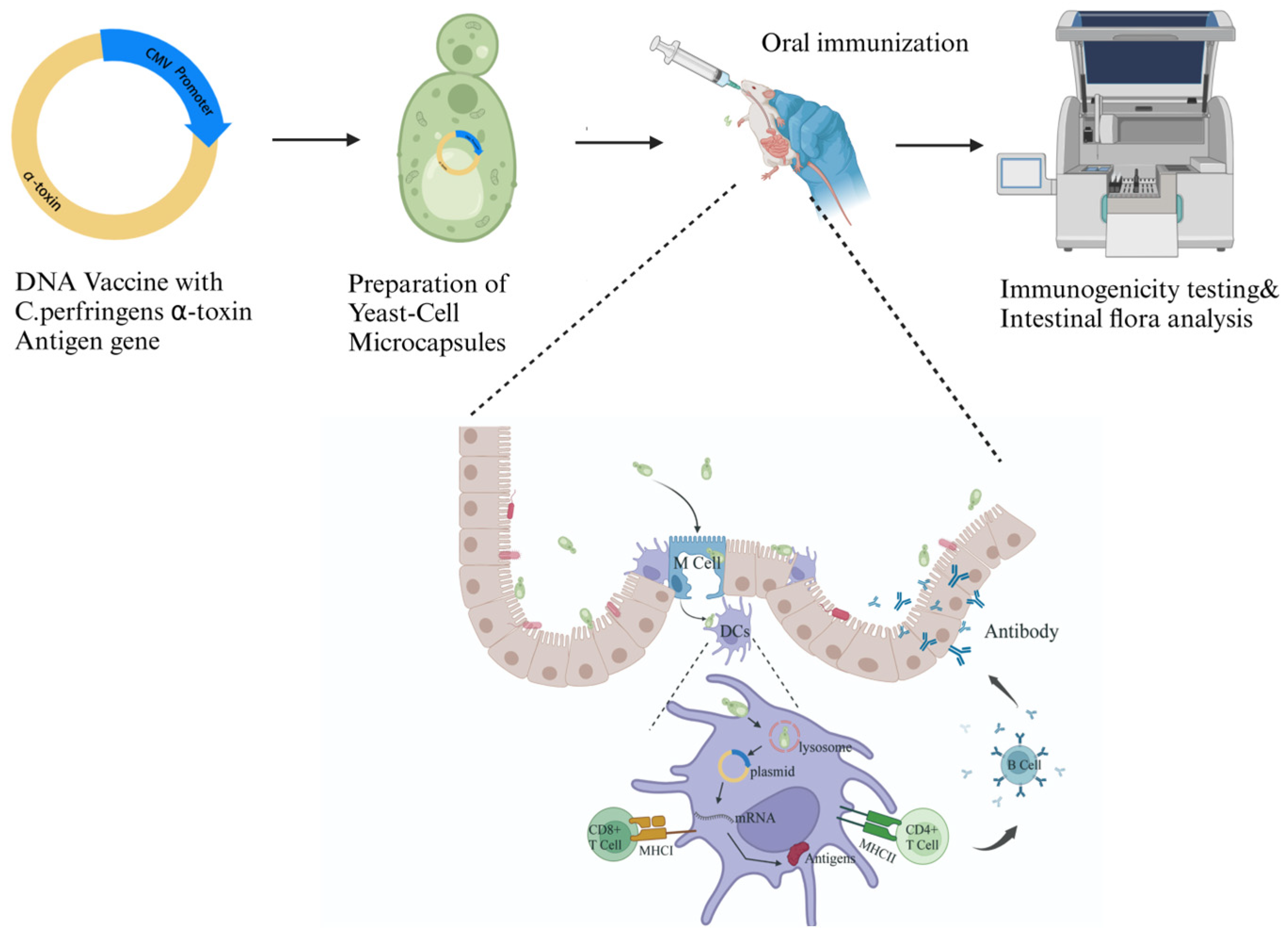
Oral Yeast-Cell Microcapsule-Mediated DNA Vaccines Against Clostridium perfringens Induce Effective Intestinal Immunity and Modulate Gut Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Testing Successful Expression of Target Vector in Eukaryotic Cells
2.3. Preparation and Screning of YCM
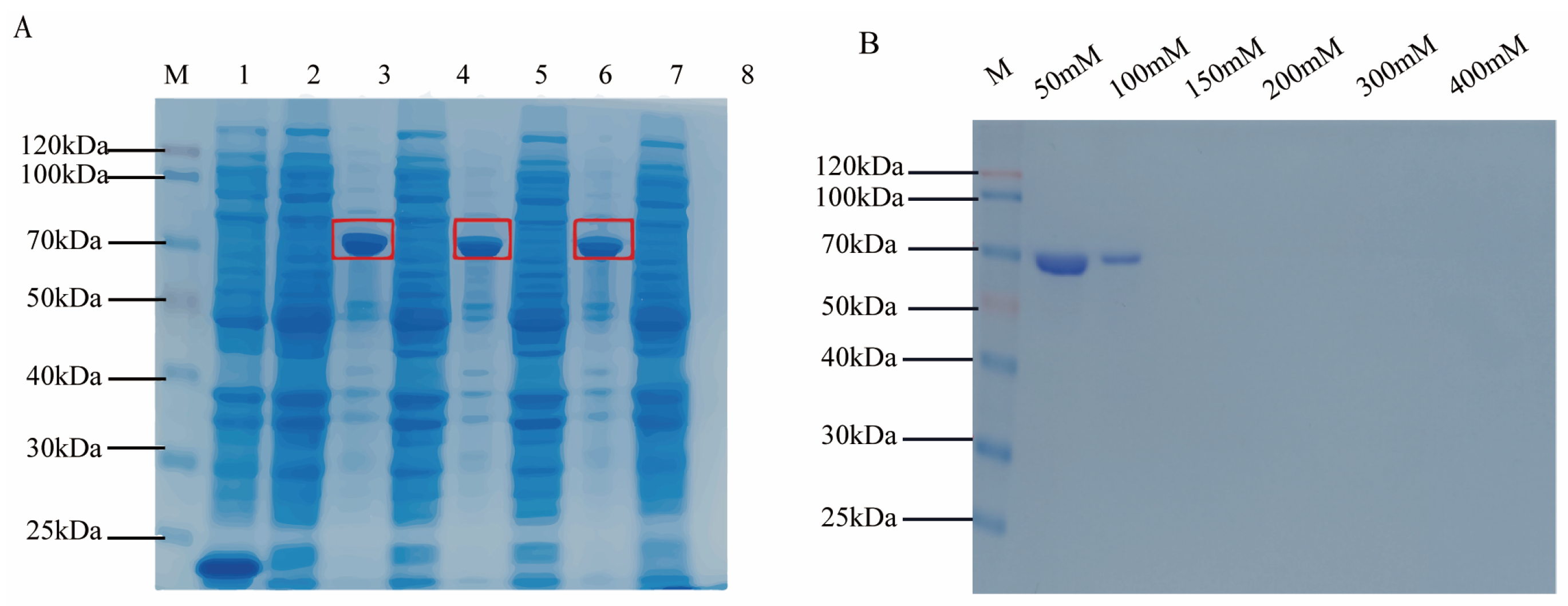
2.4. Prokaryotic Expression and Purification of Target Antigen Protein
2.5. Oral Immunization
2.6. Detection of IgG, sIgA, and Cytokines by ELISA
2.7. RNA Extraction and qRT-PCR
2.8. 16S rRNA Sequencing of Gut Microbiota
2.9. Statistical Analysis
3. Results
3.1. Construction and Expression of Eukaryotic Vectors
3.2. YCM Screening and Identification
3.3. Detection of Cpα Protein Prokaryotic Induction Expression
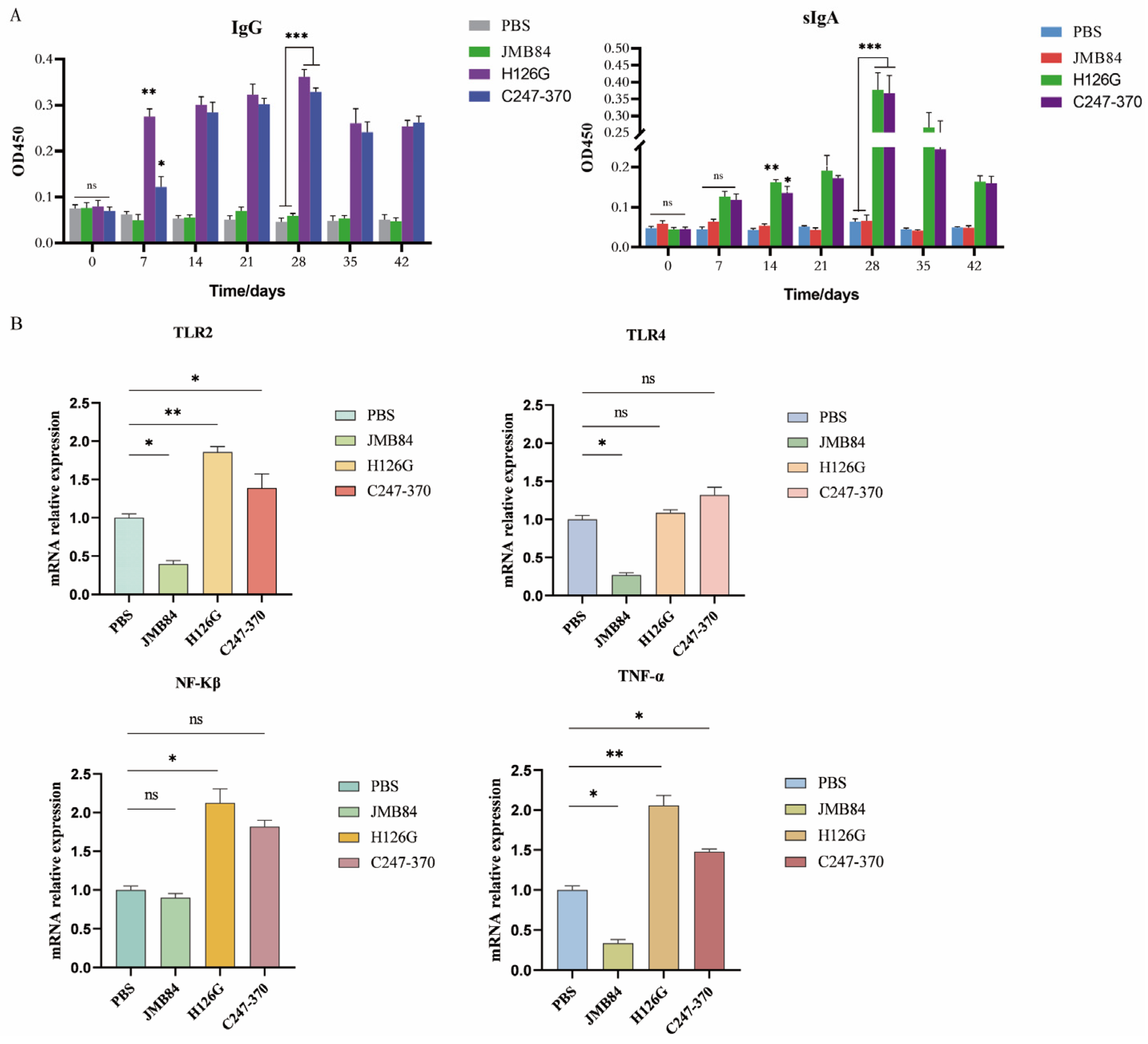
3.4. Specific Immune Response Induced by Oral Yeast Microcapsule-Mediated DNA Vaccine in Mice
3.5. Oral YCM DNA Vaccines Activate Mouse Immune Responses via the TLR2 Signaling Pathway
3.6. Gut Microbiota Composition Changes After Oral Immunization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferreira, M.R.A.; Motta, J.F.; Azevedo, M.L.; Dos Santos, L.M.; Júnior, C.M.; Rodrigues, R.R.; Donassolo, R.A.; Reis, A.D.S.B.; Barbosa, J.D.; Salvarani, F.M.; et al. Inactivated Recombinant Escherichia coli as a Candidate Vaccine Against Clostridium perfringens Alpha Toxin in Sheep. Anaerobe 2019, 59, 163–166. [Google Scholar] [CrossRef] [PubMed]
- Uzal, F.A.; Vidal, J.E.; McClane, B.A.; Gurjar, A.A. Clostridium perfringens Toxins Involved in Mammalian Veterinary Diseases. Open Toxinol. J. 2010, 2, 24–42. [Google Scholar] [CrossRef]
- Rood, J.I.; Adams, V.; Lacey, J.; Lyras, D.; McClane, B.A.; Melville, S.B.; Moore, R.J.; Popoff, M.R.; Sarker, M.R.; Songer, J.G.; et al. Expansion of the Clostridium perfringens Toxin-Based Typing Scheme. Anaerobe 2018, 53, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Adams, V.; Bannam, T.L.; Miyamoto, K.; Garcia, J.P.; Uzal, F.A.; Rood, J.I.; McClane, B.A. Toxin Plasmids of Clostridium perfringens. Microbiol. Mol. Biol. Rev. 2013, 77, 208–233. [Google Scholar] [CrossRef]
- Ohtani, K.; Shimizu, T. Regulation of Toxin Production in Clostridium perfringens. Toxins 2016, 8, 207. [Google Scholar] [CrossRef]
- Jewell, S.A.; Titball, R.W.; Huyet, J.; Naylor, C.E.; Basak, A.K.; Gologan, P.; Winlove, C.P.; Petrov, P.G. Clostridium perfringens α-Toxin Interaction with Red Cells and Model Membranes. Soft Matter 2015, 11, 7748–7761. [Google Scholar] [CrossRef]
- Wang, S.; Gao, Y.; Jin, Q.; Ji, J. Emerging Antibacterial Nanomedicine for Enhanced Antibiotic Therapy. Biomater. Sci. 2020, 8, 6825–6839. [Google Scholar] [CrossRef]
- Zeng, J.; Deng, G.; Wang, J.; Zhou, J.; Liu, X.; Xie, Q.; Wang, Y. Potential Protective Immunogenicity of Recombinant Clostridium perfringens α-Β2-Β1 Fusion Toxin in Mice, Sows and Cows. Vaccine 2011, 29, 5459–5466. [Google Scholar] [CrossRef]
- Neeson, B.N.; Clark, G.C.; Atkins, H.S.; Lingard, B.; Titball, R.W. Analysis of Protection Afforded by a Clostridium perfringens Alpha-Toxoid Against Heterologous Clostridial Phospholipases C. Microb. Pathog. 2007, 43, 161–165. [Google Scholar] [CrossRef]
- Titball, R.W. Clostridium perfringens Vaccines. Vaccine 2009, 27 (Suppl. S4), D44–D47. [Google Scholar] [CrossRef]
- Ferreira, M.R.A.; Moreira, G.M.S.G.; Cunha, C.E.P.d.; Mendonça, M.; Salvarani, F.M.; Moreira, Â.N.; Conceição, F.R. Recombinant Alpha, Beta, and Epsilon Toxins of Clostridium perfringens: Production Strategies and Applications as Veterinary Vaccines. Toxins 2016, 8, 340. [Google Scholar] [CrossRef] [PubMed]
- Williamson, E.D.; Titball, R.W. A Genetically Engineered Vaccine Against the Alpha-Toxin of Clostridium perfringens Protects Mice Against Experimental Gas Gangrene. Vaccine 1993, 11, 1253–1258. [Google Scholar] [CrossRef] [PubMed]
- Titball, R.W.; Fearn, A.M.; Williamson, E.D. Biochemical and Immunological Properties of the C-Terminal Domain of the Alpha-Toxin of Clostridium perfringens. FEMS Microbiol. Lett. 1993, 110, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Stevens, D.L.; Titball, R.W.; Jepson, M.; Bayer, C.R.; Hayes-Schroer, S.M.; Bryant, A.E. Immunization with the C-Domain of Alpha -Toxin Prevents Lethal Infection, Localizes Tissue Injury, and Promotes Host Response to Challenge with Clostridium perfringens. J. Infect. Dis. 2004, 190, 767–773. [Google Scholar] [CrossRef]
- Gao, X.; Ma, Y.; Wang, Z.; Bai, J.; Jia, S.; Feng, B.; Jiang, Y.; Cui, W.; Tang, L.; Li, Y.; et al. Oral Immunization of Mice with a Probiotic Lactobacillus casei Constitutively Expressing the α-Toxoid Induces Protective Immunity Against Clostridium perfringens α-Toxin. Virulence 2019, 10, 166–179. [Google Scholar] [CrossRef]
- Francis, M.J. Recent Advances in Vaccine Technologies. Vet. Clin. North. Am. Small Anim. Pract. 2018, 48, 231–241. [Google Scholar] [CrossRef]
- Liu, M.A. DNA Vaccines: An Historical Perspective and View to the Future. Immunol. Rev. 2011, 239, 62–84. [Google Scholar] [CrossRef]
- Coffey, J.W.; Gaiha, G.D.; Traverso, G. Oral Biologic Delivery: Advances Toward Oral Subunit, DNA, and mRNA Vaccines and the Potential for Mass Vaccination During Pandemics. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 517–540. [Google Scholar] [CrossRef]
- Tan, Y.; Chen, L.; Li, K.; Lou, B.; Liu, Y.; Liu, Z. Yeast as Carrier for Drug Delivery and Vaccine Construction. J. Control. Release 2022, 346, 358–379. Available online: https://pubmed.ncbi.nlm.nih.gov/35483637/ (accessed on 4 November 2024). [CrossRef]
- Bal, J.; Luong, N.N.; Park, J.; Song, K.-D.; Jang, Y.-S.; Kim, D.-H. Comparative Immunogenicity of Preparations of Yeast-Derived Dengue Oral Vaccine Candidate. Microb. Cell Fact. 2018, 17, 24. [Google Scholar] [CrossRef]
- Lei, H.; Jin, S.; Karlsson, E.; Schultz-Cherry, S.; Ye, K. Yeast Surface-Displayed H5N1 Avian Influenza Vaccines. J. Immunol. Res. 2016, 2016, 4131324. [Google Scholar] [CrossRef] [PubMed]
- Sauerwein, H.; Schmitz, S.; Hiss, S. Effects of a Dietary Application of a Yeast Cell Wall Extract on Innate and Acquired Immunity, on Oxidative Status and Growth Performance in Weanling Piglets and on the Ileal Epithelium in Fattened Pigs. J. Anim. Physiol. Anim. Nutr. 2007, 91, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Liang, G.; Cao, L.; Qiao, Q.; Hu, Z.; Fu, M.; Hong, B.; Wu, Q.; Liang, G.; Zhang, Z.; et al. Effects of Glucagon-Like Peptide-2-Expressing Saccharomyces cerevisiae Not Different from Empty Vector. J. Microbiol. Biotechnol. 2019, 29, 1644–1655. [Google Scholar] [CrossRef] [PubMed]
- Alugongo, G.M.; Xiao, J.; Wu, Z.; Li, S.; Wang, Y.; Cao, Z. Utilization of Yeast of Saccharomyces cerevisiae Origin in Artificially Raised Calves. J. Anim. Sci. Biotechnol. 2017, 8, 34. [Google Scholar] [CrossRef]
- Kiros, T.G.; Derakhshani, H.; Pinloche, E.; D’Inca, R.; Marshall, J.; Auclair, E.; Khafipour, E.; Van Kessel, A. Effect of Live Yeast Saccharomyces cerevisiae (Actisaf Sc 47) Supplementation on the Performance and Hindgut Microbiota Composition of Weanling Pigs. Sci. Rep. 2018, 8, 5315. [Google Scholar] [CrossRef]
- Lynn, D.J.; Benson, S.C.; Lynn, M.A.; Pulendran, B. Modulation of Immune Responses to Vaccination by the Microbiota: Implications and Potential Mechanisms. Nat. Rev. Immunol. 2022, 22, 33–46. [Google Scholar] [CrossRef]
- Borey, M.; Blanc, F.; Lemonnier, G.; Leplat, J.-J.; Jardet, D.; Rossignol, M.-N.; Ravon, L.; Billon, Y.; Bernard, M.; Estellé, J.; et al. Links Between Fecal Microbiota and the Response to Vaccination Against Influenza A Virus in Pigs. NPJ Vaccines 2021, 6, 92. [Google Scholar] [CrossRef]
- de Jong, S.E.; Olin, A.; Pulendran, B. The Impact of the Microbiome on Immunity to Vaccination in Humans. Cell Host Microbe 2020, 28, 169–179. [Google Scholar] [CrossRef]
- Liu, M.; Luo, F.; Ding, C.; Albeituni, S.; Hu, X.; Ma, Y.; Cai, Y.; McNally, L.; Sanders, M.A.; Jain, D.; et al. Dectin-1 Activation by a Natural Product β-Glucan Converts Immunosuppressive Macrophages into an M1-like Phenotype. J. Immunol. 2015, 195, 5055–5065. Available online: https://pubmed.ncbi.nlm.nih.gov/26453753/ (accessed on 3 November 2024). [CrossRef]
- Zhang, M.; Jin, X.; Yang, Y.-F. β-Glucan from Saccharomyces cerevisiae Induces SBD-1 Production in Ovine Ruminal Epithelial Cells via the Dectin-1-Syk-NF-κB Signaling Pathway. Cell Signal. 2019, 53, 304–315. [Google Scholar] [CrossRef]
- Kumar, R.; Kumar, P. Yeast-Based Vaccines: New Perspective in Vaccine Development and Application. FEMS Yeast Res. 2019, 19, foz007. [Google Scholar] [CrossRef] [PubMed]
- Cereda, V.; Vergati, M.; Huen, N.-Y.; di Bari, M.G.; Jochems, C.; Intrivici, C.; Gulley, J.L.; Apelian, D.; Schlom, J.; Tsang, K.Y. Maturation of Human Dendritic Cells with Saccharomyces cerevisiae (Yeast) Reduces the Number and Function of Regulatory T Cells and Enhances the Ratio of Antigen-Specific Effectors to Regulatory T Cells. Vaccine 2011, 29, 4992–4999. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Lee, J.Y.; Kang, H.A.; Lee, Y.; Park, E.-J.; Kim, H.-J. Oral Immunization with Whole Yeast Producing Viral Capsid Antigen Provokes a Stronger Humoral Immune Response than Purified Viral Capsid Antigen. Lett. Appl. Microbiol. 2014, 58, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Kiflmariam, M.G.; Yang, H.; Zhang, Z. Gene Delivery to Dendritic Cells by Orally Administered Recombinant Saccharomyces cerevisiae in Mice. Vaccine 2013, 31, 1360–1363. [Google Scholar] [CrossRef]
- Yan, N.; Xu, K.; Li, X.; Liu, Y.; Bai, Y.; Zhang, X.; Han, B.; Chen, Z.; Zhang, Z. Recombinant Saccharomyces cerevisiae Serves as Novel Carrier for Oral DNA Vaccines in Carassius auratus. Fish. Shellfish. Immunol. 2015, 47, 758–765. [Google Scholar] [CrossRef]
- Zakria, H.M.; Han, B.; Yue, F.; Mu, L.; Fang, Y.; Li, X.; Xu, K.; Zhang, Z. Significant Body Mass Increase by Oral Administration of a Cascade of shIL21-MSTN Yeast-Based DNA Vaccine in Mice. Biomed. Pharmacother. 2019, 118, 109147. [Google Scholar] [CrossRef]
- Zhao, L.; Guo, Z.; Liu, J.; Wang, Z.; Wang, R.; Li, Y.; Wang, L.; Xu, Y.; Tang, L.; Qiao, X. Recombinant Lactobacillus casei Expressing Clostridium perfringens Toxoids α, Β2, ε and Β1 Gives Protection Against Clostridium perfringens in Rabbits. Vaccine 2017, 35, 4010–4021. [Google Scholar] [CrossRef]
- Jayachandran, M.; Chen, J.; Chung, S.S.M.; Xu, B. A Critical Review on the Impacts of β-Glucans on Gut Microbiota and Human Health. J. Nutr. Biochem. 2018, 61, 101–110. [Google Scholar] [CrossRef]
- Roeder, A.; Kirschning, C.J.; Rupec, R.A.; Schaller, M.; Korting, H.C. Toll-like Receptors and Innate Antifungal Responses. Trends Microbiol. 2004, 12, 44–49. [Google Scholar] [CrossRef]
- Sun, S.-C. The Non-Canonical NF-κB Pathway in Immunity and Inflammation. Nat. Rev. Immunol. 2017, 17, 545–558. [Google Scholar] [CrossRef]
- Konopiński, M.K. Shannon Diversity Index: A Call to Replace the Original Shannon’s Formula with Unbiased Estimator in the Population Genetics Studies. PeerJ 2020, 8, e9391. [Google Scholar] [CrossRef] [PubMed]
- Avolio, M.L.; Carroll, I.T.; Collins, S.L.; Houseman, G.R.; Hallett, L.M.; Isbell, F.; Koerner, S.E.; Komatsu, K.J.; Smith, M.D.; Wilcox, K.R. A Comprehensive Approach to Analyzing Community Dynamics Using Rank Abundance Curves. Ecosphere 2019, 10, e02881. [Google Scholar] [CrossRef]
- Nijland, R.; Lindner, C.; van Hartskamp, M.; Hamoen, L.W.; Kuipers, O.P. Heterologous Production and Secretion of Clostridium perfringens Beta-Toxoid in Closely Related Gram-Positive Hosts. J. Biotechnol. 2007, 127, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Morrison, C. DNA Vaccines Against Zika Virus Speed into Clinical Trials. Nat. Rev. Drug Discov. 2016, 15, 521–522. [Google Scholar] [CrossRef]
- Lee, L.Y.Y.; Izzard, L.; Hurt, A.C. A Review of DNA Vaccines Against Influenza. Front. Immunol. 2018, 9, 1568. [Google Scholar] [CrossRef]
- Li, L.; Petrovsky, N. Molecular Mechanisms for Enhanced DNA Vaccine Immunogenicity. Expert. Rev. Vaccines 2016, 15, 313–329. [Google Scholar] [CrossRef]
- Remondo, C.; Cereda, V.; Mostböck, S.; Sabzevari, H.; Franzusoff, A.; Schlom, J.; Tsang, K.-Y. Human Dendritic Cell Maturation and Activation by a Heat-Killed Recombinant Yeast (Saccharomyces cerevisiae) Vector Encoding Carcinoembryonic Antigen. Vaccine 2009, 27, 987–994. [Google Scholar] [CrossRef]
- Darby, R.A.J.; Cartwright, S.P.; Dilworth, M.V.; Bill, R.M. Which Yeast Species Shall I Choose? Saccharomyces cerevisiae versus Pichia pastoris. Methods Mol. Biol. 2012, 866, 11–23. [Google Scholar] [CrossRef]
- Kurup, V.M.; Thomas, J. Edible Vaccines: Promises and Challenges. Mol. Biotechnol. 2020, 62, 79–90. [Google Scholar] [CrossRef]
- Eloe-Fadrosh, E.A.; McArthur, M.A.; Seekatz, A.M.; Drabek, E.F.; Rasko, D.A.; Sztein, M.B.; Fraser, C.M. Impact of Oral Typhoid Vaccination on the Human Gut Microbiota and Correlations with S. Typhi-Specific Immunological Responses. PLoS ONE 2013, 8, e62026. [Google Scholar] [CrossRef]
- Zhao, T.; Li, J.; Fu, Y.; Ye, H.; Liu, X.; Li, G.; Yang, X.; Yang, J. Influence of Gut Microbiota on Mucosal IgA Antibody Response to the Polio Vaccine. NPJ Vaccines 2020, 5, 47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, Q.; Zhou, M.; Luo, Z.; Lv, L.; Pei, J.; Wang, C.; Chai, B.; Sui, B.; Huang, F.; et al. Composition of the Murine Gut Microbiome Impacts Humoral Immunity Induced by Rabies Vaccines. Clin. Transl. Med. 2020, 10, e161. [Google Scholar] [CrossRef] [PubMed]
- Praharaj, I.; Parker, E.P.K.; Giri, S.; Allen, D.J.; Silas, S.; Revathi, R.; Kaliappan, S.P.; John, J.; Prasad, J.H.; Kampmann, B.; et al. Influence of Nonpolio Enteroviruses and the Bacterial Gut Microbiota on Oral Poliovirus Vaccine Response: A Study from South India. J. Infect. Dis. 2019, 219, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
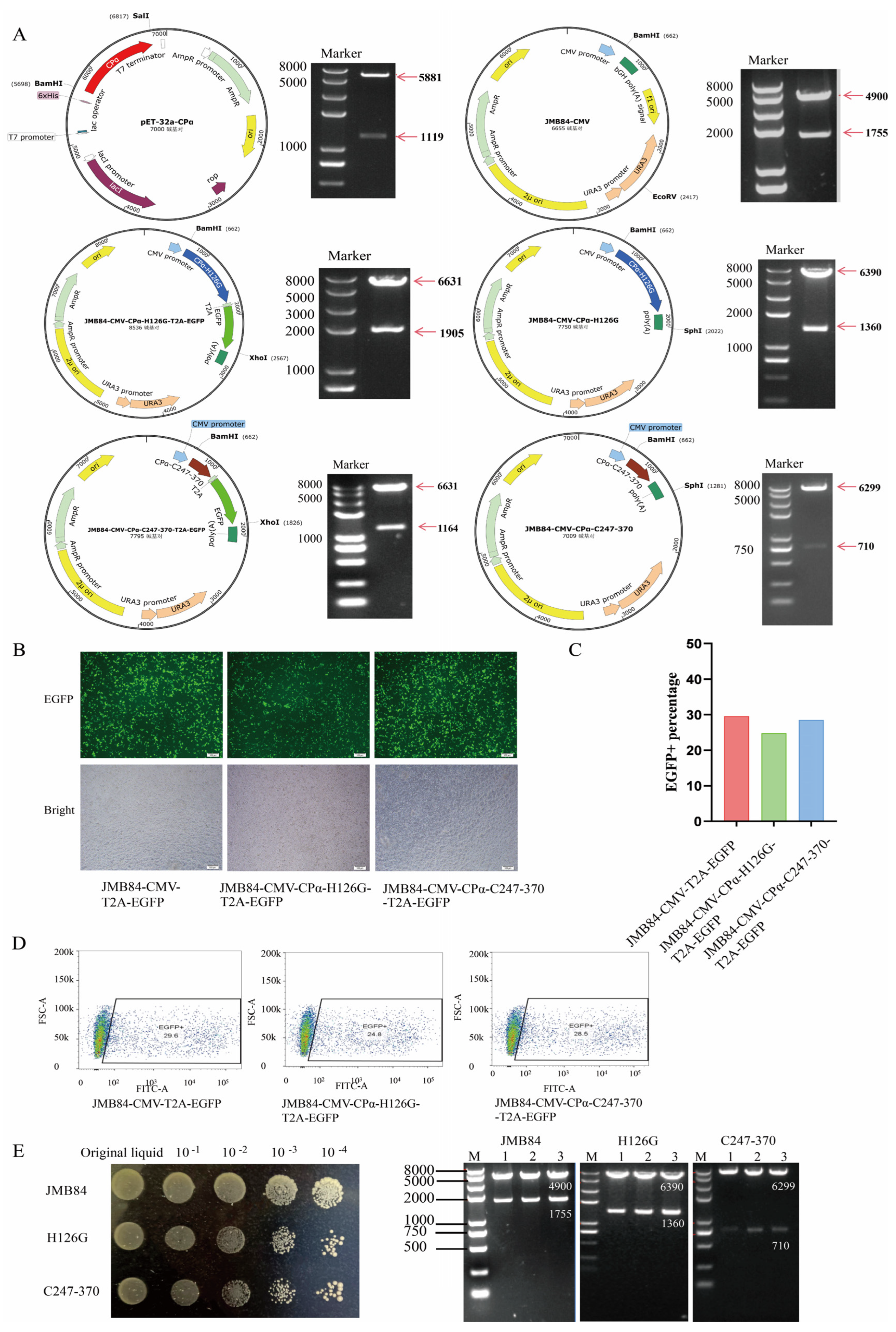

| Plasmids | Relevant Characteristic | Resistance Marker (Reference) |
|---|---|---|
| JMB84-CMV-T2A-EGFP | An empty vector carrying EGFP composed of CMV strong promoter and T2A cleavable peptide | AmpR, URA3; (In our lab) |
| JMB84-CMV-CPα- H126G-T2A-EGFP | Expressed CPα-H126G with EGFP Used to verify expression efficiency | AmpR, URA3; (In this work) |
| JMB84-CMV-CPα- C247-370-T2A-EGFP | Expressed CPα C-terminal domain With EGFP, used to verify expression Efficiency | AmpR,URA3; (In this work) |
| JMB84-CMV | Empty vector without EGFP | AmpR,URA3; (In this work) |
| JMB84-CMV-CPα-H126G | Expressed CPα-H126G without EGFP, used to construct recombinant yeast | AmpR,URA3; (In this work) |
| JMB84-CMV-CPα- C247-370 | Expressed CPα C-terminal domain without EGFP, used to construct recombinant yeast | AmpR,URA3; (In this work) |
| pET-32a | Prokaryotic expression empty vector | AmpR; (In our lab) |
| pET-32a-CPα | Prokaryotic expression vector for producing α-toxin protein | AmpR; (In this work) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, L.; Jia, S.; Zhang, W.; Cai, C.; Liu, Y.; Wang, C.; Zhu, Y.; Ma, X.; Yang, X.; Wei, Z.; et al. Oral Yeast-Cell Microcapsule-Mediated DNA Vaccines Against Clostridium perfringens Induce Effective Intestinal Immunity and Modulate Gut Microbiota. Vaccines 2024, 12, 1360. https://doi.org/10.3390/vaccines12121360
Du L, Jia S, Zhang W, Cai C, Liu Y, Wang C, Zhu Y, Ma X, Yang X, Wei Z, et al. Oral Yeast-Cell Microcapsule-Mediated DNA Vaccines Against Clostridium perfringens Induce Effective Intestinal Immunity and Modulate Gut Microbiota. Vaccines. 2024; 12(12):1360. https://doi.org/10.3390/vaccines12121360
Chicago/Turabian StyleDu, Lihong, Shaona Jia, Wenqiang Zhang, Chang Cai, Yufei Liu, Chuhan Wang, Yufei Zhu, Xiaotao Ma, Xiaojun Yang, Zehui Wei, and et al. 2024. "Oral Yeast-Cell Microcapsule-Mediated DNA Vaccines Against Clostridium perfringens Induce Effective Intestinal Immunity and Modulate Gut Microbiota" Vaccines 12, no. 12: 1360. https://doi.org/10.3390/vaccines12121360
APA StyleDu, L., Jia, S., Zhang, W., Cai, C., Liu, Y., Wang, C., Zhu, Y., Ma, X., Yang, X., Wei, Z., & Xu, K. (2024). Oral Yeast-Cell Microcapsule-Mediated DNA Vaccines Against Clostridium perfringens Induce Effective Intestinal Immunity and Modulate Gut Microbiota. Vaccines, 12(12), 1360. https://doi.org/10.3390/vaccines12121360






