AGS-v PLUS, a Mosquito Salivary Peptide Vaccine, Modulates the Response to Aedes Mosquito Bites in Humans
Abstract
1. Introduction
2. Methods
2.1. Trial Design and Participants
2.2. Ethics
2.3. Procedures
2.4. Bioinformatic Analyses
2.5. Gene Ontology Analysis
2.6. Kegg Pathway Analysis
2.7. Immune Deconvolution
2.8. Gene Nomenclature
3. Results
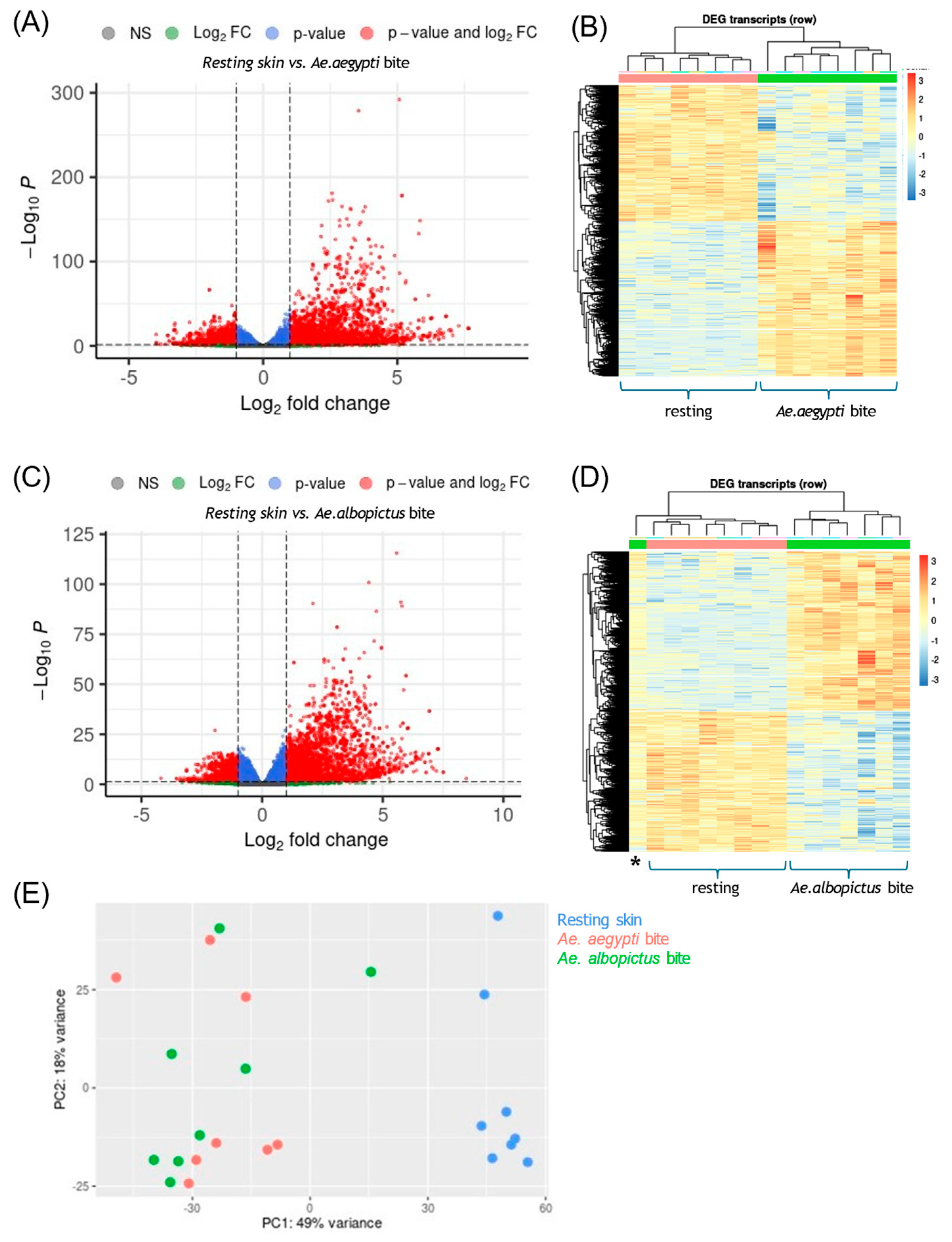
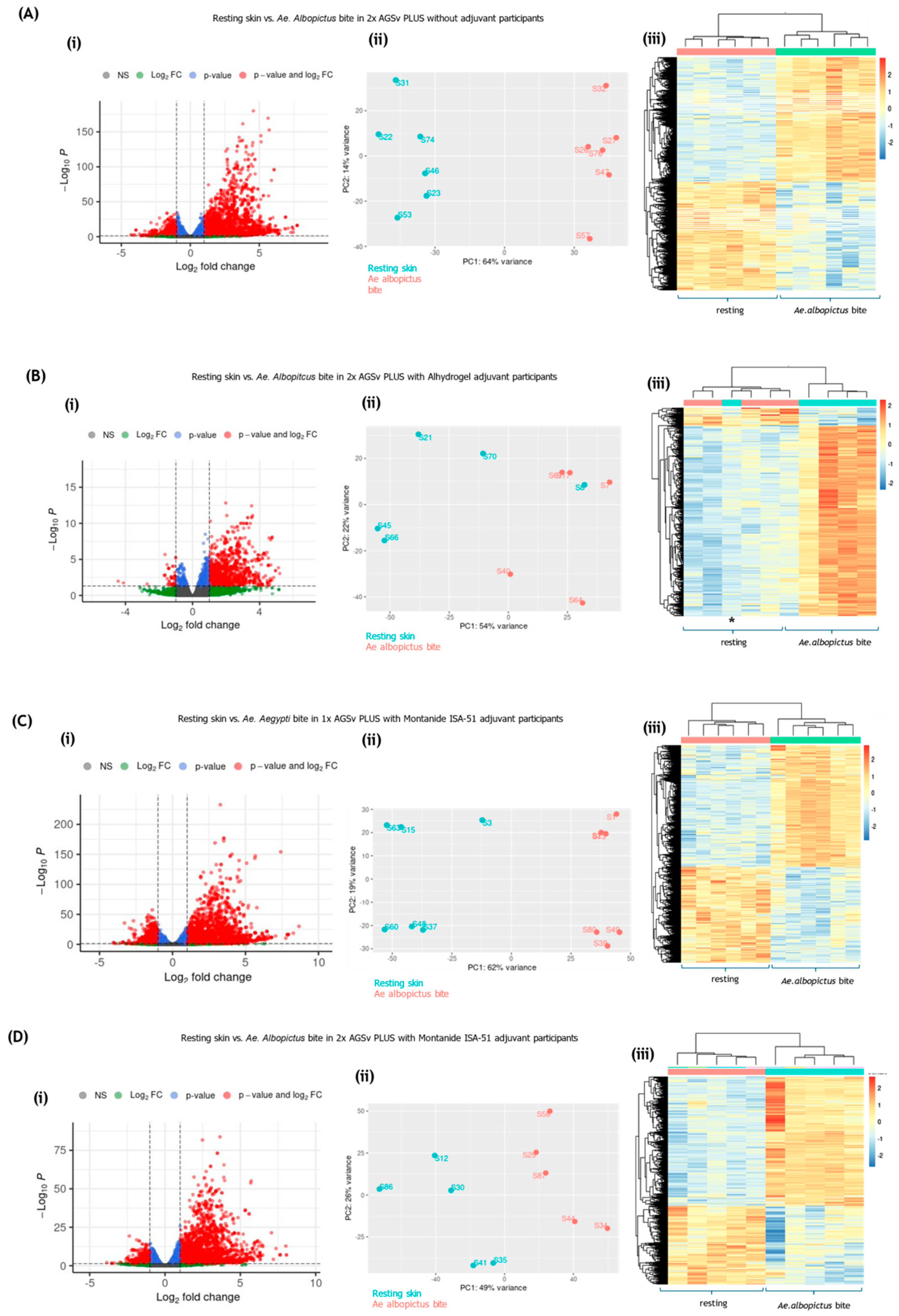
3.1. Mosquito Biting Resulted in Large Numbers of DEGs by 48 h
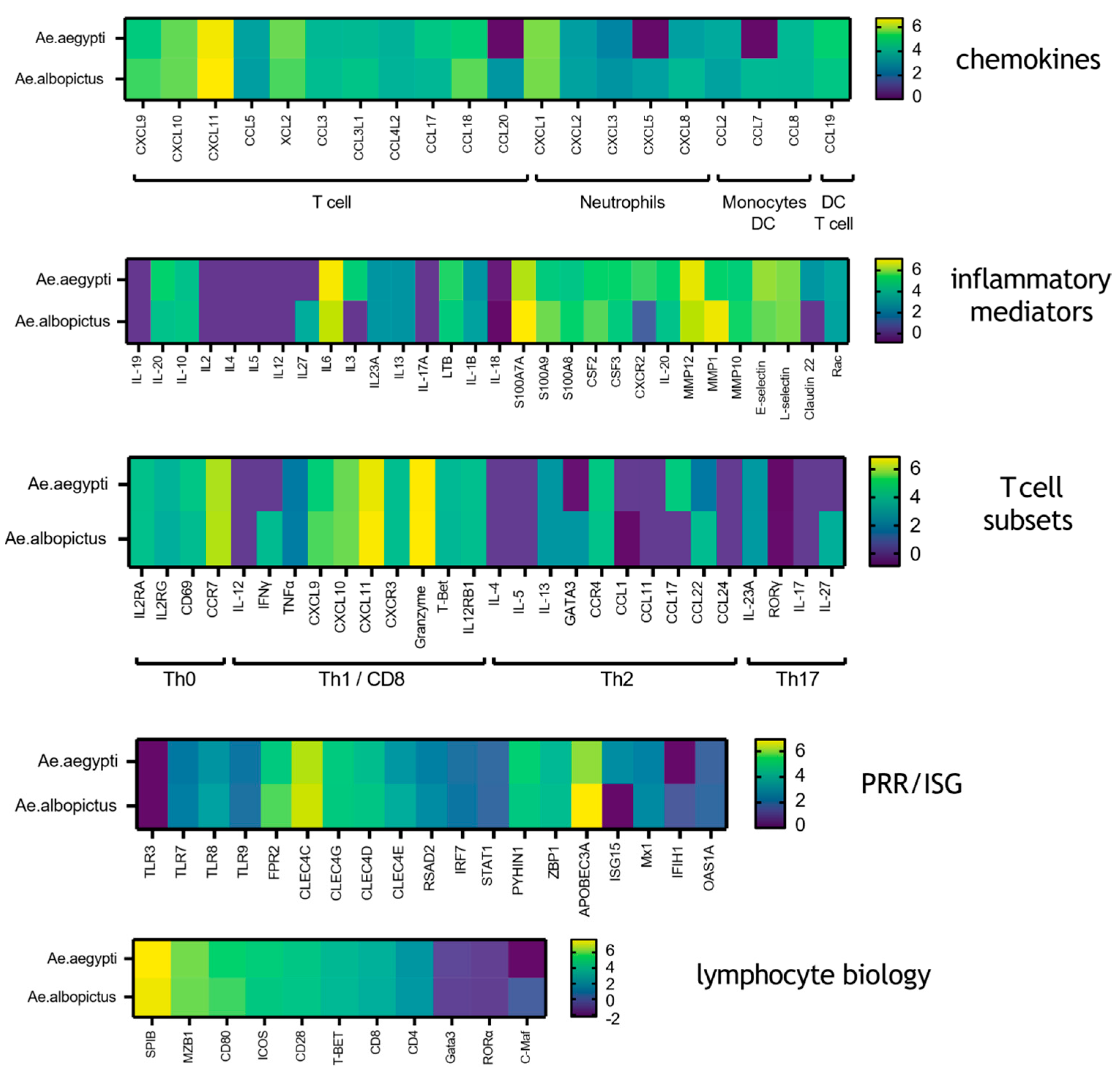
3.2. Mosquito Biting Results in Upregulation of Both Innate Immune and Adaptive Immune Genes
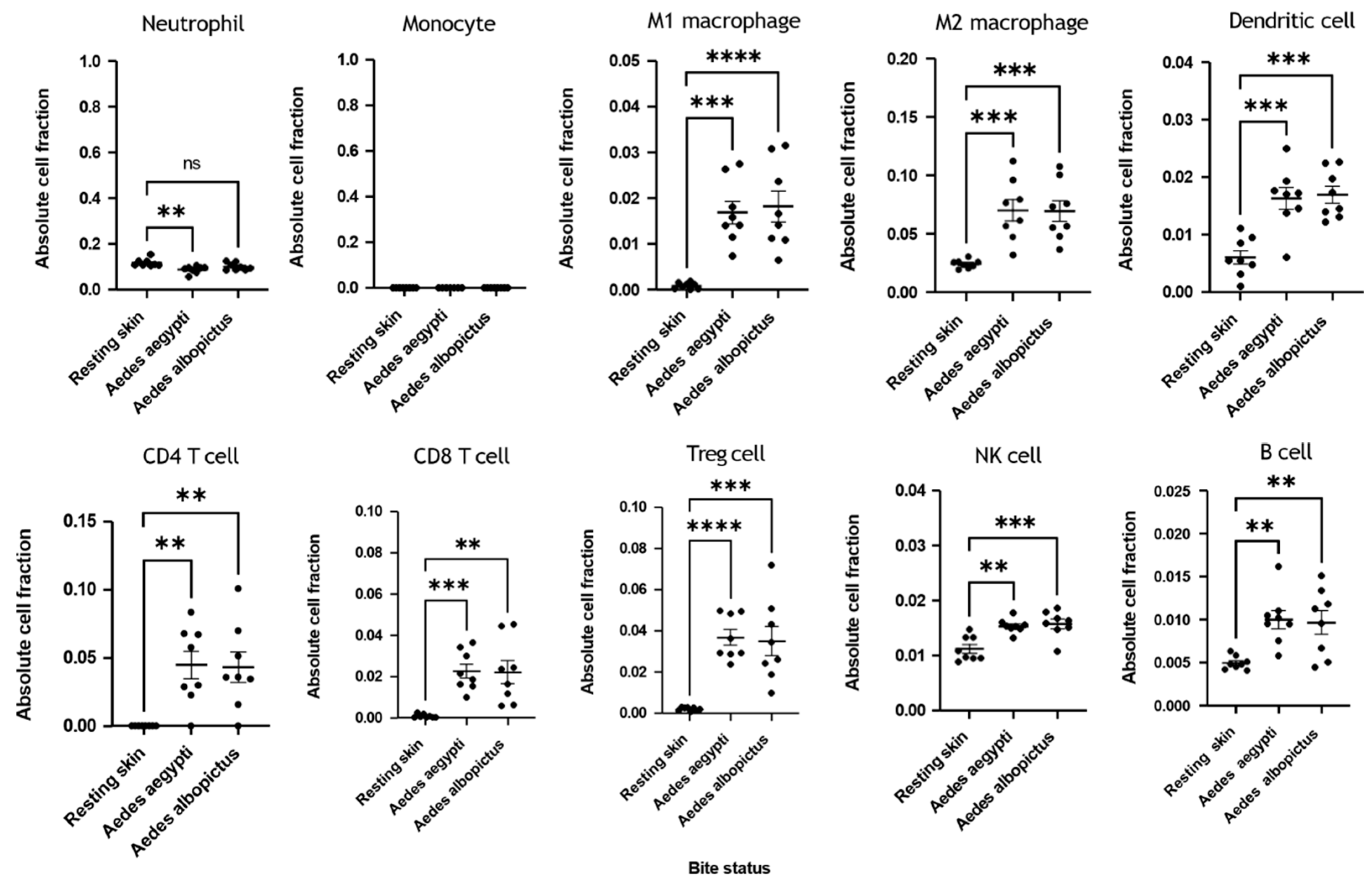
3.3. Immune Deconvolution Identifies Significant Increases in Leukocyte Signatures
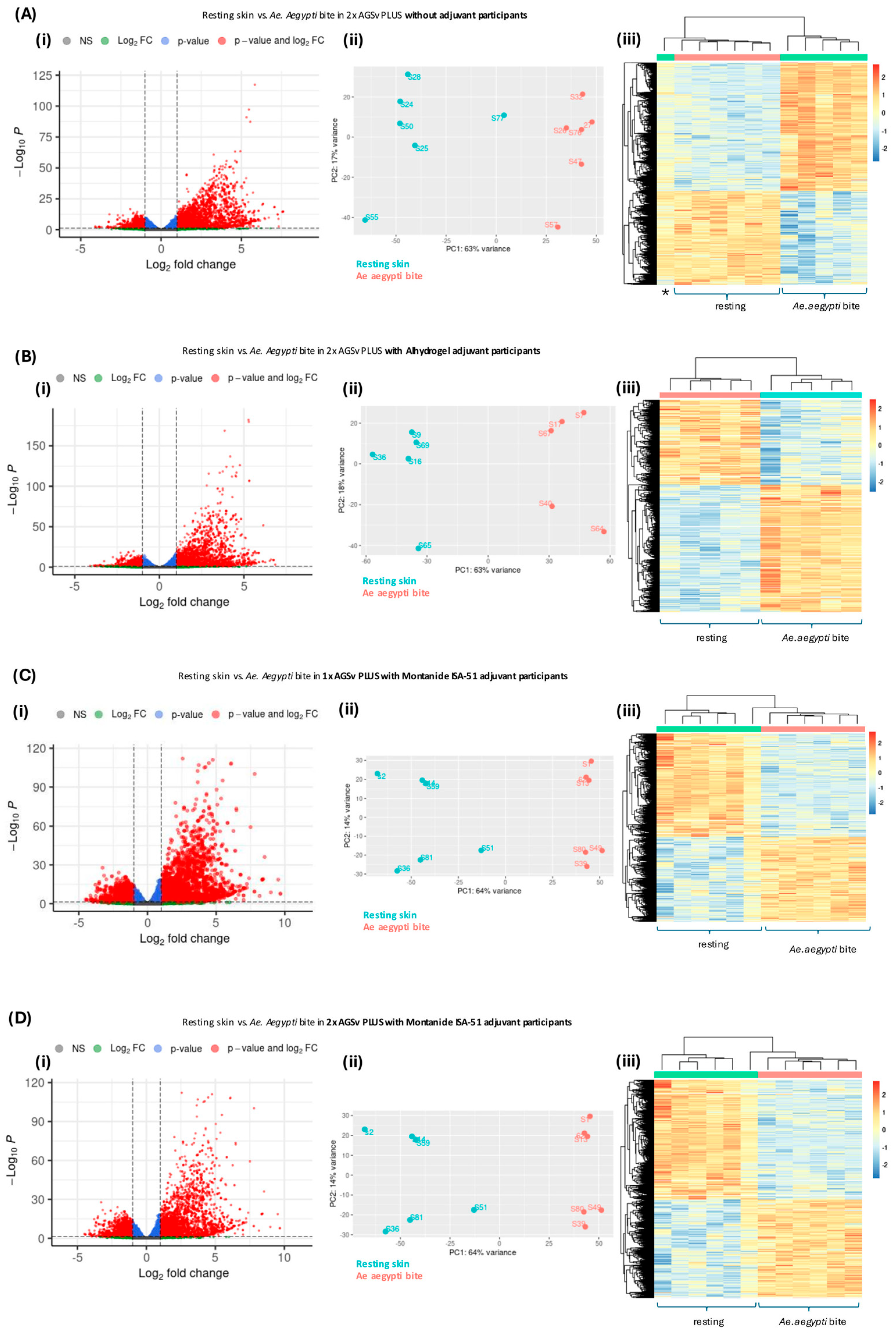
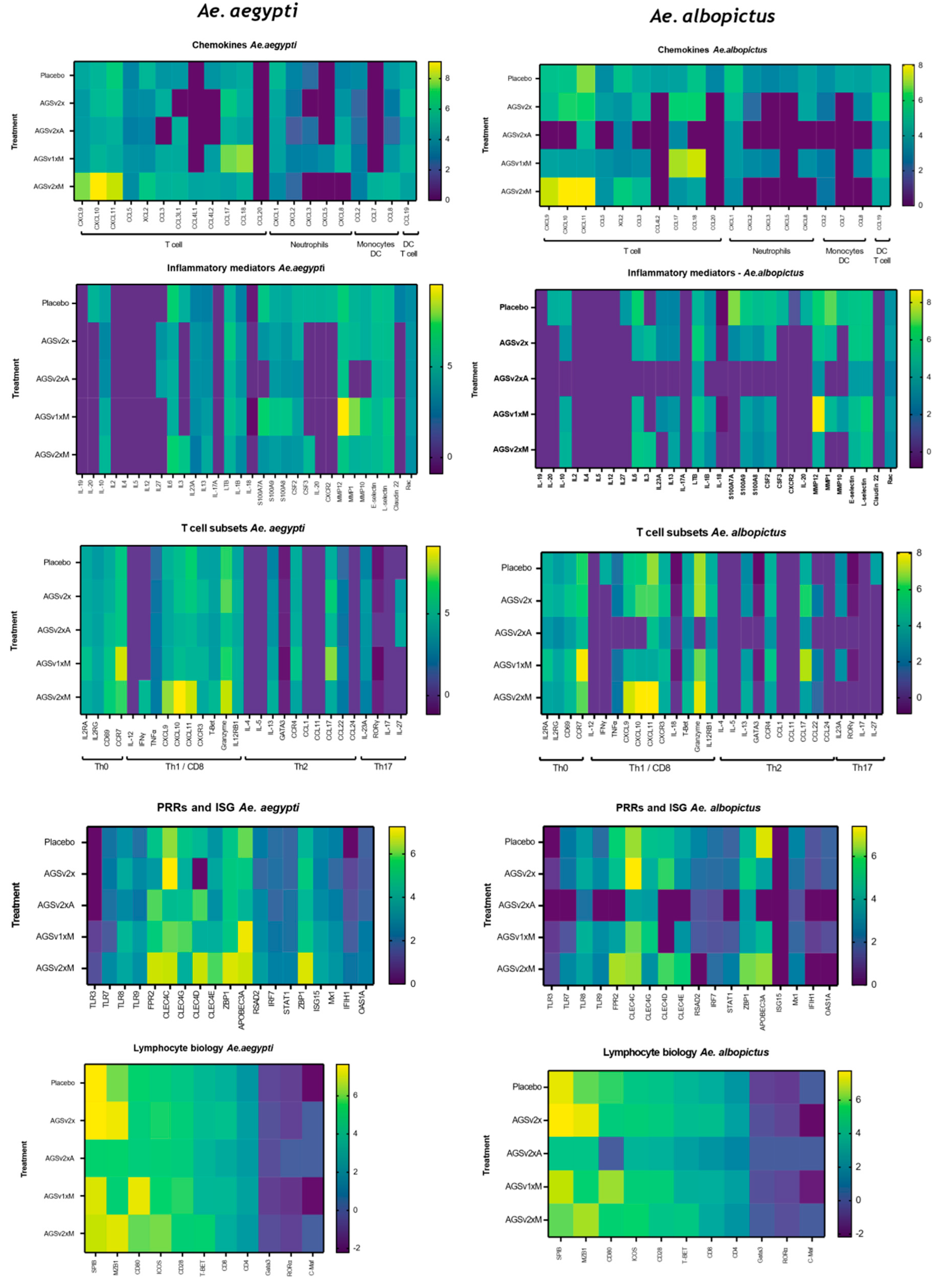
3.4. Vaccination with AGS-v PLUS Modulates the Skin’s Response to Mosquito Biting
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Impact Statement
References
- Tabachnick, W.J. Climate Change and the Arboviruses: Lessons from the Evolution of the Dengue and Yellow Fever Viruses. Annu. Rev. Virol. 2016, 3, 125–145. [Google Scholar] [CrossRef]
- Weaver, S.C.; Charlier, C.; Vasilakis, N.; Lecuit, M. Zika, Chikungunya, and Other Emerging Vector-Borne Viral Diseases. Annu. Rev. Med. 2018, 69, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Bryden, S.R.; Pingen, M.; Lefteri, D.A.; Miltenburg, J.; Delang, L.; Jacobs, S.; Abdelnabi, R.; Neyts, J.; Pondeville, E.; Major, J.; et al. Pan-Viral Protection against Arboviruses by Activating Skin Macrophages at the Inoculation Site. Sci. Transl. Med. 2020, 12, eaax2421. [Google Scholar] [CrossRef] [PubMed]
- van Bree, J.W.M.; Visser, I.; Duyvestyn, J.M.; Aguilar-Bretones, M.; Marshall, E.M.; van Hemert, M.J.; Pijlman, G.P.; van Nierop, G.P.; Kikkert, M.; Rockx, B.H.G.; et al. Novel Approaches for the Rapid Development of Rationally Designed Arbovirus Vaccines. One Health 2023, 16, 100565. [Google Scholar] [CrossRef] [PubMed]
- Manning, J.E.; Morens, D.M.; Kamhawi, S.; Valenzuela, J.G.; Memoli, M. Mosquito Saliva: The Hope for a Universal Arbovirus Vaccine? J. Infect. Dis. 2018, 218, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Marin-Lopez, A.; Wang, Y.; Jiang, J.; Ledizet, M.; Fikrig, E. AgBR1 and NeSt1 Antisera Protect Mice from Aedes aegypti-Borne Zika Infection. Vaccine 2021, 39, 1675–1679. [Google Scholar] [CrossRef] [PubMed]
- Marín-López, A.; Raduwan, H.; Chen, T.Y.; Utrilla-Trigo, S.; Wolfhard, D.P.; Fikrig, E. Mosquito Salivary Proteins and Arbovirus Infection: From Viral Enhancers to Potential Targets for Vaccines. Pathogens 2023, 12, 371. [Google Scholar] [CrossRef] [PubMed]
- Hastings, A.K.; Uraki, R.; Gaitsch, H.; Dhaliwal, K.; Stanley, S.; Sproch, H.; Williamson, E.; MacNeil, T.; Marin-Lopez, A.; Hwang, J.; et al. Aedes aegypti NeSt1 Protein Enhances Zika Virus Pathogenesis by Activating Neutrophils. J. Virol. 2019, 93, e00395-19. [Google Scholar] [CrossRef] [PubMed]
- Manning, J.E.; Oliveira, F.; Coutinho-Abreu, I.V.; Herbert, S.; Meneses, C.; Kamhawi, S.; Baus, H.A.; Han, A.; Czajkowski, L.; Rosas, L.A.; et al. Safety and Immunogenicity of a Mosquito Saliva Peptide-Based Vaccine: A Randomised, Placebo-Controlled, Double-Blind, Phase 1 Trial. Lancet 2020, 395, 1998–2007. [Google Scholar] [CrossRef]
- Reagan, K.L.; Machain-Williams, C.; Wang, T.; Blair, C.D. Immunization of Mice with Recombinant Mosquito Salivary Protein D7 Enhances Mortality from Subsequent West Nile Virus Infection via Mosquito Bite. PLoS Negl. Trop. Dis. 2012, 6, e1935. [Google Scholar] [CrossRef] [PubMed]
- Machain-Williams, C.; Reagan, K.; Wang, T.; Zeidner, N.S.; Blair, C.D. Immunization with Culex Tarsalis Mosquito Salivary Gland Extract Modulates West Nile Virus Infection and Disease in Mice. Viral. Immunol. 2013, 26, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Friedman-Klabanoff, D.A.J.; Birkhold, M.; Short, M.T.; Wilson, T.R.; Meneses, C.R.; Lacsina, J.R.; Oliveira, F.; Kamhawi, S.; Valenzuela, J.G.; Hunsberger, S.; et al. Safety and Immunogenicity of AGS-v PLUS, a Mosquito Saliva Peptide Vaccine against Arboviral Diseases: A Randomized, Double-Blind, Placebo-Controlled Phase 1 Trial. EBioMedicine 2022, 86, 104375. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.M.; Alameh, M.G.; Abouneameh, S.; Raduwan, H.; Ledizet, M.; Weissman, D.; Fikrig, E. A Mosquito AgTRIO MRNA Vaccine Contributes to Immunity against Malaria. NPJ Vaccines 2023, 8, 88. [Google Scholar] [CrossRef] [PubMed]
- Calvo, E.; Mans, B.J.; Andersen, J.F.; Ribeiro, J.M.C. Function and Evolution of a Mosquito Salivary Protein Family. J. Biol. Chem. 2006, 281, 1935–1942. [Google Scholar] [CrossRef]
- Ribeiro, J.M.C.; Arcà, B.; Lombardo, F.; Calvo, E.; Phan, V.M.; Chandra, P.K.; Wikel, S.K. An Annotated Catalogue of Salivary Gland Transcripts in the Adult Female Mosquito, Aedes aegypti. BMC Genom. 2007, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Martin-Martin, I.; Valenzuela Leon, P.C.; Amo, L.; Shrivastava, G.; Iniguez, E.; Aryan, A.; Brooks, S.; Kojin, B.B.; Williams, A.E.; Bolland, S.; et al. Aedes aegypti Sialokinin Facilitates Mosquito Blood Feeding and Modulates Host Immunity and Vascular Biology. Cell Rep. 2022, 39, 110648. [Google Scholar] [CrossRef] [PubMed]
- Pingen, M.; Schmid, M.A.; Harris, E.; McKimmie, C.S. Mosquito Biting Modulates Skin Response to Virus Infection. Trends Parasitol. 2017, 33, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Lefteri, D.A.; Bryden, S.R.; Pingen, M.; Terry, S.; McCafferty, A.; Beswick, E.F.; Georgiev, G.; Van der Laan, M.; Mastrullo, V.; Campagnolo, P.; et al. Mosquito Saliva Enhances Virus Infection through Sialokinin-Dependent Vascular Leakage. Proc. Natl. Acad. Sci. USA 2022, 119, e2114309119. [Google Scholar] [CrossRef]
- Pingen, M.; Bryden, S.R.; Pondeville, E.; Schnettler, E.; Kohl, A.; Merits, A.; Fazakerley, J.K.; Graham, G.J.; McKimmie, C.S. Host Inflammatory Response to Mosquito Bites Enhances the Severity of Arbovirus Infection. Immunity 2016, 44, 1455–1469. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Nie, K.; Zhu, Y.; Liu, Y.; Wu, P.; Liu, Z.; Du, S.; Fan, H.; Chen, C.-H.; Zhang, R.; et al. A Mosquito Salivary Protein Promotes Flavivirus Transmission by Activation of Autophagy. Nat. Commun. 2020, 11, 260. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Guo, X.; Shen, C.; Hao, X.; Sun, P.; Li, P.; Xu, T.; Hu, C.; Rose, O.; Zhou, H.; et al. Salivary Factor LTRIN from Aedes aegypti Facilitates the Transmission of Zika Virus by Interfering with the Lymphotoxin-β Receptor. Nat. Immunol. 2018, 19, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Styer, L.M.; Lim, P.-Y.; Louie, K.L.; Albright, R.G.; Kramer, L.D.; Bernard, K.A. Mosquito Saliva Causes Enhancement of West Nile Virus Infection in Mice. J. Virol. 2011, 85, 1517–1527. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Nie, K.; Liang, Y.; Niu, J.; Yu, X.; Zhang, O.; Liu, L.; Shi, X.; Wang, Y.; Feng, X.; et al. A Mosquito Salivary Protein-Driven Influx of Myeloid Cells Facilitates Flavivirus Transmission. EMBO J. 2024, 43, 1690–1721. [Google Scholar] [CrossRef]
- Schmid, M.A.; Glasner, D.R.; Shah, S.; Michlmayr, D.; Kramer, L.D.; Harris, E. Mosquito Saliva Increases Endothelial Permeability in the Skin, Immune Cell Migration, and Dengue Pathogenesis during Antibody-Dependent Enhancement. PLoS Pathog. 2016, 12, e1005676. [Google Scholar] [CrossRef]
- Guerrero, D.; Vo, H.T.M.; Lon, C.; Bohl, J.A.; Nhik, S.; Chea, S.; Man, S.; Sreng, S.; Pacheco, A.R.; Ly, S.; et al. Evaluation of Cutaneous Immune Response in a Controlled Human in Vivo Model of Mosquito Bites. Nat. Commun. 2022, 13, 7036. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R Package Rsubread Is Easier, Faster, Cheaper and Better for Alignment and Quantification of RNA Sequencing Reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Hu, E.; Cai, Y.; Xie, Z.; Luo, X.; Zhan, L.; Tang, W.; Wang, Q.; Liu, B.; Wang, R.; et al. Using ClusterProfiler to Characterize Multiomics Data. Nat. Protoc. 2024, 19, 3292–3320. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; He, Q.Y. ReactomePA: An R/Bioconductor Package for Reactome Pathway Analysis and Visualization. Mol. Biosyst. 2016, 12, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Sturm, G.; Finotello, F.; Petitprez, F.; Zhang, J.D.; Baumbach, J.; Fridman, W.H.; List, M.; Aneichyk, T. Comprehensive Evaluation of Transcriptome-Based Cell-Type Quantification Methods for Immuno-Oncology. Bioinformatics 2019, 35, i436–i445. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for Analysis of Tumor-Infiltrating Immune Cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Finotello, F.; Mayer, C.; Plattner, C.; Laschober, G.; Rieder, D.; Hackl, H.; Krogsdam, A.; Loncova, Z.; Posch, W.; Wilflingseder, D.; et al. Molecular and Pharmacological Modulators of the Tumor Immune Contexture Revealed by Deconvolution of RNA-Seq Data. Genome Med. 2019, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Conway, M.J.; Colpitts, T.M.; Fikrig, E. Role of the Vector in Arbovirus Transmission. Annu. Rev. Virol. 2014, 1, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Katebi, A.; Gholami, E.; Taheri, T.; Zahedifard, F.; Habibzadeh, S.; Taslimi, Y.; Shokri, F.; Papadopoulou, B.; Kamhawi, S.; Valenzuela, J.G.; et al. Leishmania Tarentolae Secreting the Sand Fly Salivary Antigen PpSP15 Confers Protection against Leishmania Major Infection in a Susceptible BALB/c Mice Model. Mol. Immunol. 2015, 67, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Katz, G.; Leisnham, P.T.; Ladeau, S.L. Aedes albopictus Body Size Differs Across Neighborhoods with Varying Infrastructural Abandonment. J. Med. Entomol. 2020, 57, 615–619. [Google Scholar] [CrossRef] [PubMed]
- Vogt, M.B.; Lahon, A.; Arya, R.P.; Kneubehl, A.R.; Spencer Clinton, J.L.; Paust, S.; Rico-Hesse, R. Mosquito Saliva Alone Has Profound Effects on the Human Immune System. PLoS Negl. Trop. Dis. 2018, 12, e0006439. [Google Scholar] [CrossRef] [PubMed]
- Keskek Turk, Y.; Barningham, L.D.; McKimmie, C.S. Sensing the Danger in Mosquito Spit. EMBO J. 2024, 43, 1687–1689. [Google Scholar] [CrossRef]
- Moser, L.A.; Lim, P.-Y.; Styer, L.M.; Kramer, L.D.; Bernard, K.A. Parameters of Mosquito-Enhanced West Nile Virus Infection. J. Virol. 2016, 90, 292–299. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barningham, L.; Carr, I.M.; Jossi, S.; Cole, M.; Ponce, A.; Short, M.; Meneses, C.; Lacsina, J.R.; Valenzuela, J.G.; Oliveira, F.; et al. AGS-v PLUS, a Mosquito Salivary Peptide Vaccine, Modulates the Response to Aedes Mosquito Bites in Humans. Vaccines 2025, 13, 1026. https://doi.org/10.3390/vaccines13101026
Barningham L, Carr IM, Jossi S, Cole M, Ponce A, Short M, Meneses C, Lacsina JR, Valenzuela JG, Oliveira F, et al. AGS-v PLUS, a Mosquito Salivary Peptide Vaccine, Modulates the Response to Aedes Mosquito Bites in Humans. Vaccines. 2025; 13(10):1026. https://doi.org/10.3390/vaccines13101026
Chicago/Turabian StyleBarningham, Liam, Ian M. Carr, Siân Jossi, Megan Cole, Aiyana Ponce, Mara Short, Claudio Meneses, Joshua R. Lacsina, Jesus G. Valenzuela, Fabiano Oliveira, and et al. 2025. "AGS-v PLUS, a Mosquito Salivary Peptide Vaccine, Modulates the Response to Aedes Mosquito Bites in Humans" Vaccines 13, no. 10: 1026. https://doi.org/10.3390/vaccines13101026
APA StyleBarningham, L., Carr, I. M., Jossi, S., Cole, M., Ponce, A., Short, M., Meneses, C., Lacsina, J. R., Valenzuela, J. G., Oliveira, F., Laurens, M. B., Friedman-Klabanoff, D. J., Pleguezuelos, O., Stead, L. F., & McKimmie, C. S. (2025). AGS-v PLUS, a Mosquito Salivary Peptide Vaccine, Modulates the Response to Aedes Mosquito Bites in Humans. Vaccines, 13(10), 1026. https://doi.org/10.3390/vaccines13101026





