV, T. gondii, and H. pylori. High Throughput 2017, 6, 14. [Google Scholar] [CrossRef]
- Yoshida, S.; Ono, C.; Hayashi, H.; Fukumoto, S.; Shiraishi, S.; Tomono, K.; Arase, H.; Matsuura, Y.; Nakagami, H. SARS-CoV-2-induced humoral immunity through B cell epitope analysis in COVID-19 infected individuals. Sci. Rep. 2021, 11, 5934. [Google Scholar] [CrossRef]
- Guo, J.Y.; Liu, I.J.; Lin, H.T.; Wang, M.-J.; Chang, Y.-L.; Lin, S.-C.; Liao, M.-Y.; Hsu, W.-C.; Lin, Y.-L.; Liao, J.C.; et al. Identification of COVID-19 B-cell epitopes with phage-displayed peptide library. J. Biomed. Sci. 2021, 28, 43. [Google Scholar] [CrossRef]
- Chen, Z.; Ruan, P.; Wang, L.; Nie, X.; Ma, X.; Tan, Y. T and B cell Epitope analysis of SARS-CoV-2 S protein based on immunoinformatics and experimental research. J. Cell Mol. Med. 2021, 25, 1274–1289. [Google Scholar] [CrossRef]
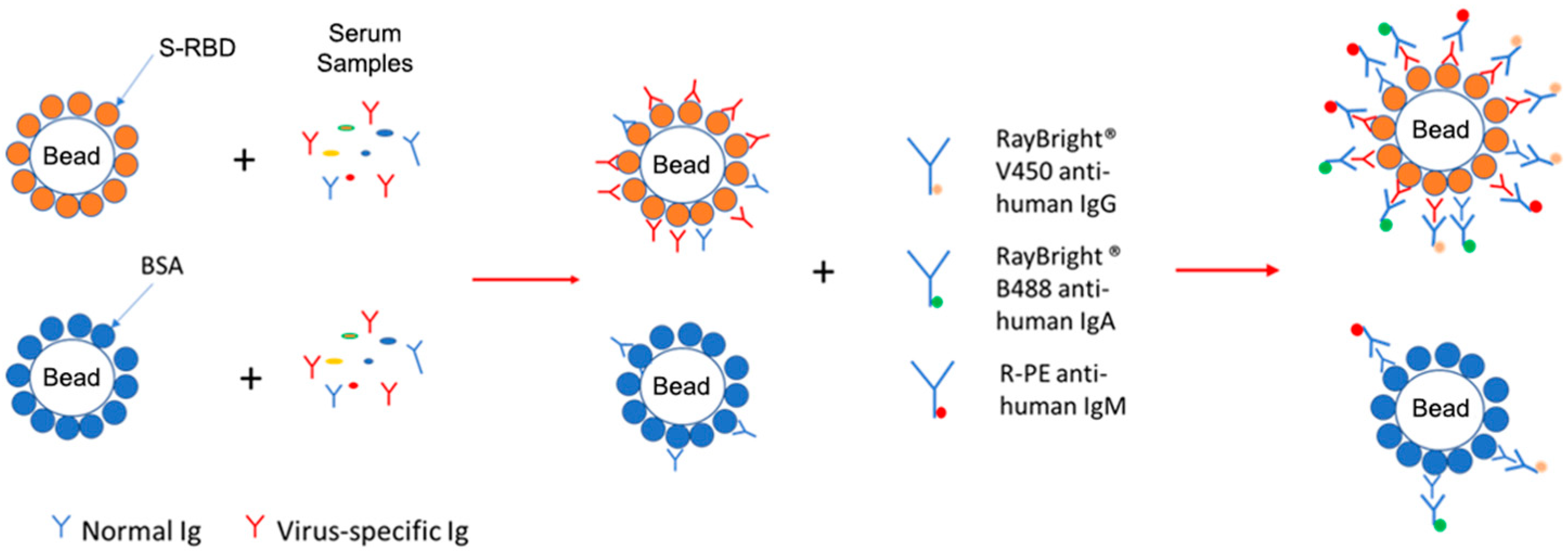
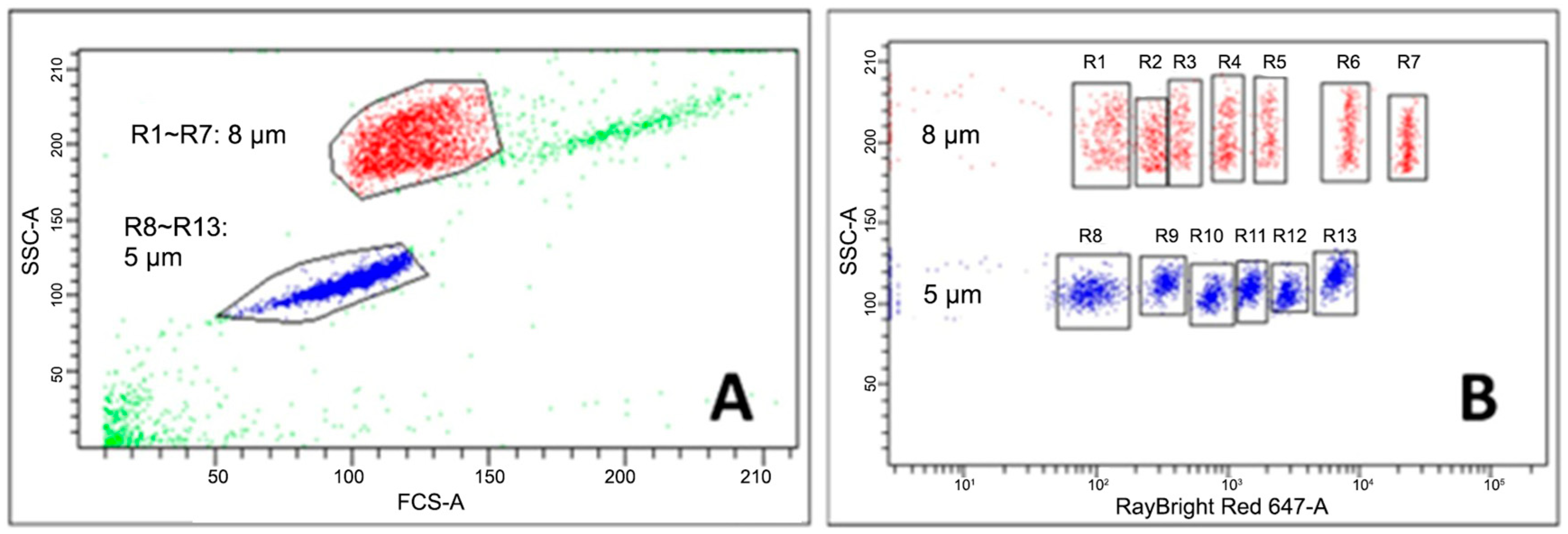
| Sample Dilution | Beads | Sample#1 | Sample#2 | Sample#3 | Sample#4 | Sample#5 | Sample#6 | Sample#7 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IgM | IgG | IgA | IgM | IgG | IgA | IgM | IgG | IgA | IgM | IgG | IgA | IgM | IgG | IgA | IgM | IgG | IgA | IgM | IgG | IgA | ||
| 1/2000 | Control Beads | 575 | 1158 | 311 | 575 | 1035 | 260 | 554 | 1538 | 300 | 638 | 1077 | 314 | 787 | 1220 | 375 | 749 | 1216 | 395 | 745 | 1378 | 419 |
| Antigen Beads | 20,168 | 59,470 | 12,889 | 20,353 | 55,955 | 12,540 | 15,711 | 51,380 | 10,132 | 22,991 | 65,758 | 14,782 | 25,891 | 70,530 | 16,445 | 23,992 | 63,205 | 14,827 | 22,301 | 53,946 | 13,166 | |
| Ratio | 35.07 | 51.36 | 41.44 | 35.40 | 54.06 | 48.23 | 28.36 | 33.41 | 33.77 | 36.04 | 61.06 | 47.08 | 32.90 | 57.81 | 43.85 | 32.03 | 51.98 | 37.54 | 29.93 | 39.15 | 31.42 | |
| 1/4000 | Control Beads | 326 | 962 | 216 | 293 | 809 | 187 | 294 | 1216 | 187 | 325 | 900 | 224 | 423 | 995 | 249 | 438 | 989 | 278 | 438 | 1231 | 357 |
| Antigen Beads | 10,194 | 32,142 | 6416 | 10,256 | 30,335 | 6436 | 8014 | 27,267 | 5090 | 11,763 | 35,976 | 7517 | 13,409 | 37,430 | 8337 | 12,464 | 34,474 | 7586 | 11,515 | 29,424 | 6840 | |
| Ratio | 31.27 | 33.41 | 29.70 | 35.00 | 37.50 | 34.42 | 27.26 | 22.42 | 27.22 | 36.19 | 39.97 | 33.56 | 31.70 | 37.62 | 33.48 | 28.46 | 34.86 | 27.29 | 26.29 | 23.90 | 19.16 | |
| 1/8000 | Control Beads | 186 | 813 | 163 | 158 | 714 | 133 | 157 | 1047 | 147 | 179 | 775 | 170 | 231 | 865 | 194 | 257 | 849 | 233 | 325 | 1124 | 327 |
| Antigen Beads | 5410 | 17,585 | 3436 | 5560 | 17,214 | 3395 | 4075 | 14,737 | 2588 | 6436 | 20,540 | 4026 | 7203 | 21,897 | 4425 | 6696 | 19,034 | 4075 | 6243 | 17,005 | 3765 | |
| Ratio | 29.09 | 21.63 | 21.08 | 35.19 | 24.11 | 25.53 | 25.96 | 14.08 | 17.61 | 35.96 | 26.50 | 23.68 | 31.18 | 25.31 | 22.81 | 26.05 | 22.42 | 17.49 | 19.21 | 15.13 | 11.51 | |
| 1/16000 | Control Beads | 91.6 | 823 | 143 | 95.9 | 688 | 119 | 89.7 | 1051 | 123 | 103 | 765 | 142 | 132 | 897 | 150 | 174 | 854 | 199 | 254 | 1103 | 304 |
| Antigen Beads | 2951 | 9798 | 1909 | 2871 | 9332 | 1801 | 2189 | 8186 | 1416 | 3468 | 11,001 | 2216 | 3870 | 11,944 | 2450 | 3619 | 10,477 | 2306 | 3426 | 9361 | 2150 | |
| Ratio | 32.22 | 11.91 | 13.35 | 29.94 | 13.56 | 15.13 | 24.40 | 7.79 | 11.51 | 33.67 | 14.38 | 15.61 | 29.32 | 13.32 | 16.33 | 20.80 | 12.27 | 11.59 | 13.49 | 8.49 | 7.07 | |
| 1/32000 | Control Beads | 58.4 | 791 | 140 | 51.3 | 655 | 109 | 47.3 | 977 | 116 | 61.5 | 720 | 122 | 89.9 | 831 | 142 | 135 | 811 | 192 | 218 | 1073 | 300 |
| Antigen Beads | 1524 | 5090 | 995 | 1528 | 4848 | 953 | 1180 | 4534 | 782 | 1807 | 5874 | 1201 | 2054 | 6281 | 1345 | 1944 | 5628 | 1246 | 1840 | 5137 | 1261 | |
| Ratio | 26.10 | 6.43 | 7.11 | 29.79 | 7.40 | 8.74 | 24.95 | 4.64 | 6.74 | 29.38 | 8.16 | 9.84 | 22.85 | 7.56 | 9.47 | 14.40 | 6.94 | 6.49 | 8.44 | 4.79 | 4.20 | |
| 1/64000 | Control Beads | 43.2 | 752 | 126 | 38.4 | 638 | 105 | 38.6 | 936 | 104 | 45.6 | 692 | 121 | 65.9 | 799 | 139 | 111 | 789 | 177 | 200 | 1057 | 293 |
| Antigen Beads | 784 | 3098 | 575 | 784 | 2880 | 566 | 594 | 2819 | 452 | 930 | 3468 | 645 | 1035 | 3800 | 720 | 1051 | 3374 | 727 | 1054 | 3204 | 770 | |
| Ratio | 18.15 | 4.12 | 4.56 | 20.42 | 4.51 | 5.39 | 15.39 | 3.01 | 4.35 | 20.39 | 5.01 | 5.33 | 15.71 | 4.76 | 5.18 | 9.47 | 4.28 | 4.11 | 5.27 | 3.03 | 2.63 | |
| 1/128000 | Control Beads | 36.7 | 736 | 124 | 30.4 | 628 | 93.6 | 36.6 | 925 | 102 | 36.8 | 688 | 116 | 62.6 | 796 | 138 | 97.3 | 784 | 181 | 197 | 1064 | 297 |
| Antigen Beads | 382 | 1835 | 345 | 398 | 1685 | 314 | 299 | 1852 | 276 | 448 | 2029 | 370 | 512 | 2196 | 428 | 494 | 1986 | 430 | 561 | 2066 | 507 | |
| Ratio | 10.41 | 2.49 | 2.78 | 13.09 | 2.68 | 3.35 | 8.17 | 2.00 | 2.71 | 12.17 | 2.95 | 3.19 | 8.18 | 2.76 | 3.10 | 5.08 | 2.53 | 2.38 | 2.85 | 1.94 | 1.71 | |
| 1/256000 | Control Beads | 29.9 | 756 | 116 | 20.7 | 634 | 103 | 31.6 | 953 | 109 | 31.9 | 705 | 117 | 53.4 | 816 | 137 | 93.3 | 811 | 185 | 191 | 1064 | 294 |
| Antigen Beads | 220 | 1296 | 241 | 223 | 1165 | 207 | 174 | 1374 | 199 | 237 | 1292 | 245 | 304 | 1464 | 291 | 302 | 1374 | 325 | 383 | 1538 | 406 | |
| Ratio | 7.36 | 1.71 | 2.08 | 10.77 | 1.84 | 2.01 | 5.51 | 1.44 | 1.83 | 7.43 | 1.83 | 2.09 | 5.69 | 1.79 | 2.12 | 3.24 | 1.69 | 1.76 | 2.01 | 1.45 | 1.38 | |
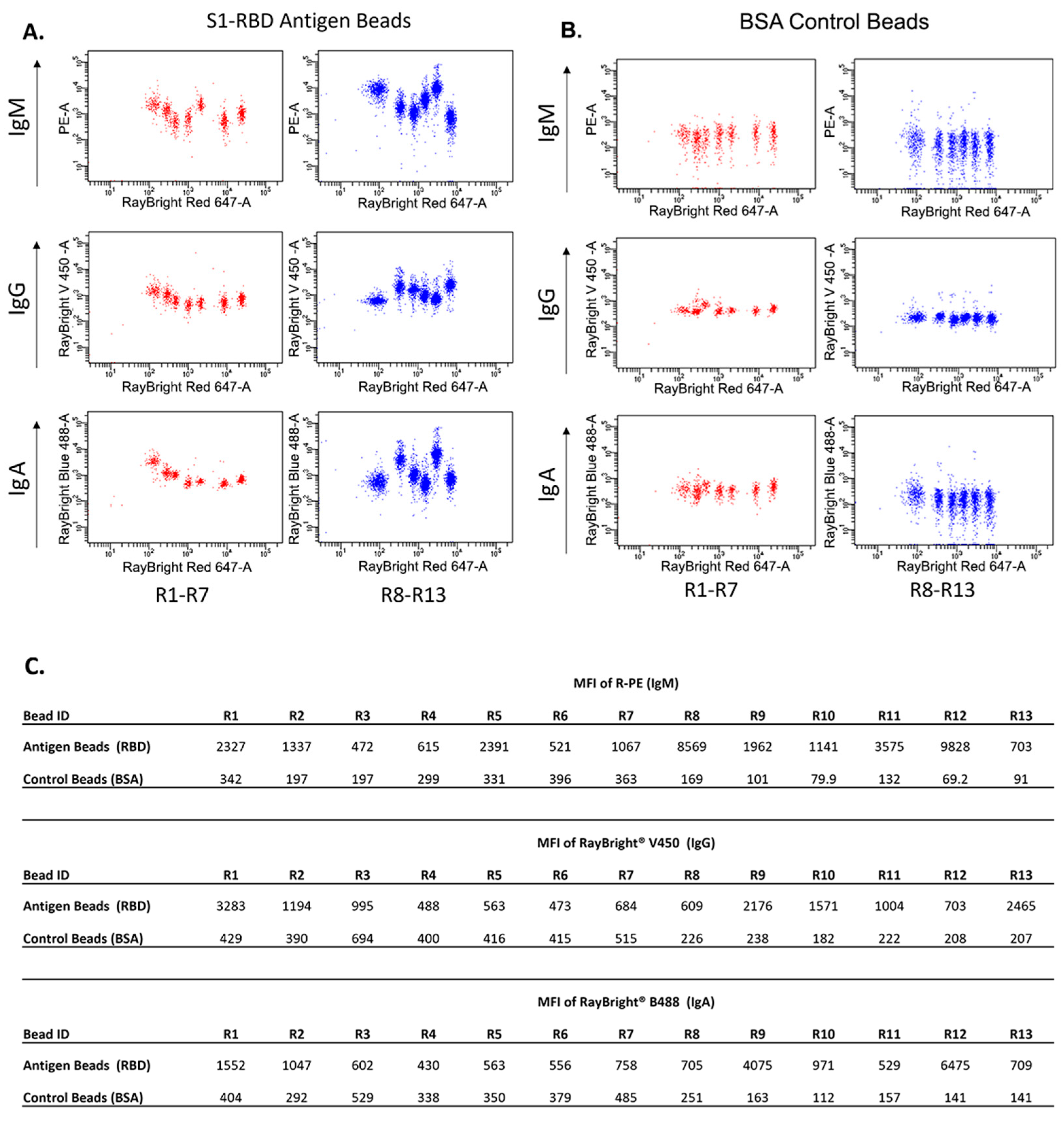
| Bead ID | MFI of R-PE (IgM) | ||||||||||||
| R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 | R9 | R10 | R11 | R12 | R13 | |
| Antigen Beads (RBD) | 2327 | 1337 | 472 | 615 | 2391 | 521 | 1067 | 8569 | 1962 | 1141 | 3575 | 9828 | 703 |
| Control Beads (BSA) | 342 | 197 | 197 | 299 | 331 | 396 | 363 | 169 | 101 | 79.9 | 132 | 69.2 | 91 |
| Ratio | 6.8 | 6.8 | 2.4 | 2.1 | 7.2 | 1.3 | 2.9 | 50.7 | 19.4 | 14.3 | 27.1 | 142.0 | 7.7 |
| Antigen Beads—Control Beads | 1985 | 1140 | 275 | 316 | 2060 | 125 | 704 | 8400 | 1861 | 1061 | 3443 | 9759 | 612 |
| Expected Positivity | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Testing Positivity | + | + | + | + | + | - | + | + | + | + | + | + | + |
| Bead ID | MFI of RayBright® V450 (IgG) | ||||||||||||
| R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 | R9 | R10 | R11 | R12 | R13 | |
| Antigen Beads (RBD) | 3283 | 1194 | 995 | 488 | 563 | 473 | 684 | 609 | 2176 | 1571 | 1004 | 703 | 2465 |
| Control Beads (BSA) | 429 | 390 | 694 | 400 | 416 | 415 | 515 | 226 | 238 | 182 | 222 | 208 | 207 |
| Ratio | 7.7 | 3.1 | 1.4 | 1.2 | 1.4 | 1.1 | 1.3 | 2.7 | 9.1 | 8.6 | 4.5 | 3.4 | 11.9 |
| Antigen Beads—Control Beads | 2854 | 804 | 301 | 88 | 147 | 58 | 169 | 383 | 1938 | 1389 | 782 | 495 | 2258 |
| Expected Positivity | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Testing Positivity | + | + | - | - | - | - | - | + | + | + | + | + | + |
| Bead ID | MFI of RayBright® B488 (IgA) | ||||||||||||
| R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 | R9 | R10 | R11 | R12 | R13 | |
| Antigen Beads (RBD) | 1552 | 1047 | 602 | 430 | 563 | 556 | 758 | 705 | 4075 | 971 | 529 | 6475 | 709 |
| Control Beads (BSA) | 404 | 292 | 529 | 338 | 350 | 379 | 485 | 251 | 163 | 112 | 157 | 141 | 141 |
| Ratio | 3.8 | 3.6 | 1.1 | 1.3 | 1.6 | 1.5 | 1.6 | 2.8 | 25.0 | 8.7 | 3.4 | 45.9 | 5.0 |
| Antigen Beads—Control Beads | 1148 | 755 | 73 | 92 | 213 | 177 | 273 | 454 | 3912 | 859 | 372 | 6334 | 568 |
| Expected Positivity | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Testing Positivity | + | + | - | - | + | - | + | + | + | + | + | + | + |
| Bead ID | IgM MFI | Bead ID | IgG MFI | Bead ID | IgA MFI | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | Intra-Plate CV | 1 | 2 | 3 | 4 | Intra-Plate CV | 1 | 2 | 3 | 4 | Intra-Plate CV | |||||
| R1 (Antigen) | 557 | 528 | 525 | 440 | 8.52% | R1 (Antigen) | 484 | 475 | 480 | 455 | 2.35% | R1 (Antigen) | 568 | 523 | 552 | 510 | 4.26% | ||
| R2 (Antigen) | 482 | 445 | 471 | 403 | 6.75% | R2 (Antigen) | 452 | 444 | 442 | 430 | 1.78% | R2 (Antigen) | 489 | 464 | 482 | 435 | 4.46% | ||
| R3 (Antigen) | 429 | 429 | 434 | 394 | 3.80% | R3 (Antigen) | 521 | 539 | 530 | 528 | 1.21% | R3 (Antigen) | 445 | 403 | 441 | 409 | 4.40% | ||
| R4 (Antigen) | 544 | 539 | 510 | 533 | 2.45% | R4 (Antigen) | 487 | 481 | 493 | 465 | 2.17% | R4 (Antigen) | 584 | 570 | 552 | 526 | 3.89% | ||
| R5 (Antigen) | 701 | 613 | 671 | 582 | 7.29% | R5 (Antigen) | 529 | 528 | 522 | 515 | 1.07% | R5 (Antigen) | 619 | 588 | 636 | 579 | 3.80% | ||
| R6 (Antigen) | 680 | 642 | 660 | 615 | 3.68% | R6 (Antigen) | 454 | 446 | 454 | 433 | 1.92% | R6 (Antigen) | 643 | 603 | 637 | 580 | 4.17% | ||
| R7 (Antigen) | 744 | 726 | 728 | 672 | 3.79% | R7 (Antigen) | 501 | 509 | 502 | 486 | 1.68% | R7 (Antigen) | 709 | 722 | 719 | 657 | 3.75% | ||
| R8 (Antigen) | 219 | 221 | 226 | 184 | 7.84% | R8 (Antigen) | 239 | 241 | 235 | 231 | 1.62% | R8 (Antigen) | 214 | 208 | 208 | 190 | 4.39% | ||
| R9 (Antigen) | 234 | 224 | 251 | 214 | 5.92% | R9 (Antigen) | 264 | 265 | 259 | 247 | 2.77% | R9 (Antigen) | 228 | 218 | 232 | 215 | 3.13% | ||
| R10 (Antigen) | 210 | 215 | 220 | 182 | 7.12% | R10 (Antigen) | 205 | 206 | 202 | 195 | 2.13% | R10 (Antigen) | 201 | 185 | 194 | 172 | 5.77% | ||
| R11 (Antigen) | 243 | 233 | 233 | 220 | 3.52% | R11 (Antigen) | 231 | 234 | 236 | 223 | 2.14% | R11 (Antigen) | 233 | 229 | 229 | 215 | 3.02% | ||
| R12 (Antigen) | 193 | 176 | 195 | 165 | 6.80% | R12 (Antigen) | 198 | 197 | 199 | 189 | 2.02% | R12 (Antigen) | 194 | 187 | 191 | 165 | 6.18% | ||
| R13 (Antigen) | 210 | 201 | 204 | 194 | 2.85% | R13 (Antigen) | 226 | 229 | 222 | 218 | 1.85% | R13 (Antigen) | 234 | 227 | 226 | 207 | 4.48% | ||
| R1 (Control) | 228 | 217 | 230 | 218 | 2.60% | R1 (Control) | 306 | 309 | 320 | 307 | 1.80% | R1 (Control) | 266 | 258 | 263 | 250 | 2.34% | ||
| R2 (Control) | 175 | 182 | 168 | 198 | 6.15% | R2 (Control) | 279 | 268 | 288 | 272 | 2.74% | R2 (Control) | 219 | 208 | 205 | 202 | 3.08% | ||
| R3 (Control) | 200 | 203 | 206 | 187 | 3.64% | R3 (Control) | 418 | 409 | 418 | 415 | 0.89% | R3 (Control) | 234 | 219 | 222 | 226 | 2.50% | ||
| R4 (Control) | 227 | 209 | 210 | 192 | 5.91% | R4 (Control) | 288 | 282 | 310 | 287 | 3.69% | R4 (Control) | 248 | 248 | 250 | 250 | 0.40% | ||
| R5 (Control) | 259 | 267 | 275 | 293 | 4.61% | R5 (Control) | 346 | 318 | 335 | 311 | 4.21% | R5 (Control) | 280 | 264 | 267 | 266 | 2.34% | ||
| R6 (Control) | 289 | 300 | 293 | 286 | 1.80% | R6 (Control) | 285 | 283 | 281 | 281 | 0.59% | R6 (Control) | 317 | 302 | 316 | 306 | 2.07% | ||
| R7 (Control) | 391 | 407 | 409 | 391 | 2.14% | R7 (Control) | 347 | 328 | 355 | 345 | 2.86% | R7 (Control) | 419 | 418 | 403 | 406 | 1.72% | ||
| R8 (Control) | 107 | 90 | 103 | 102 | 6.31% | R8 (Control) | 176 | 169 | 176 | 171 | 1.78% | R8 (Control) | 107 | 102 | 108 | 106 | 2.15% | ||
| R9 (Control) | 93 | 93 | 102 | 94 | 3.95% | R9 (Control) | 180 | 169 | 181 | 182 | 2.95% | R9 (Control) | 98 | 104 | 105 | 101 | 2.68% | ||
| R10 (Control) | 88 | 86 | 79 | 71 | 8.24% | R10 (Control) | 140 | 136 | 141 | 142 | 1.63% | R10 (Control) | 81 | 84 | 83 | 85 | 1.78% | ||
| R11 (Control) | 96 | 109 | 103 | 111 | 5.58% | R11 (Control) | 170 | 161 | 168 | 169 | 2.12% | R11 (Control) | 119 | 116 | 115 | 110 | 2.82% | ||
| R12 (Control) | 62 | 61 | 73 | 64 | 7.30% | R12 (Control) | 135 | 127 | 136 | 134 | 2.66% | R12 (Control) | 91 | 85 | 86 | 85 | 2.87% | ||
| R13 (Control) | 77 | 75 | 86 | 77 | 5.42% | R13 (Control) | 169 | 159 | 164 | 163 | 2.18% | R13 (Control) | 119 | 114 | 117 | 117 | 1.53% | ||
| Average Intra-plate MFI CV: | 5.15% | Average Intra-plate MFI CV: | 2.11% | Average Intra-plate MFI CV: | 3.23% | ||||||||||||||
| Average Intra-plate MFI Ratio CV: | 6.71% | Average Intra-plate MFI Ratio CV: | 3.16% | Average Intra-plate MFI Ratio CV: | 3.82% | ||||||||||||||
| Bead ID | IgM MFI | Bead ID | IgG MFI | Bead ID | IgA MFI | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | Inter-Plate CV | 1 | 2 | 3 | 4 | Inter-Plate CV | 1 | 2 | 3 | 4 | Inter-Plate CV | |||||
| R1 (Antigen) | 2852 | 2886 | 2894 | 2911 | 0.74% | R1 (Antigen) | 1333 | 1632 | 1361 | 1395 | 8.29% | R1 (Antigen) | 1014 | 1116 | 1051 | 1036 | 3.60% | ||
| R2 (Antigen) | 2619 | 2673 | 2689 | 2705 | 1.21% | R2 (Antigen) | 1177 | 1338 | 1207 | 1238 | 4.88% | R2 (Antigen) | 900 | 927 | 892 | 930 | 1.81% | ||
| R3 (Antigen) | 2116 | 2201 | 2145 | 2226 | 2.01% | R3 (Antigen) | 1141 | 1375 | 1158 | 1203 | 7.61% | R3 (Antigen) | 761 | 774 | 783 | 777 | 1.04% | ||
| R4 (Antigen) | 3209 | 3376 | 3435 | 3306 | 2.53% | R4 (Antigen) | 1487 | 1723 | 1537 | 1553 | 5.64% | R4 (Antigen) | 1121 | 1187 | 1205 | 1166 | 2.68% | ||
| R5 (Antigen) | 3376 | 3567 | 3554 | 3601 | 2.48% | R5 (Antigen) | 1504 | 1792 | 1615 | 1685 | 6.35% | R5 (Antigen) | 1201 | 1225 | 1334 | 1243 | 4.02% | ||
| R6 (Antigen) | 3212 | 3353 | 3320 | 3313 | 1.60% | R6 (Antigen) | 1357 | 1556 | 1436 | 1459 | 4.89% | R6 (Antigen) | 1161 | 1205 | 1168 | 1245 | 2.80% | ||
| R7 (Antigen) | 3010 | 3054 | 3147 | 3171 | 2.13% | R7 (Antigen) | 1308 | 1430 | 1342 | 1377 | 3.31% | R7 (Antigen) | 1150 | 1202 | 1190 | 1198 | 1.74% | ||
| R8 (Antigen) | 1019 | 1011 | 1066 | 1095 | 3.29% | R8 (Antigen) | 526 | 561 | 530 | 562 | 3.09% | R8 (Antigen) | 368 | 358 | 364 | 391 | 3.38% | ||
| R9 (Antigen) | 1149 | 1140 | 1166 | 1192 | 1.70% | R9 (Antigen) | 558 | 620 | 568 | 580 | 4.05% | R9 (Antigen) | 388 | 403 | 406 | 411 | 2.13% | ||
| R10 (Antigen) | 1059 | 1055 | 1093 | 1072 | 1.39% | R10 (Antigen) | 456 | 513 | 485 | 496 | 4.25% | R10 (Antigen) | 351 | 362 | 361 | 365 | 1.46% | ||
| R11 (Antigen) | 1175 | 1213 | 1180 | 1213 | 1.49% | R11 (Antigen) | 524 | 572 | 528 | 551 | 3.55% | R11 (Antigen) | 388 | 421 | 396 | 422 | 3.69% | ||
| R12 (Antigen) | 936 | 936 | 932 | 995 | 2.76% | R12 (Antigen) | 414 | 471 | 427 | 437 | 4.83% | R12 (Antigen) | 325 | 339 | 336 | 336 | 1.60% | ||
| R13 (Antigen) | 1032 | 1079 | 975 | 1051 | 3.68% | R13 (Antigen) | 450 | 535 | 450 | 478 | 7.26% | R13 (Antigen) | 360 | 372 | 365 | 374 | 1.52% | ||
| R1 (Control) | 201 | 210 | 217 | 183 | 6.28% | R1 (Control) | 384 | 410 | 395 | 391 | 2.41% | R1 (Control) | 266 | 278 | 259 | 265 | 2.58% | ||
| R2 (Control) | 147 | 162 | 167 | 154 | 4.85% | R2 (Control) | 305 | 336 | 326 | 316 | 3.59% | R2 (Control) | 201 | 198 | 189 | 202 | 2.59% | ||
| R3 (Control) | 183 | 166 | 173 | 164 | 4.33% | R3 (Control) | 446 | 477 | 467 | 464 | 2.41% | R3 (Control) | 215 | 226 | 225 | 217 | 2.18% | ||
| R4 (Control) | 177 | 217 | 200 | 176 | 8.88% | R4 (Control) | 350 | 366 | 363 | 351 | 1.98% | R4 (Control) | 245 | 238 | 248 | 241 | 1.57% | ||
| R5 (Control) | 218 | 263 | 240 | 255 | 7.02% | R5 (Control) | 386 | 419 | 403 | 398 | 2.95% | R5 (Control) | 270 | 266 | 254 | 266 | 2.27% | ||
| R6 (Control) | 276 | 287 | 270 | 281 | 2.25% | R6 (Control) | 361 | 384 | 374 | 365 | 2.39% | R6 (Control) | 305 | 297 | 303 | 300 | 1.01% | ||
| R7 (Control) | 375 | 394 | 372 | 377 | 2.26% | R7 (Control) | 447 | 467 | 462 | 458 | 1.61% | R7 (Control) | 399 | 421 | 414 | 437 | 3.27% | ||
| R8 (Control) | 90 | 94 | 99 | 90 | 3.97% | R8 (Control) | 209 | 221 | 211 | 212 | 2.16% | R8 (Control) | 108 | 115 | 109 | 115 | 2.93% | ||
| R9 (Control) | 76 | 92 | 82 | 99 | 10.17% | R9 (Control) | 204 | 222 | 211 | 216 | 3.10% | R9 (Control) | 108 | 107 | 112 | 102 | 3.32% | ||
| R10 (Control) | 70 | 71 | 82 | 78 | 6.60% | R10 (Control) | 158 | 173 | 166 | 164 | 3.24% | R10 (Control) | 82 | 86 | 85 | 86 | 1.93% | ||
| R11 (Control) | 92 | 86 | 91 | 94 | 3.25% | R11 (Control) | 198 | 212 | 204 | 204 | 2.43% | R11 (Control) | 109 | 121 | 122 | 111 | 5.01% | ||
| R12 (Control) | 55 | 64 | 62 | 63 | 5.80% | R12 (Control) | 156 | 166 | 160 | 159 | 2.27% | R12 (Control) | 80 | 93 | 87 | 86 | 5.33% | ||
| R13 (Control) | 82 | 74 | 76 | 85 | 5.60% | R13 (Control) | 192 | 204 | 199 | 200 | 2.18% | R13 (Control) | 131 | 115 | 130 | 121 | 5.32% | ||
| Average Inter-plate MFI CV: | 3.78% | Average Inter-plate MFI CV: | 3.87% | Average Inter-plate MFI CV: | 2.72% | ||||||||||||||
| Average Inter-plate MFI Ratio CV: | 5.49% | Average Inter-plate MFI Ratio CV: | 3.33% | Average Inter-plate MFI Ratio CV: | 3.55% | ||||||||||||||
| IgM, IgG, IgA Multiplex Bead-based Array | Combined IgM, IgG, IgA S1-RBD ELISA | IgM S1-RBD ELISA | IgG S1-RBD ELISA | IgA S1-RBD ELISA | ||
|---|---|---|---|---|---|---|
| Positive Population | Total Samples Tested | 121 | 116 | 116 | 116 | 116 |
| False Negative (#) | 2 | 2 | 8 | 20 | 11 | |
| Control Population | Total Samples Tested | 299 | 259 | 259 | 259 | 259 |
| False Positive (#) | 2 | 182 | 109 | 18 | 126 | |
| Sensitivity (%) | 98.3% | 98.0% | 93.2% | 88.9% | 90.5% | |
| Specificity (%) | 99.3% | 30.0% | 57.9% | 93.0% | 51.4% | |
| Accuracy (%) | 99.0% | 51.0% | 51.4% | 90.0% | 63.0% |
| Disease | Patient # | IgM MFI | IgG MFI | IgA MFI | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Antigen Beads | Control Beads | Ratio | Antigen Beads | Control Beads | Ratio | Antigen Beads | Control Beads | Ratio | ||
| ANA | 1 | 81 | 75 | 1.08 | 784 | 793 | 0.99 | 139 | 137 | 1.01 |
| 2 | 77 | 67 | 1.15 | 716 | 694 | 1.03 | 116 | 110 | 1.05 | |
| 3 | 37 | 44 | 0.84 | 919 | 967 | 0.95 | 112 | 111 | 1.01 | |
| 4 | 74 | 67 | 1.10 | 705 | 729 | 0.97 | 129 | 132 | 0.98 | |
| 5 | 121 | 115 | 1.05 | 825 | 821 | 1.00 | 161 | 148 | 1.09 | |
| HCV | 1 | 134 | 124 | 1.08 | 817 | 835 | 0.98 | 204 | 183 | 1.11 |
| 2 | 218 | 212 | 1.03 | 1116 | 1161 | 0.96 | 356 | 342 | 1.04 | |
| 3 | 20 | 20 | 1.00 | 412 | 414 | 1.00 | 57 | 55 | 1.04 | |
| 4 | 78 | 68 | 1.15 | 449 | 441 | 1.02 | 60 | 57 | 1.05 | |
| 5 | 48 | 63 | 0.76 | 329 | 329 | 1.00 | 48 | 42 | 1.14 | |
| RSV | 1 | 72 | 55 | 1.31 | 435 | 435 | 1.00 | 57 | 56 | 1.02 |
| 2 | 88 | 82 | 1.07 | 334 | 345 | 0.97 | 56 | 59 | 0.95 | |
| 3 | 66 | 54 | 1.22 | 434 | 443 | 0.98 | 86 | 85 | 1.01 | |
| 4 | 95 | 77 | 1.23 | 189 | 180 | 1.05 | 74 | 69 | 1.07 | |
| 5 | 105 | 103 | 1.02 | 185 | 160 | 1.16 | 98 | 84 | 1.17 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, B.; Zhang, Z.; Zhao, Y.; Lu, J.; Fang, J.; Petritis, B.; Whittaker, K.; Huang, R.; Huang, R.-P. A Novel Flow Cytometry Array for High Throughput Detection of SARS-CoV-2 Antibodies. Vaccines 2025, 13, 1063. https://doi.org/10.3390/vaccines13101063
Zhang B, Zhang Z, Zhao Y, Lu J, Fang J, Petritis B, Whittaker K, Huang R, Huang R-P. A Novel Flow Cytometry Array for High Throughput Detection of SARS-CoV-2 Antibodies. Vaccines. 2025; 13(10):1063. https://doi.org/10.3390/vaccines13101063
Chicago/Turabian StyleZhang, Benyue, Zhuo Zhang, Yichao Zhao, Jingqiao Lu, Jianmin Fang, Brianne Petritis, Kelly Whittaker, Rani Huang, and Ruo-Pan Huang. 2025. "A Novel Flow Cytometry Array for High Throughput Detection of SARS-CoV-2 Antibodies" Vaccines 13, no. 10: 1063. https://doi.org/10.3390/vaccines13101063
APA StyleZhang, B., Zhang, Z., Zhao, Y., Lu, J., Fang, J., Petritis, B., Whittaker, K., Huang, R., & Huang, R.-P. (2025). A Novel Flow Cytometry Array for High Throughput Detection of SARS-CoV-2 Antibodies. Vaccines, 13(10), 1063. https://doi.org/10.3390/vaccines13101063





