Molecular Events in Immune Responses to Sublingual Influenza Vaccine with Hemagglutinin Antigen and Poly(I:C) Adjuvant in Nonhuman Primates, Cynomolgus Macaques
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents, Wares, and Antibodies
2.2. Animals
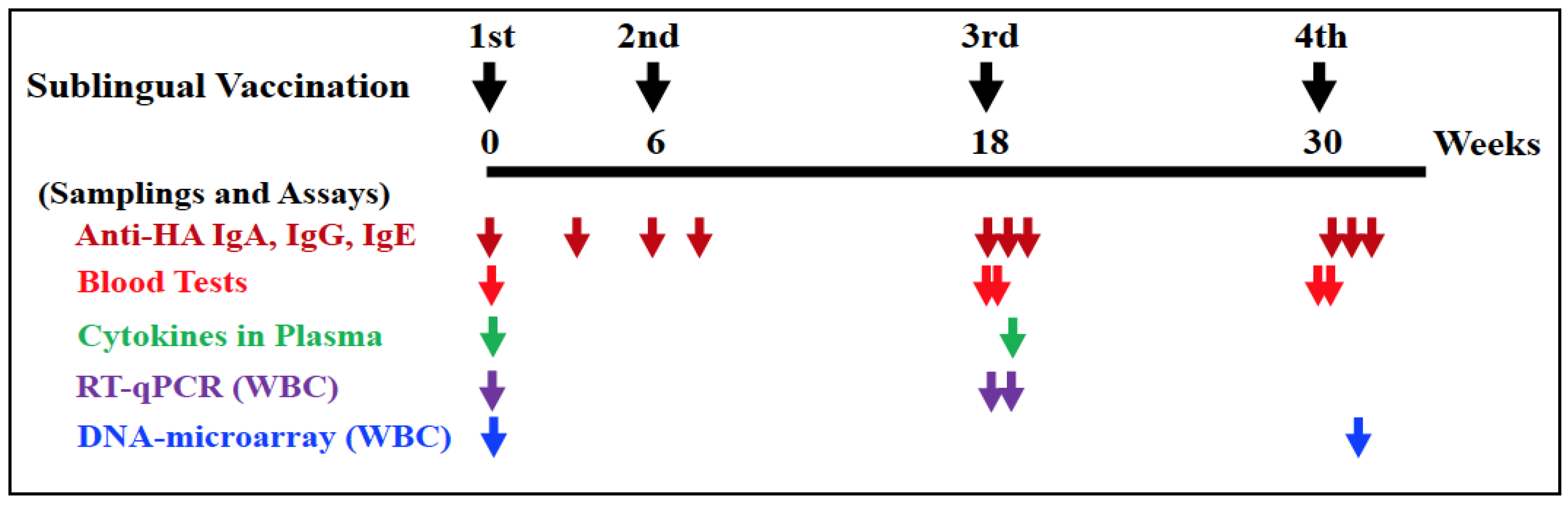
2.3. Vaccination and Sampling
2.4. ELISA for HA-Specific Antibodies
2.5. Blood Testing
2.6. Cytokine Testing
2.7. Isolation of WBCs
2.8. Isolation of RNAs
2.9. Quantitative Reverse Transcription PCR for Gene Expression Analyses
2.10. DNA Microarray for High-Throughput Gene Expression Analyses
2.11. Bioinformatic for Microarray Data Analyses
2.12. Data Expression and Statistical Analysis
3. Results
3.1. Complete Blood Count, Biochemical Blood Test, and Plasma CRP
3.2. HA-Specific Antibodies
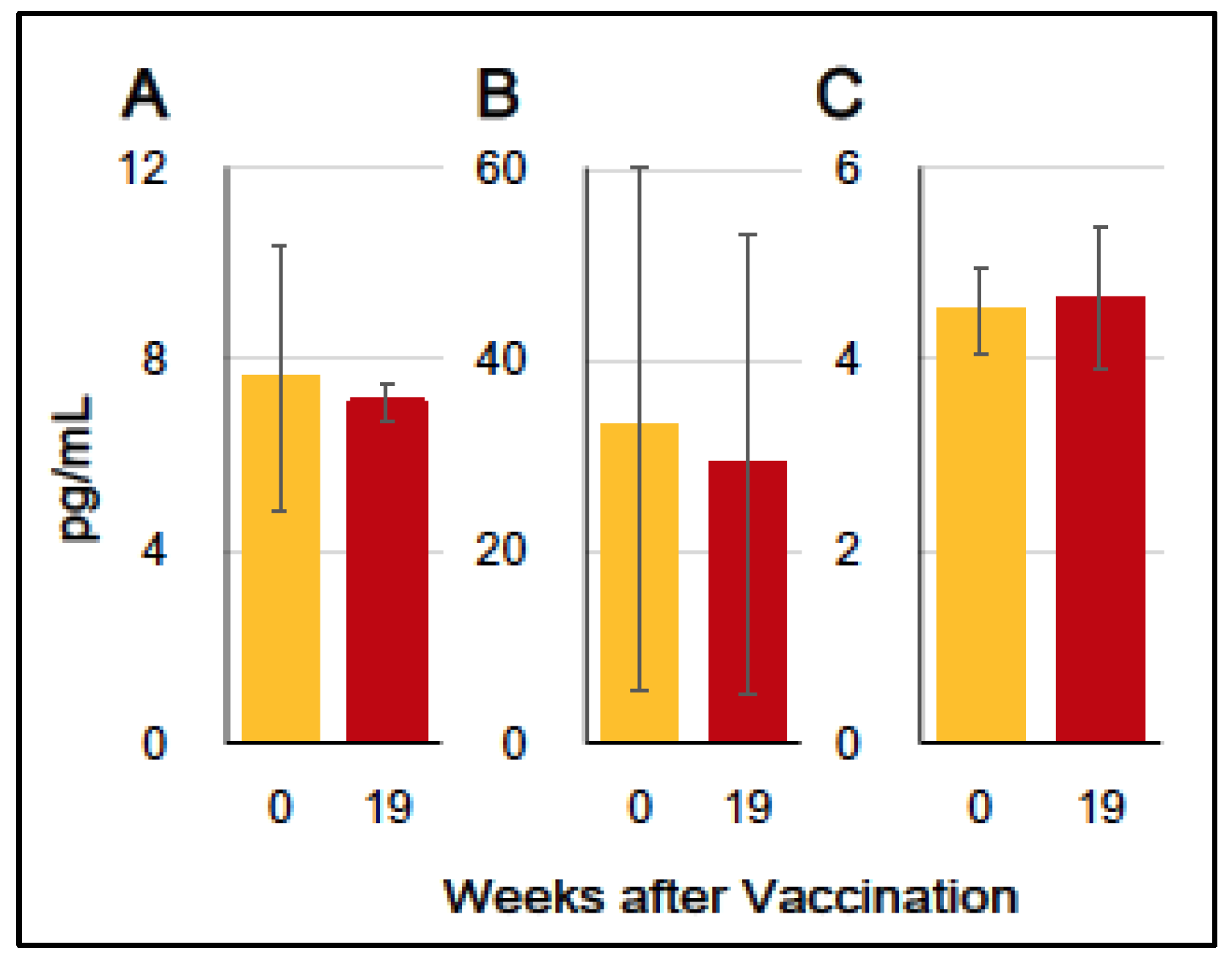
3.3. Production of Inflammation-Associating Cytokines
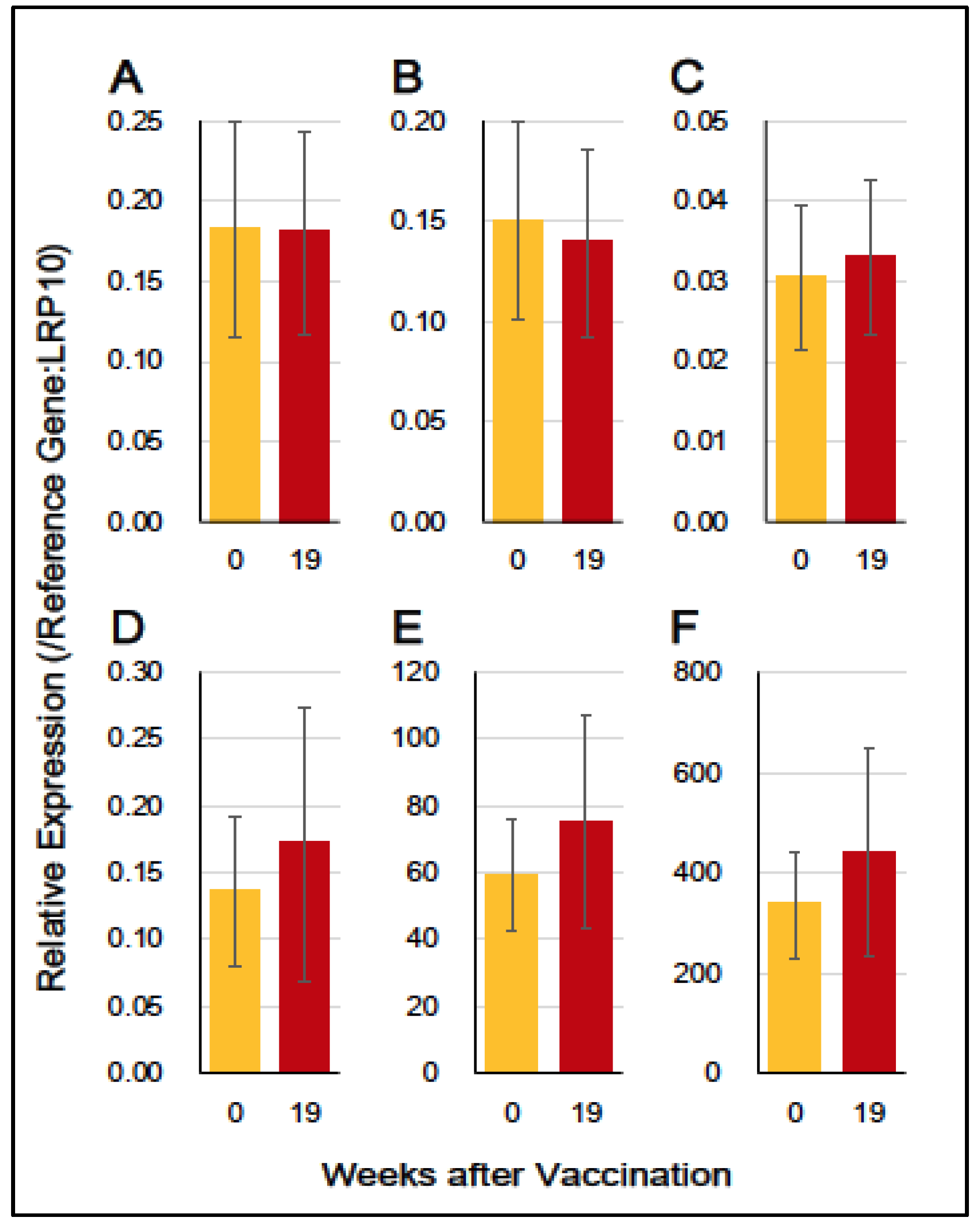
3.4. Gene Expression of Immuno-Proinflammatory Factors
3.5. Microarray Analyses and Bioinformatics
3.5.1. Upregulated Genes
3.5.2. Downregulated Genes
4. Discussion
4.1. Poly(I:C) Adjuvant
4.2. Sublingual Poly(I:C)-Adjuvanted Vaccination
4.3. Genes Upregulated by Sublingual Vaccine with HA Antigen and Poly(I:C) Adjuvant
4.4. Genes Downregulated by Sublingual Vaccine with HA Antigen and Poly(I:C) Adjuvant
4.5. A Balance State of Immune-Enhancing and -Suppressive Response Induced by the Sublingual Vaccine
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Influenza (Seasonal). 2023. Available online: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (accessed on 9 April 2024).
- World Health Organization. Vaccines Against Influenza: WHO Position Paper—May 2022. Available online: https://www.who.int/publications/i/item/who-wer9719 (accessed on 9 April 2024).
- Colombo, L.; Hadigal, S.; Nauta, J.; Kondratenko, A.; Rogoll, J.; van de Witte, S. Influvac Tetra: Clinical experience on safety, efficacy, and immunogenicity. Expert Rev. Vaccines 2024, 23, 88–101. [Google Scholar] [CrossRef]
- Mudgal, R.; Nehul, S.; Tomar, S. Prospects for mucosal vaccine: Shutting the door on SARS-CoV-2. Hum. Vaccines Immunother. 2020, 16, 2921–2931. [Google Scholar] [CrossRef]
- Lemiale, F.; Kong, W.; Akyurek, L.M.; Ling, X.; Huang, Y.; Chakrabarti, B.K.; Eckhaus, M.; Nabel, G.J. Enhanced mucosal immunoglobulin a response of intranasal adenoviral vector human immunodeficiency virus vaccine and localization in the central nervous system. J. Virol. 2003, 77, 10078–10087. [Google Scholar] [CrossRef]
- Sasaki, E.; Momose, H.; Hiradate, Y.; Mizukami, T.; Hamaguchi, I. Establishment of a novel safety assessment method for vaccine adjuvant development. Vaccine 2018, 36, 7112–7118. [Google Scholar] [CrossRef]
- Yamamoto, T.; Tanji, M.; Mitsunaga, F.; Nakamura, S. SARS-CoV-2 sublingual vaccine with RBD antigen and poly(I:C) adjuvant: Preclinical study in cynomolgus macaques. Biol. Methods Protoc. 2023, 8, bpad017. [Google Scholar] [CrossRef]
- Yamamoto, T.; Mitsunaga, F.; Wasaki, K.; Kotani, A.; Tajima, K.; Tanji, M.; Nakamura, S. Mechanism Underlying the Immune Responses of a Sublingual Vaccine for SARS-CoV-2 with RBD Antigen and Adjuvant, Poly(I:C) or AddaS03, in Non-human Primates. Arch. Microbiol. Immunol. 2023, 07, 150–164. [Google Scholar] [CrossRef]
- Iwasaki, A.; Omer, S.B. Why and How Vaccines Work. Cell 2020, 15, 290–295. [Google Scholar] [CrossRef]
- Zhao, T.; Cai, Y.; Jiang, Y.; He, X.; Wei, Y.; Yu, Y.; Tian, X. Vaccine adjuvants: Mechanisms and platforms. Signal Transduct. Target. Ther. 2023, 8, 283. [Google Scholar] [CrossRef]
- Ainai, A.; van Riet, E.; Ito, R.; Ikeda, K.; Senchi, K.; Suzuki, T.; Tamura, S.; Asanuma, H.; Odagiri, T.; Tashiro, M.; et al. Human immune responses elicited by an intranasal inactivated H5 influenza vaccine. Microbiol. Immunol. 2020, 64, 313–325. [Google Scholar] [CrossRef]
- A Hoffman, R.; Kung, P.C.; Hansen, W.P.; Goldstein, G. Simple and rapid measurement of human T lymphocytes and their subclasses in peripheral blood. Proc. Natl. Acad. Sci. USA 1980, 77, 4914–4917. [Google Scholar] [CrossRef]
- Jeong, A.R.; Nakamura, S.; Mitsunaga, F. Gene expression profile of Th1 and Th2 cytokines and their receptors in human and nonhuman primates. J. Med. Primatol. 2008, 37, 290–296. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Gabrielsson, B.G.; Olofsson, L.E.; Sjögren, A.; Jernås, M.; Elander, A.; Lönn, M.; Rudemo, M.; Carlsson, L.M.S. Evaluation of Reference Genes for Studies of Gene Expression in Human Adipose Tissue. Obes. Res. 2005, 13, 649–652. [Google Scholar] [CrossRef]
- Hoffmann, D.; Mereiter, S.; Oh, Y.J.; Monteil, V.; Elder, E.; Zhu, R.; Canena, D.; Hain, L.; Laurent, E.; Grünwald-Gruber, C.; et al. Identification of lectin receptors for conserved SARS-CoV-2 glycosylation sites. EMBO J. 2021, 40, e108375. [Google Scholar] [CrossRef]
- Lu, Q.; Liu, J.; Zhao, S.; Castro, M.F.G.; Laurent-Rolle, M.; Dong, J.; Ran, X.; Damani-Yokota, P.; Tang, H.; Karakousi, T.; et al. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2. Immunity 2021, 54, 1304–1319.e9. [Google Scholar] [CrossRef]
- Dominguez-Soto, A.; Aragoneses-Fenoll, L.; Martin-Gayo, E.; Martinez-Prats, L.; Colmenares, M.; Naranjo-Gomez, M.; Borras, F.E.; Munoz, P.; Zubiaur, M.; Toribio, M.L.; et al. The DC-SIGN–related lectin LSECtin mediates antigen capture and pathogen binding by human myeloid cells. Blood 2007, 109, 5337–5345. [Google Scholar] [CrossRef] [PubMed]
- Struyf, S.; Salogni, L.; Burdick, M.D.; Vandercappellen, J.; Gouwy, M.; Noppen, S.; Proost, P.; Opdenakker, G.; Parmentier, M.; Gerard, C.; et al. Angiostatic and chemotactic activities of the CXC chemokine CXCL4L1 (platelet factor-4 variant) are mediated by CXCR3. Blood 2011, 117, 480–488. [Google Scholar] [CrossRef]
- Teruya, S.; Okamura, T.; Komai, T.; Inoue, M.; Iwasaki, Y.; Sumitomo, S.; Shoda, H.; Yamamoto, K.; Fujio, K. Egr2-independent, Klf1-mediated induction of PD-L1 in CD4+ T cells. Sci. Rep. 2018, 8, 7021. [Google Scholar] [CrossRef]
- Shi, W.; Ye, Z.; Zhuang, L.; Li, Y.; Shuai, W.; Zuo, Z.; Mao, X.; Liu, R.; Wu, J.; Chen, S.; et al. Olfactomedin 1 negatively regulates NF-κB signalling and suppresses the growth and metastasis of colorectal cancer cells. J. Pathol. 2016, 240, 352–365. [Google Scholar] [CrossRef]
- Ruiz-Ballesteros, E.; Mollejo, M.; Rodriguez, A.; Camacho, F.I.; Algara, P.; Martinez, N.; Pollán, M.; Sanchez-Aguilera, A.; Menarguez, J.; Campo, E.; et al. Splenic marginal zone lymphoma: Proposal of new diagnostic and prognostic markers identified after tissue and cDNA microarray analysis. Blood 2005, 106, 1831–1838. [Google Scholar] [CrossRef]
- Rullinkov, G.; Tamme, R.; Sarapuu, A.; Laurén, J.; Sepp, M.; Palm, K.; Timmusk, T. Neuralized-2: Expression in human and rodents and interaction with Delta-like ligands. Biochem. Biophys. Res. Commun. 2009, 389, 420–425. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Z.; Zhang, X.; Yan, Y.; Shao, S.; Yao, D.; Gong, T. Aberrant methylation and microRNA-target regulation are associated with downregulated NEURL1B: A diagnostic and prognostic target in colon cancer. Cancer Cell Int. 2020, 20, 342. [Google Scholar] [CrossRef]
- Verkoczy, L.K.; Marsden, P.A.; Berinstein, N.L. hBRAG, a novel B cell lineage cDNA encoding a type II transmembrane glycoprotein potentially involved in the regulation of recombination activating gene 1 (RAG1). Eur. J. Immunol. 1998, 28, 2839–2853. [Google Scholar] [CrossRef]
- Dong, L.; Cao, Y.; Yang, H.; Hou, Y.; He, Y.; Wang, Y.; Yang, Q.; Bi, Y.; Liu, G. The hippo kinase MST1 negatively regulates the differentiation of follicular helper T cells. Immunology 2023, 168, 511–525. [Google Scholar] [CrossRef]
- Cornely, R.; Pollock, A.H.; Rentero, C.; E Norris, S.; Alvarez-Guaita, A.; Grewal, T.; Mitchell, T.; Enrich, C.; E Moss, S.; Parton, R.G.; et al. Annexin A6 regulates interleukin-2-mediated T-cell proliferation. Immunol. Cell Biol. 2016, 94, 543–553. [Google Scholar] [CrossRef]
- Momiuchi, Y.; Kumada, K.; Kuraishi, T.; Takagaki, T.; Aigaki, T.; Oshima, Y.; Kurata, S. The Role of the Phylogenetically Conserved Cochaperone Protein Droj2/DNAJA3 in NF-κB Signaling. J. Biol. Chem. 2015, 290, 23816–23825. [Google Scholar] [CrossRef]
- Bracher, A.; Verghese, J. The nucleotide exchange factors of Hsp70 molecular chaperones. Front. Mol. Biosci. 2015, 2, 10. [Google Scholar] [CrossRef]
- Boudesco, C.; Verhoeyen, E.; Martin, L.; Chassagne-Clement, C.; Salmi, L.; Mhaidly, R.; Pangault, C.; Fest, T.; Ramla, S.; Jardin, F.; et al. HSP110 sustains chronic NF-κB signaling in activated B-cell diffuse large B-cell lymphoma through MyD88 stabilization. Blood 2018, 132, 510–520. [Google Scholar] [CrossRef]
- Sasaki, E.; Momose, H.; Hiradate, Y.; Furuhata, K.; Takai, M.; Asanuma, H.; Ishii, K.J.; Mizukami, T.; Hamaguchi, I. Modeling for influenza vaccines and adjuvants profile for safety prediction system using gene expression profiling and statistical tools. PLoS ONE 2018, 13, e0191896. [Google Scholar] [CrossRef]
- Sasaki, E.; Asanuma, H.; Momose, H.; Furuhata, K.; Mizukami, T.; Hamaguchi, I. Immunogenicity and Toxicity of Different Adjuvants Can Be Characterized by Profiling Lung Biomarker Genes After Nasal Immunization. Front. Immunol. 2020, 11, 2171. [Google Scholar] [CrossRef]
- Dotiwala, F.; Upadhyay, A.K. Next Generation Mucosal Vaccine Strategy for Respiratory Pathogens. Vaccines 2023, 11, 1585. [Google Scholar] [CrossRef]
- Mestas, J.; Hughes, C.C.W. Of mice and not men: Differences between mouse and human immunology. J. Immunol. 2004, 172, 2731–2738. [Google Scholar] [CrossRef]
- Le Naour, J.; Galluzzi, L.; Zitvogel, L.; Kroemer, G.; Vacchelli, E. Trial watch: TLR3 agonists in cancer therapy. OncoImmunology 2020, 9, 1771143. [Google Scholar] [CrossRef]
- De Waele, J.; Verhezen, T.; van der Heijden, S.; Berneman, Z.N.; Peeters, M.; Lardon, F.; Wouters, A.; Smits, E.L.J.M. A systematic review on poly(I:C) and poly-ICLC in glioblastoma: Adjuvants coordinating the unlocking of immunotherapy. J. Exp. Clin. Cancer Res. 2021, 40, 213. [Google Scholar] [CrossRef]
- Tewari, K.; Flynn, B.J.; Boscardin, S.B.; Kastenmueller, K.; Salazar, A.M.; Anderson, C.A.; Soundarapandian, V.; Ahumada, A.; Keler, T.; Hoffman, S.L.; et al. Poly(I:C) is an effective adjuvant for antibody and multi-functional CD4+ T cell responses to Plasmodium falciparum circumsporozoite protein (CSP) and αDEC-CSP in non human primates. Vaccine 2010, 28, 7256–7266. [Google Scholar] [CrossRef]
- Fučíková, J.; Rožková, D.; Ulčová, H.; Budinský, V.; Sochorová, K.; Pokorná, K.; Bartůňková, J.; Špíšek, R. Poly I: C-activated dendritic cells that were generated in CellGro for use in cancer immunotherapy trials. J. Transl. Med. 2011, 9, 223. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef]
- Sultan, H.; Wu, J.; Kumai, T.; Salazar, A.M.; Celis, E. Role of MDA5 and interferon-I in dendritic cells for T cell expansion by anti-tumor peptide vaccines in mice. Cancer Immunol. Immunother. 2018, 67, 1091–1103. [Google Scholar] [CrossRef]
- Kraan, H.; Vrieling, H.; Czerkinsky, C.; Jiskoot, W.; Kersten, G.; Amorij, J.-P. Buccal and sublingual vaccine delivery. J. Control. Release 2014, 190, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Mascarell, L.; Lombardi, V.; Louise, A.; Saint-Lu, N.; Chabre, H.; Moussu, H.; Betbeder, D.; Balazuc, A.-M.; Van Overtvelt, L.; Moingeon, P. Oral dendritic cells mediate antigen-specific tolerance by stimulating TH1 and regulatory CD4+ T cells. J. Allergy Clin. Immunol. 2008, 122, 603–609.e5. [Google Scholar] [CrossRef] [PubMed]
- Hervouet, C.; Luci, C.; Bekri, S.; Juhel, T.; Bihl, F.; Braud, V.M.; Czerkinsky, C.; Anjuère, F. Antigen-bearing dendritic cells from the sublingual mucosa recirculate to distant systemic lymphoid organs to prime mucosal CD8 T cells. Mucosal Immunol. 2014, 7, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Paris, A.; Colomb, E.; Verrier, B.; Anjuère, F.; Monge, C. Sublingual vaccination and delivery systems. J. Control. Release 2021, 332, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Hillaire, M.L.B.; Nieuwkoop, N.J.; Boon, A.C.M.; de Mutsert, G.; Trierum, S.E.V.-V.; Fouchier, R.A.M.; Osterhaus, A.D.M.E.; Rimmelzwaan, G.F. Binding of DC-SIGN to the hemagglutinin of influenza a viruses supports virus replication in DC-SIGN expressing cells. PLoS ONE 2013, 8, e56164. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Yang, J.; Liu, W.; Tang, X.; Chen, J.; Zhao, D.; Wang, M.; Xu, F.; Lu, Y.; Liu, B.; et al. Liver sinusoidal endothelial cell lectin, LSECtin, negatively regulates hepatic T-cell immune response. Gastroenterology 2009, 137, 1498–1508.e5. [Google Scholar] [CrossRef] [PubMed]
- Moingeon, P.; Mascarell, L. Induction of tolerance via the sublingual route: Mechanisms and applications. Clin. Dev. Immunol. 2011, 2012, 623474. [Google Scholar] [CrossRef] [PubMed]
- Hibbert, R.G.; Teriete, P.; Grundy, G.J.; Beavil, R.L.; Reljić, R.; Holers, V.M.; Hannan, J.P.; Sutton, B.J.; Gould, H.J.; McDonnell, J.M. The structure of human CD23 and its interactions with IgE and CD21. J. Exp. Med. 2005, 202, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Brandhofer, M.; Hoffmann, A.; Blanchet, X.; Siminkovitch, E.; Rohlfing, A.-K.; El Bounkari, O.; Nestele, J.A.; Bild, A.; Kontos, C.; Hille, K.; et al. Heterocomplexes between the atypical chemokine MIF and the CXC-motif chemokine CXCL4L1 regulate inflammation and thrombus formation. Cell. Mol. Life Sci. 2022, 79, 512. [Google Scholar] [CrossRef]
- Dutchak, K.; Garnett, S.; Nicoll, M.; de Bruyns, A.; Dankort, D. MOB3A Bypasses BRAF and RAS Oncogene-Induced Senescence by Engaging the Hippo Pathway. Mol. Cancer Res. 2022, 20, 770–781. [Google Scholar] [CrossRef]
 ) indicate the control group, and solid brown circles (
) indicate the control group, and solid brown circles ( ) indicate the experiment/HA + Poly(I:C)) group. Red arrows (
) indicate the experiment/HA + Poly(I:C)) group. Red arrows ( ) indicate vaccination. Relative titers were estimated using the conditions mentioned in Section 2.
) indicate vaccination. Relative titers were estimated using the conditions mentioned in Section 2.
 ) indicate the control group, and solid brown circles (
) indicate the control group, and solid brown circles ( ) indicate the experiment/HA + Poly(I:C)) group. Red arrows (
) indicate the experiment/HA + Poly(I:C)) group. Red arrows ( ) indicate vaccination. Relative titers were estimated using the conditions mentioned in Section 2.
) indicate vaccination. Relative titers were estimated using the conditions mentioned in Section 2.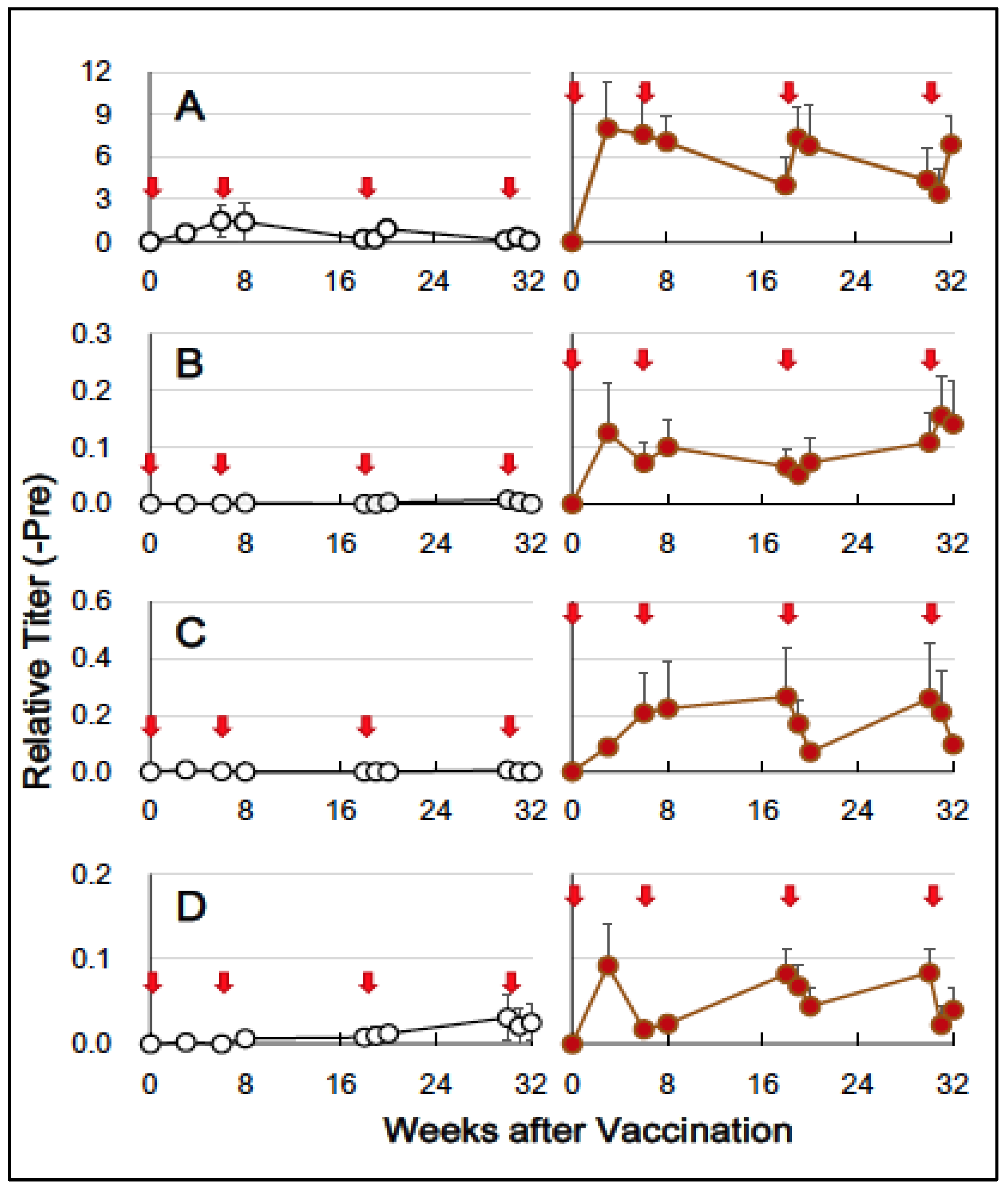
| Gene Symbol | Fold Change | Product. Description. Function [Reference]. | Expected Effect * |
|---|---|---|---|
| CLEC4G | 2.2 | C-type lectin. Receptor of PAMP [16]. Activation of inflammatory reactions [17]. Transcript variants [18]. | ↑ |
| PF4V1 | 2.2 | Also known as CXCL4L1. Chemoattractant of T and NK cells [19]. | ↑ |
| KLF1 | 2.1 | Zn-finger transcription factor. Transcription of γ-globin gene. Upregulation of CD274 in Tregs [20]. | ↓ |
| OLFM1 | 2.1 | Nervous system. Inhibition of non-canonical NF-κB pathway in CRC [21]. | ↓ |
| GNG11 | 2.0 | G protein gamma family. Downregulated in splenic marginal zone lymphomas [22]. | ↓ |
| Gene Symbol | Fold Change | Product. Description. Function [Reference]. | Expected Effect * |
|---|---|---|---|
| NEURL1B | 0.45 | Ubiquitin protein ligase. Development of the nervous system [23]. Downregulated in CRC tissues [24]. Downregulated by Poly(I:C) adjuvant [8]. | ↑ |
| CHST15 | 0.45 | Sulfotransferase. Expressed in B-cell lineage. Upregulation of RAG1 [25]. | ↓ |
| MOB3A | 0.48 | Protein kinase activator. Inhibition of GC through the Hippo pathway [26]. | ↑ |
| ANXA6 | 0.48 | Expressed in T cells. Component of T-cell plasma membrane. Stimulation of helper T cells [27]. | ↓ |
| DNAJA3 | 0.48 | Mitochondrial HSP. Stimulation of ATPase activity of Hsp70. Activation of NF-κB [28]. | ↓ |
| HSPH1 | 0.49 | HSP. Replacement of Hsp70-bound ADP with ATP. Holdase and disaggregase activity [29]. Stimulation of NF-κB signaling in activated B-cell diffuse large B-cell lymphoma [30]. | ↓ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamoto, T.; Hirano, M.; Mitsunaga, F.; Wasaki, K.; Kotani, A.; Tajima, K.; Nakamura, S. Molecular Events in Immune Responses to Sublingual Influenza Vaccine with Hemagglutinin Antigen and Poly(I:C) Adjuvant in Nonhuman Primates, Cynomolgus Macaques. Vaccines 2024, 12, 643. https://doi.org/10.3390/vaccines12060643
Yamamoto T, Hirano M, Mitsunaga F, Wasaki K, Kotani A, Tajima K, Nakamura S. Molecular Events in Immune Responses to Sublingual Influenza Vaccine with Hemagglutinin Antigen and Poly(I:C) Adjuvant in Nonhuman Primates, Cynomolgus Macaques. Vaccines. 2024; 12(6):643. https://doi.org/10.3390/vaccines12060643
Chicago/Turabian StyleYamamoto, Tetsuro, Makoto Hirano, Fusako Mitsunaga, Kunihiko Wasaki, Atsushi Kotani, Kazuki Tajima, and Shin Nakamura. 2024. "Molecular Events in Immune Responses to Sublingual Influenza Vaccine with Hemagglutinin Antigen and Poly(I:C) Adjuvant in Nonhuman Primates, Cynomolgus Macaques" Vaccines 12, no. 6: 643. https://doi.org/10.3390/vaccines12060643
APA StyleYamamoto, T., Hirano, M., Mitsunaga, F., Wasaki, K., Kotani, A., Tajima, K., & Nakamura, S. (2024). Molecular Events in Immune Responses to Sublingual Influenza Vaccine with Hemagglutinin Antigen and Poly(I:C) Adjuvant in Nonhuman Primates, Cynomolgus Macaques. Vaccines, 12(6), 643. https://doi.org/10.3390/vaccines12060643





