Exigency of Plant-Based Vaccine against COVID-19 Emergence as Pandemic Preparedness
Abstract
1. Introduction
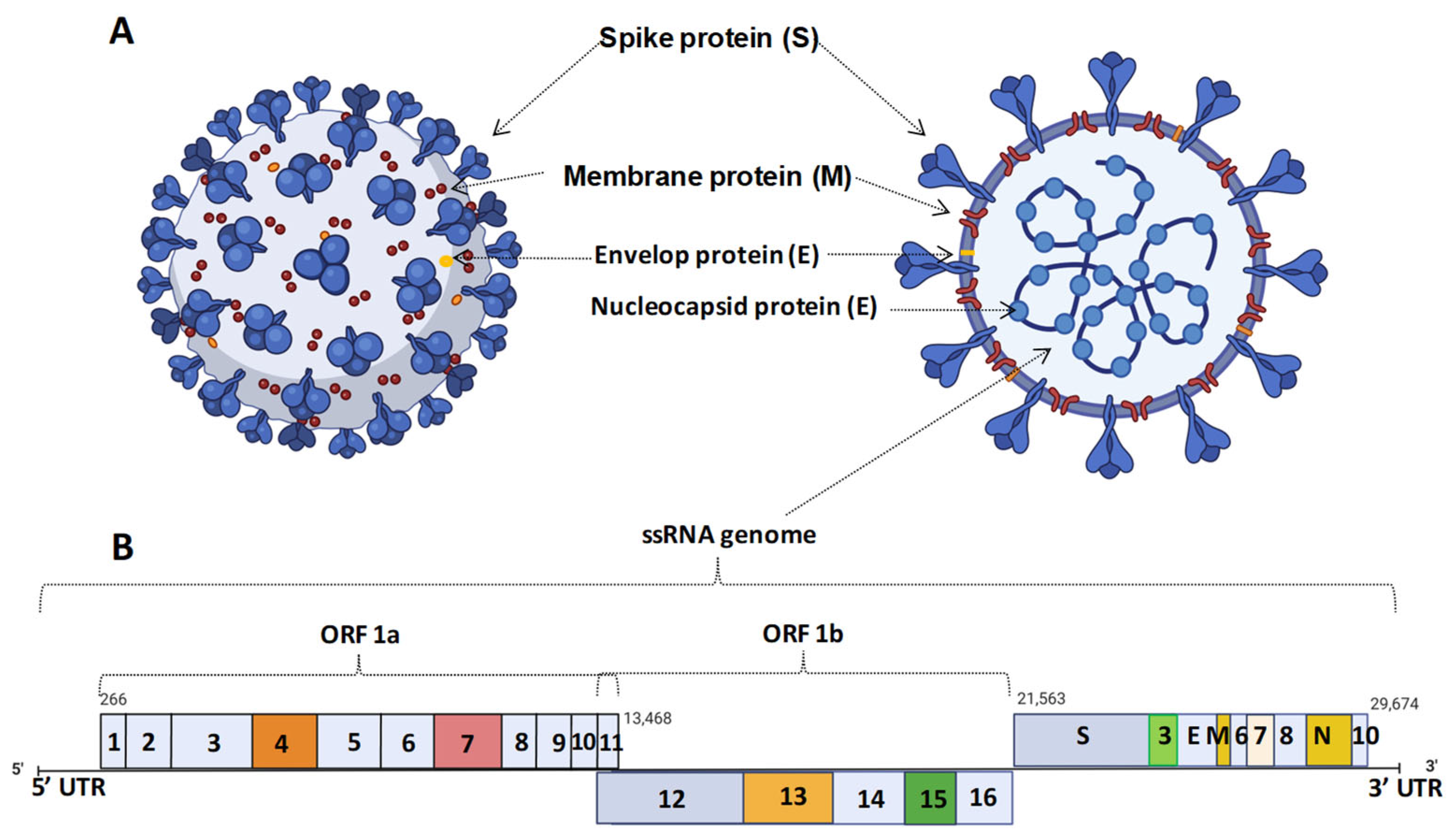
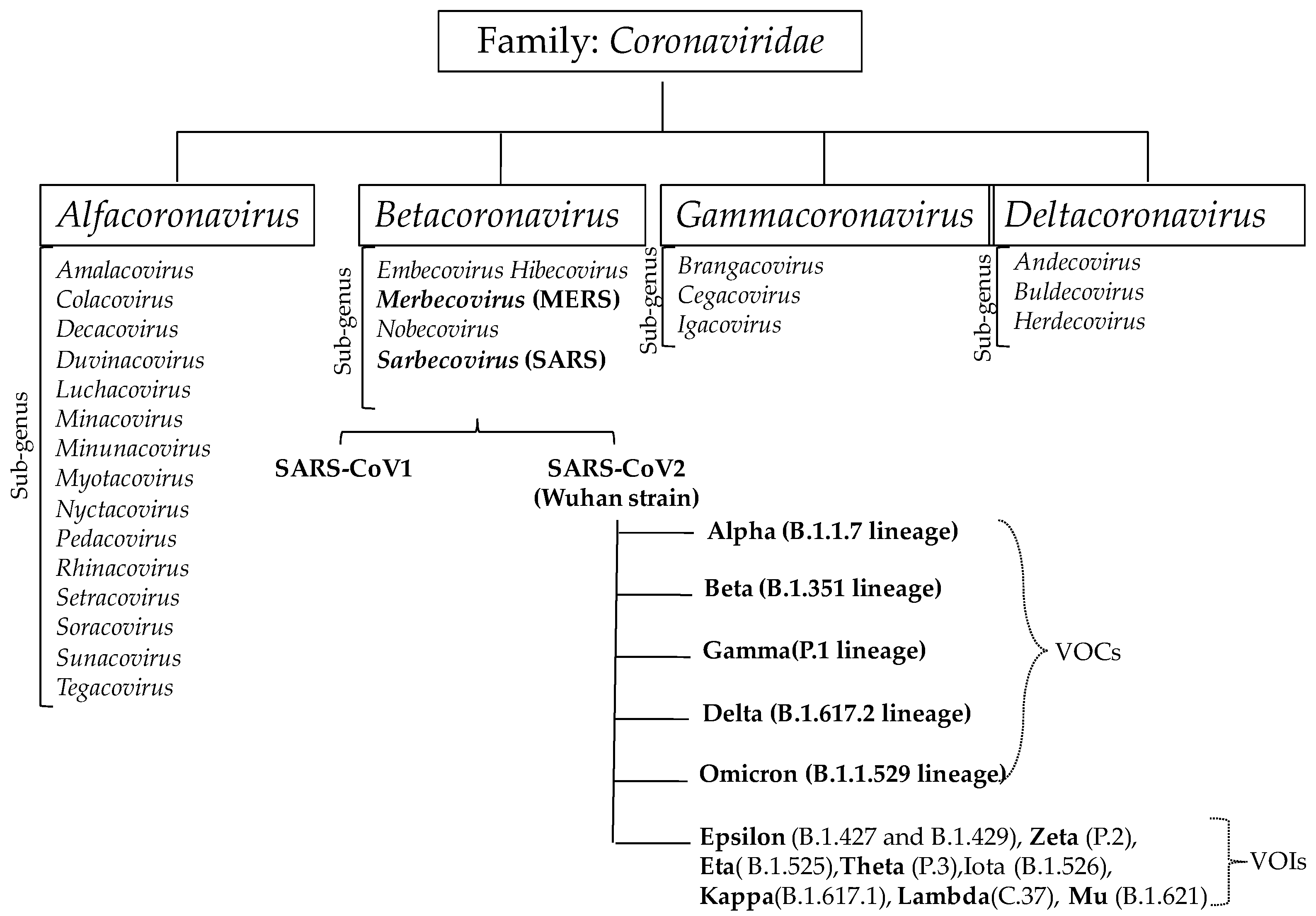
2. Coronavirus Infection and Their Strain Variation
3. Challenges in Vaccine Development during COVID-19 Outbreak
4. Developmental Status of Various Vaccines against COVID-19
4.1. Pathogen-Based Vaccines
4.1.1. Inactivated Pathogen Vaccines
4.1.2. Live-Attenuated Pathogen Vaccine
4.2. Recombinant Vector Vaccines
4.2.1. Replicating Viral Vector-Based Vaccine
4.2.2. Non-Replicating Viral Vectors
4.3. Nucleic Acid-Based Vaccines
4.3.1. DNA Vaccine
4.3.2. RNA Vaccine
4.4. Protein-Based Vaccines
4.4.1. Recombinant Protein Subunit
4.4.2. VLP-Based Vaccines
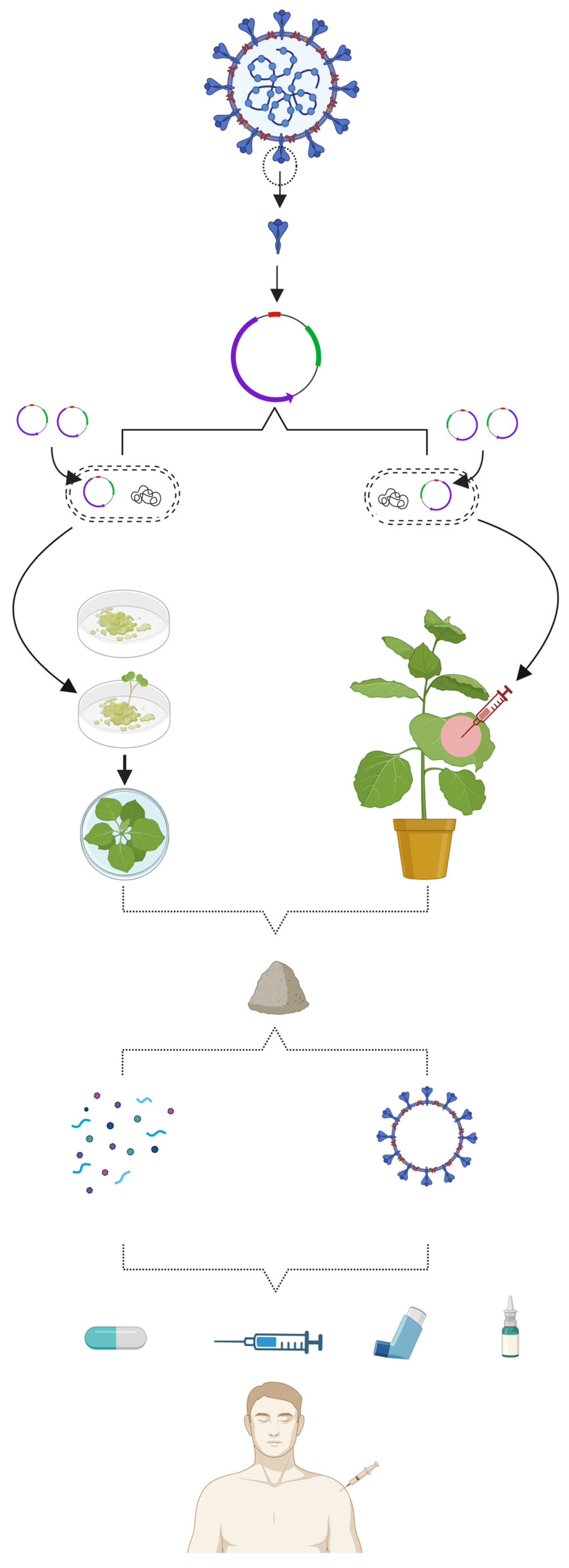
5. Why Is a Plant-Based Platform Preferred for Vaccine Production against COVID-19?
6. Status of Plant-Based Vaccines against COVID-19
6.1. Production of Subunit Vaccines in Plants
6.2. Production of VLP Vaccines in Plants
| Type of Vaccine | Name of Vaccine | Epitope | Production Strategy * | Formulation | Administration | Immune Response | Developer | Development Phase | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Recombinant protein subunit | Subunit protein | Spike (S) protein | iBio’s Fast-Pharming facility (VLPExpress™) | - | - | - | iBio Inc. (Bryan, TX, USA) and CC-Pharming Ltd. (Beijing, China) | Pre-Clinical | [234] |
| IBIO-201 | Spike (S)protein | - | Formulated with LicKMTM adjuvant | Intramuscular and intranasal | Immune response to SARS-CoV-2 | iBio Inc. (Bryan, TX, USA) | Pre-Clinical | [236,237] | |
| Baiya SARS-CoV-2 Vax 1 | - | - | Formulated with alum adjuvant | - | Immune responsein mice and monkeys after two doses | Baiya Phytopharm Co., Ltd. (Bangkok, Thailand) | Clinical trial (Phase I) | [238] | |
| Subunit protein | Antigen cocktail (S, N, N+RBD protein) | - | Antigen cocktailinAlhydrogel adjuvant | IM immunization at0 and 21days | Elicitation of antibodies in mice | Akdeniz University, (Antalya, Turkey) | Pre-clinical | [191] | |
| Subunit protein | S-protein | NXS/T Generation™ platform | glycan-enhanced (GE) Spike protein | Intranasal delivery | Broad spectrum immunity in hamsters against SARS-CoV-2 variants | University of Cape Town, (Cape Town, South Africa) | Pre-clinical | [187] | |
| Recombinant Protein | S1 protein with the Fc domain of human immunoglobulin G1 (IgG1) | Geminiviral expression vector | S1-Fc protein formulated withaluminum hydroxide gel(adjuvant) | IM injection at0 and 21 days | RBD specific antibodies and T cell immune responses in mice | Chulalongkorn University (Bangkok, Thailand) | Pre-clinical | [189] | |
| Recombinant vaccine | Receptor-binding domain | - | - | - | Formation of neutralizing antibodies against Wuhan strain | G+Plus Life Science Co., Ltd. (Seoul, Republic of Korea) | Pre-clinical | [239] | |
| Virus like particle (VLPs) | Covifenz | S protein | Proficia® Technology | Recombinant VLP adjuvanted withAS03 | IM injection with 2 dosesin 21 days apart | Interferon gamma (IFN-γ) and IL-4 responses | Medicago Inc., (Quebec, Canada) in collaboration with Glaxo-Smith Kline (Middlesex, United Kingdom) | Approved in canada; Clinical trial in Argentina, Brazil, U.K.Ireland, USA, Japan | [188] |
| IBIO-200 (glycosylated/non-glycosylatedVLPs) | S protein | FastPharming System | Special adjuvant (of IDRI) | - | Specific immunity in mice | iBio, Inc. (Bryan, TX, USA) & Infectious Disease Research Institute (Seattle, WA, USA) | Pre-Clinical | [234] | |
| IBIO-202 | N protein | FastPharming System | Special adjuvant | Intramuscular and Intranasal | broader range defense against SARS-CoV2variants | iBio Inc. (Bryan, TX, USA) | Pre-Clinical | [237] | |
| KBP 201 | RBD of S protein fused to a human IgG1 Fc domain | Tobacco mosaic virus (TMV)-based expression systm | Chimeric VLPsconjugated with CpG 1018 (adjuvant) | Intramuscular injection at an interval of 21 days | Specific immunity in pre-clinical trials | Kentucky Bioprocessing Inc. (Owensboro, KY, USA) | Clinicaltrial (Phase II) | [240] | |
| VLPcandidates displaying the S-protein | spike (S) surface glycoprotein | - | VLPs mixed with three adjuvants i.e., oil-in-water, synthetic oligodeoxynucleotide (ODN) | Two doses at 0 and 21 days | Elicitation of antibodies to cross-neutralise Delta (B.1.617.2) and Omicron variant in white rabbits | Council for Scientific and Industrial Research, (Pretoria, South Africa) | Pre-clinicaltrial | [190] |
7. Limitations of Plant-Based Vaccine Production
8. Path Ahead for the Improvement of Plant-Based Platform
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, Y.C.; Chen, C.S.; Chan, Y.J. The outbreak of COVID-19: An overview. J. Chin. Med. Assoc. 2020, 83, 217–220. [Google Scholar] [PubMed]
- World Health Organization. WHO COVID-19 Dashboard; WHO: Geneva, Switzerland, 2020; Available online: https://covid19.who.int/ (accessed on 1 June 2023).
- Pak, A.; Adegboye, O.A.; Adekunle, A.I.; Rahman, K.M.; McBryde, E.S.; Eisen, D.P. Economic consequences of the COVID-19 outbreak: The need for epidemic preparedness. Front. Public Health 2020, 8, 241. [Google Scholar] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel Coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [PubMed]
- Guo, Y.R.; Cao, Q.D.; Hong, Z.S.; Tan, Y.Y.; Chen, S.D.; Jin, H.J.; Tan, K.S.; Wang, D.Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—An update on the status. Mil. Med. Res. 2020, 7, 11. [Google Scholar]
- World Health Organization. Draft Landscape of COVID-19 Candidate Vaccines—11 April 2020. 2020. Available online: https://www.who.int/blueprint/priority-diseases/keyaction/Novel_Coronavirus_Landscape_nCoV_11April,2020.PDF?ua=1 (accessed on 1 April 2020).
- Majumder, J.; Minko, T. Recent developments on therapeutic and diagnostic approaches for COVID-19. AAPS J. 2021, 23, 14. [Google Scholar]
- Kumari, M.; Lu, R.M.; Li, M.C.; Huang, J.-L.; Hsu, F.-F.; Ko, S.-H.; Ke, F.-Y.; Su, S.-C.; Liang, K.-H.; Yuan, J.P.-Y.; et al. A critical overview of current progress for COVID-19: Development of vaccines, antiviral drugs, and therapeutic antibodies. J. Biomed. Sci. 2022, 29, 68. [Google Scholar]
- Vetter, V.; Denizer, G.; Friedland, L.R.; Krishnan, J.; Shapiro, M. Understanding modern-day vaccines: What you need to know. Ann. Med. 2018, 50, 110–120. [Google Scholar] [CrossRef]
- Travieso, T.; Li, J.; Mahesh, S.; Mello, J.D.F.R.E.; Blasi, M. The use of viral vectors in vaccine development. NPJ Vaccines 2022, 7, 75. [Google Scholar]
- Qin, F.; Xia, F.; Chen, H.; Cui, B.; Feng, Y.; Zhang, P.; Chen, J.; Luo, M. A guide to nucleic acid vaccines in the prevention and treatment of infectious diseases and cancers: From basic principles to current applications. Front. Cell Dev. Biol. 2021, 25, 9–633776. [Google Scholar]
- Shang, L.; Cao, B. Protein-based vaccine as the booster dose for adults: Evidence and beyond. Lancet Infect. Dis. 2022, 22, 1515–1517. [Google Scholar] [CrossRef]
- Rauch, S.; Jasny, E.; Schmidt, K.E.; Petsch, B. New vaccine technologies to combat outbreak situations. Front. Immunol. 2018, 9, 1963. [Google Scholar] [CrossRef] [PubMed]
- Brisse, M.; Vrba, S.M.; Kirk, N.; Liang, Y.; Ly, H. Emerging concepts and technologies in vaccine development. Front. Immunol. 2020, 11, 583077. [Google Scholar] [CrossRef]
- Ghattas, M.; Dwivedi, G.; Lavertu, M.; Alameh, M.G. Vaccine technologies and platforms for infectious diseases: Current progress, challenges, and opportunities. Vaccines 2021, 9, 1490. [Google Scholar] [CrossRef] [PubMed]
- Nagy, A.; Alhatlani, B. An overview of current COVID-19 vaccine platforms. Comput. Struct. Biotechnol. J. 2021, 19, 2508–2517. [Google Scholar] [CrossRef]
- Chen, Q.; Lai, H. Plant-derived virus-like particles as vaccines. Hum. Vaccines Immunother. 2013, 9, 26–49. [Google Scholar] [CrossRef] [PubMed]
- Plchova, H.; Cerovska, N.; Vaculik, P.; Moravec, T. Plant viruses as scaffolds for the presentation of vaccine epitopes. Biol. Plant. 2017, 61, 1–12. [Google Scholar] [CrossRef]
- Thuenemann, E.C.; Meyers, A.E.; Verwey, J.; Rybicki, E.P.; Lomonossoff, G.P. A method for rapid production of heteromultimeric protein complexes in plants: Assembly of Protective Bluetongue Virus-Like Particles. Plant Biotechnol. J. 2013, 11, 839–846. [Google Scholar] [CrossRef]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar]
- Shereen, M.A.; Khan, S.; Kazmi, A.; Bashir, N.; Siddique, R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020, 24, 91–98. [Google Scholar] [CrossRef]
- Pal, M.; Berhanu, G.; Desalegn, C.; Kandi, V. Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): An update. Cureus 2020, 12, e7423. [Google Scholar] [CrossRef]
- Hulswit, R.J.G.; de Haan, C.A.M.; Bosch, B.-J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016, 96, 29–57. [Google Scholar]
- Monchatre-Leroy, E.; Boué, F.; Boucher, J.M.; Renault, C.; Moutou, F.; ArGouilh, M.; Umhang, G. Identification of Alpha and Beta Coronavirus in wildlife species in France: Bats, Rodents, Rabbits, and Hedgehogs. Viruses 2017, 9, 364. [Google Scholar] [CrossRef]
- Marchenko, V.; Danilenko, A.; Kolosova, N.; Bragina, M.; Molchanova, M.; Bulanovich, Y.; Gorodov, V.; Leonov, S.; Gudymo, A.; Onkhonova, G.; et al. Diversity of gammacoronaviruses and deltacoronaviruses in wild birds and poultry in Russia. Sci. Rep. 2022, 12, 19412. [Google Scholar] [CrossRef]
- Dhama, K.; Patel, S.K.; Sharun, K.; Pathak, M.; Tiwari, R.; Yatoo, M.I.; Malik, Y.S.; Sah, R.; Rabaan, A.A.; Panwar, P.K.; et al. SARS-CoV-2 jumping the species barrier: Zoonotic lessons from SARS, MERS and recent advances to combat this pandemic virus. Travel Med. Infect. Dis. 2020, 37, 101830. [Google Scholar] [CrossRef] [PubMed]
- Hui, D.S.C.; Zumla, A. Severe Acute Respiratory Syndrome: Historical, epidemiologic, and clinical features. Infect. Dis. Clin. N. Am. 2019, 33, 869–889. [Google Scholar] [CrossRef]
- Peiris, M.; Poon, L.L.M. Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) (Coronaviridae). In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 2, pp. 814–824. [Google Scholar]
- Zhong, N.; Zheng, B.; Li, Y.; Poon, L.; Xie, Z.; Chan, K.; Li, P.; Tan, S. Epidemiology and cause of Severe Acute Respiratory Syndrome (SARS) in Guangdong, People’s Republic of China, in February. Lancet 2003, 362, 1353–1358. [Google Scholar] [CrossRef]
- Wang, N.; Shi, X.; Jiang, L.; Zhang, S.; Wang, D.; Tong, P.; Guo, D.; Fu, L.; Cui, Y.; Liu, X.; et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013, 23, 986. [Google Scholar] [CrossRef] [PubMed]
- AlTakarli, N.S. Emergence of COVID-19 infection: What is known and what is to be expected—Narrative review article. Dubai Med. J. 2020, 3, 13–18. [Google Scholar] [CrossRef]
- Yee, J.; Unger, L.; Zadravecz, F.; Cariello, P.; Seibert, A.; Johnson, M.A.; Fuller, M.J. Novel Coronavirus 2019 (COVID-19): Emergence and Implications for Emergency Care. JACEP Open 2020, 1, 63–69. [Google Scholar] [CrossRef] [PubMed]
- JevšnikVirant, M.; Černe, D.; Petrovec, M.; Paller, T.; Toplak, I. Genetic characterisation and comparison of three human Coronaviruses (HKU1, OC43, 229E) from patients and Bovine Coronavirus (BCoV) from cattle with respiratory disease in Slovenia. Viruses 2021, 13, 676. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Temmam, S.; Vongphayloth, K.; Baquero, E.; Munier, S.; Bonomi, M.; Regnault, B.; Douangboubpha, B.; Karami, Y.; Chrétien, D.; Sanamxay, D.; et al. Bat Coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature 2022, 604, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Lytras, S.; Hughes, J.; Martin, D.; Swanepoel, P.; de Klerk, A.; Lourens, R.; Kosakovsky Pond, S.L.; Xia, W.; Jiang, X.; Robertson, D.L. Exploring the Natural Origins of SARS-CoV-2 in the Light of Recombination. Genome Biol. Evol. 2022, 14, evac018. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E.; Cyranoski, D. China Coronavirus: Six questionsscientists are asking. Nature 2020, 577, 605–607. [Google Scholar] [CrossRef]
- Li, F.; Du, L. MERS Coronavirus: An emerging zoonotic virus. Viruses 2019, 11, 663. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early transmission dynamics in Wuhan, China, of Novel Coronavirus-infected pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.-Y. Genomic Characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef]
- Khailany, R.A.; Safdar, M.; Ozaslan, M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020, 19, 100682. [Google Scholar] [CrossRef]
- Laamarti, M.; Alouane, T.; Kartti, S.; Chemao-Elfihri, M.W.; Hakmi, M.; Essabbar, A.; Laamarti, M.; Hlali, H.; Bendani, H.; Boumajdi, N.; et al. Large scale genomic analysis of 3067 SARS-CoV-2 genomes reveals a clonal geo-distribution and a rich genetic variations of hotspots mutations. PLoS ONE 2020, 15, e0240345. [Google Scholar] [CrossRef]
- Tang, X.; Wu, C.; Li, X.; Song, Y.; Yao, X.; Wu, X.; Duan, Y.; Zhang, H.; Wang, Y.; Qian, Z.; et al. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020, 7, 1012–1023. [Google Scholar] [CrossRef]
- Forster, P.; Forster, L.; Renfrew, C.; Forster, M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. USA 2020, 117, 9241–9243. [Google Scholar] [CrossRef] [PubMed]
- Biswas, N.; Majumder, P. Analysis of RNA sequences of 3636 SARS-CoV-2 collected from 55 countries reveals selective sweep of one virus type. Indian J. Med. Res. 2020, 151, 450–458. [Google Scholar] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef] [PubMed]
- Cochérie, T.; Zafilaza, K.; Leducq, V.; Marot, S.; Calvez, V.; Marcelin, A.G.; Todesco, E. Epidemiology and characteristics of SARS-CoV-2 variants of concern: The impacts of the spike mutations. Microorganisms 2023, 11, 30. [Google Scholar] [CrossRef]
- Yao, H.; Lu, X.; Chen, Q.; Xu, K.; Chen, Y.; Cheng, M.; Chen, K.; Cheng, L.; Weng, T.; Shi, D.; et al. Patient-derived SARS-CoV-2 mutations impact viral replication dynamics and infectivity in vitro and with clinical implications in vivo. Cell Discov. 2020, 6, 76. [Google Scholar] [CrossRef]
- Peka, M.; Balatsky, V. The impact of mutation sets in receptor-binding domain of SARS-CoV-2 variants on the stability of RBD-ACE2 complex. Future Virol. 2023, 18, 225–242. [Google Scholar] [CrossRef]
- Hill, V.; Du Plessis, L.; Peacock, T.P.; Aggarwal, D.; Colquhoun, R.; Carabelli, A.M.; Ellaby, N.; Gallagher, E.; Groves, N.; Jackson, B.; et al. The origins and molecular evolution of SARS-CoV-2 lineage B.1.1.7 in the UK. Virus Evol. 2022, 8, veac080. [Google Scholar] [CrossRef] [PubMed]
- Akkız, H. The biological functions and clinical significance of SARS-CoV-2 variants of concern. Front. Med. 2022, 9, 849217. [Google Scholar] [CrossRef]
- Ahmad, A.; Fawaz, M.A.M.; Aisha, A. A comparative overview of SARS-CoV-2 and its variants of concern. Infect. Med. 2022, 30, 328–343. [Google Scholar]
- Aleem, A.; Akbar Samad, A.B.; Vaqar, S. Emerging Variants of SARS-CoV-2 and Novel Therapeutics against Coronavirus (COVID-19); StatPearls Publishing: St. Petersburg, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK590851 (accessed on 1 June 2023).
- Banho, C.A.; Sacchetto, L.; Campos, G.R.F.; da Silva, R.; Christoff, A.P.; Cabreira, C.C.; Barbosa, J.G.; Rocha, A.P.; Dalmolin, T.V.; Barros, G.B.S.; et al. Impact of SARS-CoV-2 Gamma lineage introduction and COVID-19 vaccination on the epidemiological landscape of a Brazilian City. Commun. Med. 2022, 2, 41. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Plante, K.S.; Plante, J.A.; Xie, X.; Zhang, X.; Ku, Z.; An, Z.; Scharton, D.; Schindewolf, C.; et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 2022, 602, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Yin, H.; Yin, J.Y. B.1.617.2 (Delta) Variant of SARS-CoV-2: Features, transmission and potential strategies. Int. J. Biol. Sci. 2022, 18, 1844–1851. [Google Scholar] [CrossRef] [PubMed]
- Dhawan, M.; Sharma, A.; Priyanka, P.; Thakur, N.; Rajkhowa, T.K.; Choudhary, O.P. Delta variant (B.1.617.2) of SARS-CoV-2: Mutations, impact, challenges and possible solutions. Hum. Vaccin. Immunother. 2022, 18, 2068883. [Google Scholar] [CrossRef]
- Xia, S.; Wang, L.; Zhu, Y.; Lu, L.; Jiang, S. Origin, virological features, immune evasion and intervention of SARS-CoV-2 Omicron sublineages. Signal Transduct. Target. Ther. 2022, 7, 241. [Google Scholar] [CrossRef]
- MohseniAfshar, Z.; TavakoliPirzaman, A.; Karim, B.; Anaraki, S.R.; Hosseinzadeh, R.; Pireivatlou, E.S.; Babazadeh, A.; Hosseinzadeh, D.; Miri, S.R.; Sio, T.T.; et al. SARS-CoV-2 Omicron (B.1.1.529) variant: A challenge with COVID-19. Diagnostics 2023, 13, 559. [Google Scholar] [CrossRef]
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- Callaway, E. ‘A Bloody Mess’: Confusion reigns over naming of new COVID variants. Nature 2021, 589, 339. [Google Scholar] [CrossRef]
- Duong, D. Alpha, Beta, Delta, Gamma: What’s important to know about SARS-CoV-2 variants of concern? CMAJ 2021, 193, E1059–E1060. [Google Scholar] [CrossRef] [PubMed]
- Iketani, S.; Liu, L.; Guo, Y.; Liu, L.; Chan, J.F.; Huang, Y.; Wang, M.; Luo, Y.; Yu, J.; Chu, H.; et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature 2022, 604, 553–556. [Google Scholar] [CrossRef]
- Wang, L.; Møhlenberg, M.; Wang, P.; Zhou, H. Immune evasion of neutralizing antibodies by SARS-CoV-2 Omicron. Cytokine Growth Factor Rev. 2023, 70, 13–25. [Google Scholar] [CrossRef]
- Plotkin, S. History of vaccination. Proc. Natl. Acad. Sci. USA 2014, 111, 12283–12287. [Google Scholar] [CrossRef]
- Hajj Hussein, I.; Chams, N.; Chams, S.; El Sayegh, S.; Badran, R.; Raad, M.; Gerges-Geagea, A.; Leone, A.; Jurjus, A. Vaccines through centuries: Major cornerstones of global health. Front. Public Health 2015, 3, 269. [Google Scholar] [CrossRef]
- Pollard, A.J.; Bijker, E.M. A guide to vaccinology: From basic principles to new developments. Nat. Rev. Immunol. 2021, 21, 83–100. [Google Scholar] [CrossRef]
- Sanders, B.; Koldijk, M.; Schuitemaker, H. Inactivated viral vaccines. In Vaccine Analysis: Strategies, Principles, and Control; Nunnally, B.K., Turula, V.E., Sitrin, R.D., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 45–80. [Google Scholar]
- Shah, S.; Baral, B.; Chamlagain, R.; Murarka, H.; Raj Adhikari, Y.; Sharma Paudel, B. Reactivation of herpes zoster after vaccination with an inactivated vaccine: A case report from Nepal. Clin. Case Rep. 2021, 9, e05188. [Google Scholar] [CrossRef]
- Excler, J.L.; Saville, M.; Berkley, S.; Kim, J.H. Vaccine development for emerging infectious diseases. Nat. Med. 2021, 27, 591–600. [Google Scholar] [CrossRef]
- Palatnik-de-Sousa, C.B.; Soares, I.S.; Rosa, D.S. Editorial: Epitope discovery and synthetic vaccine design. Front. Immunol. 2018, 9, 826. [Google Scholar] [CrossRef]
- Havasi, A.; Visan, S.; Cainap, C.; Cainap, S.S.; Mihaila, A.A.; Pop, L.A. Influenza A, Influenza B, and SARS-CoV-2 similarities and differences—A focus on diagnosis. Front. Microbiol. 2022, 13, 908525. [Google Scholar] [CrossRef]
- Egidi, V.; Manfredi, P. Population dynamics and demography of COVID-19 introduction. Genus 2021, 77, 36. [Google Scholar] [CrossRef]
- Ou, J.; Zhou, Z.; Dai, R.; Zhang, J.; Zhao, S.; Wu, X.; Lan, W.; Ren, Y.; Cui, L.; Lan, Q.; et al. V367F Mutation in SARS-CoV-2 spike RBD emerging during the early transmission phase enhances viral infectivity through increased human ACE2 receptor binding affinity. J. Virol. 2021, 95, e0061721. [Google Scholar] [CrossRef]
- Gillim-Ross, L.; Subbarao, K. Emerging respiratory viruses: Challenges and vaccine strategies. Clin. Microbiol. Rev. 2006, 19, 614–636. [Google Scholar] [CrossRef]
- Kanekiyo, M.; Ellis, D.; King, N.P. New vaccine design and delivery technologies. J. Infect. Dis. 2019, 219 (Suppl. 1), S88–S96. [Google Scholar] [CrossRef]
- Du, L.; Jiang, S. Middle East Respiratory Syndrome: Current status and future prospects for vaccine development. Expert Opin. Biol. Ther. 2015, 15, 1647–1651. [Google Scholar] [CrossRef]
- Enjuanes, L.; Zuñiga, S.; Castaño-Rodriguez, C.; Gutierrez-Alvarez, J.; Canton, J.; Sola, I. Molecular basis of coronavirus virulence and vaccine development. Adv. Virus Res. 2016, 96, 245–286. [Google Scholar]
- Zhang, N.; Jiang, S.; Du, L. Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev. Vaccines 2014, 13, 761–774. [Google Scholar] [CrossRef]
- Khoshnood, S.; Arshadi, M.; Akrami, S.; Ranjbar, R.; Erfani, A.; Kooshavar, H.; Poormoghadam, D.; Ghasemnejad, A.; Dadashi, A.; Hassani, A.; et al. An overview on inactivated and live attenuated SARS-CoV-2 vaccines. J. Clin. Lab. Anal. 2022, 36, e24418. [Google Scholar] [CrossRef]
- Elveborg, S.; Monteil, V.M.; Mirazimi, A. Methods of inactivation of highly pathogenic viruses for molecular, serology or vaccine development purposes. Pathogens 2022, 11, 271. [Google Scholar] [CrossRef] [PubMed]
- Lopez, S.M.C.; Sato, A.I.; Chatterjee, A. Vaccines: An overview. In Viral, Parasitic, Bacterial, and Fungal Infections: Antimicrobial, Host Defense, and Therapeutic Strategies; Bagchi, D., Das, A., Downs, B.W., Eds.; Elsevier Science: Amsterdam, The Netherlands, 2022; pp. 699–717. [Google Scholar]
- Shouval, D. Immunization against Hepatitis A. Cold Spring Harb. Perspect. Med. 2019, 9, a031682. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.R.; Flannery, B.; Ambrose, C.S.; Bégué, R.E.; Caspard, H.; DeMarcus, L.; Fowlkes, A.L.; Kersellius, G.; Steffens, A.; Fry, A.M. Influenza Clinical Investigation for Children Study Team; Influenza Incidence Surveillance Project; US Influenza Vaccine Effectiveness Network. Live Attenuated and Inactivated Influenza Vaccine Effectiveness. Pediatrics 2019, 143, e20182094. [Google Scholar] [CrossRef] [PubMed]
- Moreira, B.L.C.; Gimenez, A.P.L.; Inagaki, J.M.F.; Raboni, S.M. Inactivated rabies vaccines: Standardization of an in vitro assay for residual viable virus detection. PLoS Neglected Trop. Dis. 2020, 14, e0008142. [Google Scholar] [CrossRef]
- Gao, Q.; Bao, L.; Mao, H.; Wang, L.; Xu, K.; Yang, M.; Li, Y.; Zhu, L.; Wang, N.; Lv, Z.; et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020, 369, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.B.; Lai, D.Y.; Jiang, H.W.; Zheng, X.K.; Wang, W.X.; Zhao, Y.; Luo, Y.; Yang, X.P.; Liu, Y.Q.; Zhang, B.; et al. Landscape of the RBD-Specific IgG, IgM, and IgA Responses triggered by the inactivated virus vaccine against the Omicron variant. Cell Discov. 2022, 8, 15. [Google Scholar] [CrossRef]
- Nouailles, G.; Adler, J.M.; Pennitz, P.; Peidli, S.; Alves, L.G.T.; Baumgardt, M.; Bushe, J.; Voss, A.; Langenhagen, A.; Langner, C.; et al. Live-attenuated vaccine sCPD9 elicits superior mucosal and systemic immunity to SARS-CoV-2 variants in hamsters. Nat. Microbiol. 2023, 8, 860–874. [Google Scholar] [CrossRef]
- Minor, P.D. Live attenuated vaccines: Historical successes and current challenges. Virology 2015, 479–480, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Mehla, R.; Kokate, P.; Bhosale, S.R.; Kumar, P.; Patil, S.; Shelar, B.; Yadav, P.; Deshmukh, M.; Gangakhedkar, R.R. A live attenuated COVID-19 candidate vaccine for children: Protection against SARS-CoV-2 challenge in Hamsters. Vaccines 2023, 11, 255. [Google Scholar] [CrossRef]
- Menachery, V.D.; Dinnon, K.H.; Yount, B.L.; McAnarney, E.T.; Gralinski, L.E.; Hale, A.; Graham, R.L.; Scobey, T.; Anthony, S.J.; Wang, L.; et al. MERS-CoV accessory ORFs play key role for infection and pathogenesis. mBio 2017, 8, e00665-17. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Gralinski, L.E.; Mitchell, H.D.; Dinnon, K.H.; Leist, S.R.; Yount, B.L.; McAnarney, E.T.; Graham, R.L.; Waters, K.M.; Baric, R.S. Middle East Respiratory Syndrome Coronavirus nonstructural protein 16 is necessary for interferon resistance and viral pathogenesis. mSphere 2017, 2, e00346-17. [Google Scholar] [CrossRef] [PubMed]
- Myeni, S.K.; Bredenbeek, P.J.; Knaap, R.C.M.; Linster, M.; Koopmans, M.P.G.; van Dinten, L.C.; de Graaf, M.; Rockx, B.; Beer, M.; Al Abdulla, F.; et al. Engineering potent live attenuated coronavirus vaccines by targeted inactivation of the immune evasive viral deubiquitinase. Nat. Commun. 2023, 14, 1141. [Google Scholar] [CrossRef]
- Chen, J.; Wang, P.; Yuan, L.; Li, S.; Yang, H.; Wang, C.; Zhang, X.; Ma, D.; Li, W.; Ma, L.; et al. A live attenuated virus-based intranasal COVID-19 vaccine provides rapid, prolonged, and broad protection against SARS-CoV-2. Sci. Bull. 2022, 67, 1372–1387. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, C.; Song, Y.; Coleman, J.R.; Stawowczyk, M.; Tafrova, J.; Tasker, S.; Boltz, D.; Baker, R.; Garcia, L.; et al. Scalable live-attenuated SARS-CoV-2 vaccine candidate demonstrates preclinical safety and efficacy. Proc. Natl. Acad. Sci. USA 2021, 118, e2102775118. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.-H.; Jang, Y. Cold-adapted live attenuated SARS-CoV-2 vaccine completely protects human ACE2 transgenic mice from SARS-CoV-2 infection. Vaccines 2020, 8, 584. [Google Scholar] [CrossRef]
- Xu, J.; Liu, M.; Niu, X.; Chen, X.; Wang, D.; Zhang, J.; Zhang, Y.; Li, Y.; Yao, G.; Zhao, P.; et al. The cold-adapted, temperature-sensitive SARS-CoV-2 strain TS11 is attenuated in Syrian Hamsters and a candidate attenuated vaccine. Viruses 2022, 15, 95. [Google Scholar] [CrossRef]
- Tang, P.C.H.; Ng, W.H.; King, N.J.C.; Mahalingam, S. Can live-attenuated SARS-CoV-2 vaccine contribute to stopping the pandemic? PLoS Pathog. 2022, 18, e1010821. [Google Scholar] [CrossRef]
- Pacheco-García, U.; Serafín-López, J. Indirect dispersion of SARS-CoV-2 live-attenuated vaccine and its contribution to herd immunity. Vaccines 2023, 11, 655. [Google Scholar] [CrossRef]
- Haddad, D.; John, S.E.; Mohammad, A.; Hammad, M.M.; Hebbar, P.; Channanath, A.; Deeb, A.; Abubaker, J.; Abu-Farha, M.; Al-Mulla, F. SARS-CoV-2: Possible recombination and emergence of potentially more virulent strains. PLoS ONE 2021, 16, e0251368. [Google Scholar] [CrossRef]
- He, D.C.; He, C.Q. Discovery of Vaccine-Like Recombinant SARS-CoV-2 Circulating in Humans. Virol. J. 2022, 19, 209. [Google Scholar] [CrossRef]
- Halstead, S.B.; Marchette, N.J.; Diwan, A.R.; Palumbo, N.E.; Putvatana, R.; Larsen, L.K. Selection of Attenuated Dengue 4 Viruses by Serial Passage in Primary Kidney Cells. III. Reversion to Virulence by Passage of Cloned Virus in Fetal Rhesus Lung Cells. Am. J. Trop. Med. Hyg. 1984, 33, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.J.; Nuismer, S.L.; Antia, R. Recombinant Vector Vaccine Evolution. PLoS Comput. Biol. 2019, 15, e1006857. [Google Scholar] [CrossRef] [PubMed]
- Shirley, J.L.; de Jong, Y.P.; Terhorst, C.; Herzog, R.W. Immune Responses to Viral Gene Therapy Vectors. Mol. Ther. 2020, 28, 709–722. [Google Scholar] [CrossRef]
- Rabinowitz, J.; Chan, Y.K.; Samulski, R.J. Adeno-Associated Virus (AAV) versus Immune Response. Viruses 2019, 11, 102. [Google Scholar] [CrossRef] [PubMed]
- Robert-Guroff, M. Replicating and Non-Replicating Viral Vectors for Vaccine Development. Curr. Opin. Biotechnol. 2007, 18, 546–556. [Google Scholar] [CrossRef]
- Cai, X.; Bai, H.; Zhang, X. Vaccines and Advanced Vaccines: A Landscape for Advanced Vaccine Technology against Infectious Disease, COVID-19, and Tumor. OSF Prepr. 2020. [Google Scholar] [CrossRef]
- Saxena, M.; Van, T.T.H.; Baird, F.J.; Coloe, P.J.; Smooker, P.M. Pre-existing immunity against vaccine vectors-friend or foe? Microbiology 2013, 159, 1–11. [Google Scholar] [CrossRef]
- Liniger, M.; Zuniga, A.; Naim, H.Y. Use of Viral Vectors for the Development of Vaccines. Expert Rev. Vaccines 2007, 6, 255–266. [Google Scholar] [CrossRef]
- Chavda, V.P.; Bezbaruah, R.; Athalye, M.; Parikh, P.K.; Chhipa, A.S.; Patel, S.; Apostolopoulos, V. Replicating Viral Vector-Based Vaccines for COVID-19: Potential Avenue in Vaccination Arena. Viruses 2022, 14, 759. [Google Scholar] [CrossRef]
- Catanzaro, A.T.; Koup, R.A.; Roederer, M.; Bailer, R.T.; Enama, M.E.; Moodie, Z.; Gu, L.; Martin, J.E.; Novik, L.; Chakrabarti, B.K.; et al. Phase I Safety and Immunogenicity Evaluation of a Multiclade HIV-1 Candidate Vaccine Delivered by a Replication-Defective Recombinant Adenovirus Vector. J. Infect. Dis. 2006, 194, 1638–1649. [Google Scholar] [CrossRef]
- García-Arriaza, J.; Garaigorta, U.; Pérez, P.; Lázaro-Frías, A.; Zamora, C.; Gastaminza, P.; Del Fresno, C.; Casasnovas, J.M.; Sorzano, C.Ó.S.; Sancho, D.; et al. COVID-19 Vaccine Candidates Based on Modified Vaccinia Virus Ankara Expressing the SARS-CoV-2 Spike Protein Induce Robust T- and B-Cell Immune Responses and Full Efficacy in Mice. J. Virol. 2021, 95, e02260-20. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liang, B.; Wang, W.; Li, L.; Feng, N.; Zhao, Y.; Wang, T.; Yan, F.; Yang, S.; Xia, X. Viral vectored vcaccines: Design, development, preventive and therapeutic applications in human diseases. Signal Transduct. Target. Ther. 2023, 8, 149. [Google Scholar] [CrossRef]
- Mendonça, S.A.; Lorincz, R.; Boucher, P.; Zhang, C.; Penninger, J.M. Adenoviral Vector Vaccine Platforms in the SARS-CoV-2 Pandemic. NPJ Vaccines 2021, 6, 97. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Liang, H.; Chen, P.; Li, Y.; Li, Z.; Fan, S.; Wu, K.; Li, X.; Chen, W.; Qin, Y.; et al. Viral Vector Vaccine Development and Application during the COVID-19 Pandemic. Microorganisms 2022, 10, 1450. [Google Scholar] [CrossRef]
- Cattaneo, M. Thrombosis with thrombocytopenia syndrome associated with viral vector COVID-19 vaccines. Eur. J. Intern. Med. 2021, 89, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ye, Q. Nucleic acid vaccines against SARS-CoV-2. Vaccines 2022, 10, 1849. [Google Scholar] [CrossRef]
- Callendret, B.; Lorin, V.; Charneau, P.; Marianneau, P.; Contamin, H.; Betton, J.M.; van der Werf, S.; Escriou, N. Heterologous viral RNA export elements improve expression of Severe Acute Respiratory Syndrome (SARS) coronavirus spike protein and protective efficacy of DNA vaccines against SARS. Virology 2007, 363, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Maruggi, G.; Shan, H.; Li, J. Advances in mRNA vaccines for infectious diseases. Front. Immunol. 2019, 10, 594. [Google Scholar] [CrossRef]
- Kutzler, M.A.; Weiner, D.B. DNA vaccines: Ready for prime time? Nat. Rev. Genet. 2008, 9, 776–788. [Google Scholar] [CrossRef]
- Zhao, X.; Long, J.; Liang, F.; Liu, N.; Sun, Y.; Xi, Y. Dynamic profiles, biodistribution and integration evaluation after intramuscular/intravenous delivery of a novel therapeutic DNA vaccine encoding chicken type II collagen for rheumatoid arthritis in vaccinated normal rodent. J. Nanobiotechnol. 2019, 17, 94. [Google Scholar] [CrossRef]
- Silveira, M.M.; Moreira, G.M.S.G.; Mendonça, M. DNA vaccines against COVID-19: Perspectives and challenges. Life Sci. 2021, 267, 118919. [Google Scholar] [CrossRef] [PubMed]
- Chavda, V.P.; Pandya, R.; Apostolopoulos, V. DNA vaccines for SARS-CoV-2: Toward third-generation vaccination era. Expert Rev. Vaccines 2021, 20, 1549–1560. [Google Scholar] [CrossRef] [PubMed]
- Ledesma-Feliciano, C.; Chapman, R.; Hooper, J.W.; Elma, K.; Zehrung, D.; Brennan, M.B.; Spiegel, E.K. Improved DNA vaccine delivery with needle-free injection systems. Vaccines 2023, 11, 280. [Google Scholar] [CrossRef]
- Bennett, R.P.; Dalby, B.; Guy, P.M. Protein delivery using VP22. Nat. Biotechnol. 2002, 20, 20. [Google Scholar] [CrossRef]
- Yang, Z.Y.; Kong, W.P.; Huang, Y.; Roberts, A.; Murphy, B.R.; Subbarao, K.; Nabel, G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature 2004, 428, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Khoshnood, S.; Ghanavati, R.; Shirani, M.; Ghahramanpour, H.; Sholeh, M.; Shariati, A.; Sadeghifard, N.; Heidary, M. Viral vector and nucleic acid vaccines against COVID-19: A narrative review. Front. Microbiol. 2022, 13, 984536. [Google Scholar] [CrossRef]
- BioNTech. mRNA Therapeutics. Available online: https://biontech.de/how-we-translate/mrna-therapeutics (accessed on 17 April 2020).
- Deering, R.P.; Kommareddy, S.; Ulmer, J.B.; Brito, L.A.; Geall, A.J. Nucleic Acid vaccines: Prospects for non-viral delivery of mRNA vaccines. Expert Opin. Drug Deliv. 2014, 11, 885–899. [Google Scholar] [CrossRef] [PubMed]
- Tews, B.A.; Meyers, G. Self-replicating RNA. Methods Mol. Biol. 2017, 1499, 15–35. [Google Scholar] [PubMed]
- Lundstrom, K. Replicon RNA viral vectors as vaccines. Vaccines 2016, 4, 39. [Google Scholar] [CrossRef]
- Schmidt, C.; Schnierle, B.S. Self-amplifying RNA vaccine candidates: Alternative platforms for mRNA vaccine development. Pathogens 2023, 12, 138. [Google Scholar] [CrossRef]
- Fang, E.; Liu, X.; Li, M.; Zhang, Z.; Song, L.; Zhu, B.; Wu, X.; Liu, J.; Zhao, D.; Li, Y. Advances in COVID-19 mRNA vaccine development. Signal Transduct. Target. Ther. 2022, 7, 94. [Google Scholar] [CrossRef]
- Fraiman, J.; Erviti, J.; Jones, M.; Greenland, S.; Whelan, P.; Kaplan, R.M.; Doshi, P. Serious adverse events of special interest following mRNA COVID-19 vaccination in randomized trials in adults. Vaccine 2022, 40, 5798–5805. [Google Scholar] [CrossRef]
- Freise, N.F.; Kivel, M.; Grebe, O.; Andree, M.; Röhmel, J.; Debus, O.; Hausler, S.F.M. Acute cardiac side effects after COVID-19 mRNA vaccination: A case series. Eur. J. Med. Res. 2022, 27, 80. [Google Scholar] [CrossRef] [PubMed]
- Hotez, P.J.; Bottazzi, M.E. Whole inactivated virus and protein-based COVID-19 vaccines. Annu. Rev. Med. 2022, 73, 55–64. [Google Scholar] [CrossRef]
- Cid, R.; Bolívar, J. Platforms for production of protein-based vaccines: From classical to next-generation strategies. Biomolecules 2021, 11, 1072. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, S.; Wang, Y. Recent advances in the production of recombinant subunit vaccines in Pichia pastoris. Bioengineered 2016, 7, 155–165. [Google Scholar] [CrossRef]
- Bill, R.M. Recombinant protein subunit vaccine synthesis in microbes: A role for yeast? J. Pharm. Pharmacol. 2015, 67, 319–328. [Google Scholar] [CrossRef]
- Liu, X.; Song, H.; Jiang, J.; Gao, X.; Yi, Y.; Shang, Y.; Li, J.; Li, D.; Zeng, Z.; Li, Y.; et al. Baculovirus-expressed self-assembling SARS-CoV-2 nanoparticle vaccines targeting the S protein induce protective immunity in mice. Process Biochem. 2023, 129, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Bisht, H.; Roberts, A.; Vogel, L.; Subbarao, K.; Moss, B. Neutralizing antibody and protective immunity to SARS coronavirus infection of mice induced by a soluble recombinant polypeptide containing an N-terminal segment of the spike glycoprotein. Virology 2005, 334, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Parums, D.V. Editorial: First approval of the protein-based adjuvanted Nuvaxovid (NVX-CoV2373) Novavax vaccine for SARS-CoV-2 could increase vaccine uptake and provide immune protection from viral variants. Med. Sci. Monit. 2022, 28, e936523-1–e936523-3. [Google Scholar] [CrossRef]
- Zeltins, A. Construction and characterization of virus-like particles: A review. Mol. Biotechnol. 2013, 53, 92–107. [Google Scholar] [CrossRef]
- Urakami, A.; Sakurai, A.; Ishikawa, M.; Yap, M.L.; Flores-Garcia, Y.; Haseda, Y.; Aoshi, T.; Zavala, F.P.; Rossmann, M.G.; Kuno, S.; et al. Development of a novel virus-like particle vaccine platform that mimics the immature form of Alphavirus. Vaccines 2017, 24, e00090-17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L. Multi-epitope vaccines: A promising strategy against tumors and viral infections. Vaccines 2018, 15, 182–184. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Chen, J.Y.; Chen, H.W.; Hu, C.J. Nanoparticle vaccines adopting virus-like features for enhanced immune potentiation. Vaccines 2017, 1, 244–260. [Google Scholar] [CrossRef]
- Chen, H.W.; Huang, C.Y.; Lin, S.Y.; Fang, Z.-S.; Hsu, C.-H.; Lin, J.-C.; Chen, Y.-I.; Yao, B.-Y.; Hu, C.-M.J. Synthetic virus-like particles prepared via protein corona formation enable effective vaccination in an avian model of coronavirus infection. Vaccines 2016, 106, 111–118. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, Z.Y.; Kong, W.P.; Nabel, G.J. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: Implications for assembly and vaccine production. Vaccines 2004, 78, 12557–12565. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Huang, Y.; Huang, L.; Li, B.; Zheng, Z.; Chen, Z.; Chen, J.; Hu, Q.; Wang, H. Effect of mucosal and systemic immunization with virus-like particles of severe acute respiratory syndrome coronavirus in mice. Vaccines 2010, 130, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Tariq, H.; Batool, S.; Asif, S.; Ali, M.; Abbasi, B.H. Virus-like particles: Revolutionary platforms for developing vaccines against emerging infectious diseases. Vaccines 2022, 12, 790121. [Google Scholar] [CrossRef]
- Wong, T.Y.; Russ, B.P.; Lee, K.S.; Miller, O.A.; Kang, J.; Cooper, M.; Winters, M.T.; Rodriguez-Aponte, S.A.; Dalvie, N.C.; Johnston, R.S.; et al. RBD-VLP vaccines adjuvanted with Alum or SWE protect K18-hACE2 mice against SARS-CoV-2 VOC challenge. Vaccines 2022, 7, e0024322. [Google Scholar] [CrossRef]
- Prates-Syed, W.A.; Chaves, L.C.S.; Crema, K.P.; Vuitika, L.; Lira, A.; Côrtes, N.; Kersten, V.; Guimarães, F.E.G.; Sadraeian, M.; Barroso da Silva, F.L.; et al. VLP-Based COVID-19 Vaccines: An Adaptable Technology against the Threat of New Variants. Vaccines 2021, 9, 1409. [Google Scholar] [CrossRef] [PubMed]
- Mandal, H. Achievements of the COVID-19 Turkey Platform in vaccine and drug development with an approach of “co-creation and succeeding together”. Vaccines 2021, 51, 3139–3149. [Google Scholar] [CrossRef]
- Sharifzadeh, M.; Mottaghi-Dastjerdi, N.; SoltanyRezaeRaad, M. A review of virus-like particle-based SARS-CoV-2 vaccines in clinical trial phases. Vaccines 2022, 21, e127042. [Google Scholar] [CrossRef]
- Gupta, V.; Sengupta, M.; Prakash, J.; Tripathy, B.C. Production of recombinant pharmaceutical proteins. In Basic and Applied Aspects of Biotechnology; Springer: Singapore, 2017; pp. 77–101. ISBN 978-981-10-0873-3. [Google Scholar]
- Venkataraman, S.; Hefferon, K.; Makhzoum, A.; Abouhaidar, M. Combating human viral diseases: Will plant-based vaccines be the answer? Vaccines 2021, 9, 761. [Google Scholar] [CrossRef] [PubMed]
- Geraghty, R.J.; Capes-Davis, A.; Davis, J.M.; Downward, J.; Freshney, R.I.; Knezevic, I.; Lovell-Badge, R.; Masters, J.R.W.; Meredith, J.; Stacey, G.N.; et al. Guidelines for the use of cell lines in biomedical research. Br. J. Cancer 2014, 111, 1021–1046. [Google Scholar] [CrossRef]
- Shanmugaraj, B.; Bulaon, C.J.I.; Phoolcharoen, W. Plant molecular farming: A viable platform for recombinant biopharmaceutical production. Plants 2020, 9, 842. [Google Scholar] [CrossRef]
- Margolin, E.; Chapman, R.; Williamson, A.-L.; Rybicki, E.P.; Meyers, A.E. Production of complex viral glycoproteins in plants as vaccine immunogens. Vaccines 2018, 16, 1531–1545. [Google Scholar] [CrossRef]
- Shahid, N.; Daniell, H. Plant-based oral vaccines against zoonotic and non-zoonotic diseases. Vaccines 2016, 14, 2079–2099. [Google Scholar] [CrossRef]
- Capell, T.; Twyman, R.M.; Armario-Najera, V.; Ma, J.K.; Schillberg, S.; Christou, P. Potential applications of plant biotechnology against SARS-CoV-2. Vaccines 2020, 25, 635–643. [Google Scholar] [CrossRef]
- LeBlanc, Z.; Waterhouse, P.; Bally, J. Plant-based vaccines: The way ahead? Viruses 2020, 13, 5. [Google Scholar] [CrossRef]
- Buyel, J.F.; Twyman, R.M.; Fischer, R. Extraction and downstream processing of plant-derived recombinant proteins. Biotechnol. Adv. 2015, 33, 902–913. [Google Scholar] [CrossRef] [PubMed]
- Monreal-Escalante, E.; Ramos-Vega, A.; Angulo, C.; Bañuelos-Hernández, B. Plant-Based Vaccines: Antigen design, diversity, and strategies for high level production. Vaccines 2022, 10, 100. [Google Scholar] [CrossRef]
- Minor, P. Considerations for setting the specifications of vaccines. Vaccines 2012, 11, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Kurup, V.M.; Thomas, J. Edible vaccines: Promises and challenges. Mol. Biotechnol. 2020, 62, 79–90. [Google Scholar] [CrossRef]
- Strasser, R. Plant protein glycosylation. Vaccines 2016, 26, 926–939. [Google Scholar] [CrossRef]
- Chan, H.-T.; Daniell, H. Plant-made oral vaccines against human infectious diseases-are we there yet? Plant Biotechnol. J. 2015, 13, 1056–1070. [Google Scholar] [CrossRef] [PubMed]
- Laere, E.; Ling, A.P.K.; Wong, Y.P.; Koh, R.Y.; Mohd Lila, M.A.; Hussein, S. Plant-based vaccines: Production and challenges. J. Bot. 2016, 2016, 4928637. [Google Scholar] [CrossRef]
- Streatfield, S.J.; Howard, J.A. Plant-based vaccines. Vaccines 2003, 33, 479–493. [Google Scholar] [CrossRef]
- Nosaki, S.; Hoshikawa, K.; Ezura, H.; Miura, K. Transient protein expression systems in plants and their applications. Plant Biotechnol. 2021, 38, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Parvathy, S.T. Engineering plants as platforms for production of vaccines. Am. J. Plant Sci. 2020, 11, 707–735. [Google Scholar] [CrossRef]
- Merlin, M.; Gecchele, E.; Capaldi, S.; Pezzotti, M.; Avesani, L. Comparative evaluation of recombinant protein production in different biofactories: The green perspective. Biomed. Res. Int. 2014, 2014, 136419. [Google Scholar] [CrossRef] [PubMed]
- Nandi, S.; Kwong, A.T.; Holtz, B.R.; Erwin, R.L.; Marcel, S.; McDonald, K.A. Techno-economic analysis of a transient plant-based platform for monoclonal antibody production. mAbs 2016, 8, 1456–1466. [Google Scholar] [CrossRef]
- Bendandi, M.; Marillonnet, S.; Kandzia, R.; Thieme, F.; Nickstadt, A.; Herz, S.; Fröde, R.; Inogés, S.; de Cerio, A.L.-D.; Soria, E.; et al. Rapid, high-yield production in plants of individualized idiotype vaccines for non-Hodgkin’s lymphoma. Ann. Oncol. 2010, 21, 2420–2427. [Google Scholar] [CrossRef]
- Rosenberg, Y.; Sack, M.; Montefiori, D.; Forthal, D.; Mao, L.; Hernandez-Abanto, S.; Urban, L.; Landucci, G.; Fischer, R.; Jiang, X. Rapid high-level production of functional HIV broadly neutralizing monoclonal antibodies in transient plant expression systems. PLoS ONE 2013, 8, e58724. [Google Scholar] [CrossRef] [PubMed]
- Shoji, Y.; Farrance, C.E.; Bautista, J.; Bi, H.; Musiychuk, K.; Horsey, A.; Park, H.; Jaje, J.; Green, B.J.; Shamloul, M.; et al. A plant-based system for rapid production of influenza vaccine antigens. Influ. Other Respir. Viruses 2012, 6, 204–210. [Google Scholar] [CrossRef]
- Joensuu, J.J.; Conley, A.J.; Lienemann, M.; Brandle, J.E.; Linder, M.B.; Menassa, R. Hydrophobin fusions for high-level transient protein expression and purification in Nicotiana Benthamiana. Plant Physiol. 2010, 152, 622–633. [Google Scholar] [CrossRef]
- Gou, J.; Debnath, S.; Sun, L.; Flanagan, A.; Tang, Y.; Jiang, Q.; Wen, J.; Wang, Z.-Y. From model to crop: Functional characterization of SPL8 in M. Truncatula led to genetic improvement of biomass yield and abiotic stress tolerance in Alfalfa. Plant Biotechnol. J. 2018, 16, 951–962. [Google Scholar] [CrossRef] [PubMed]
- Rattanapisit, K.; Shanmugaraj, B.; Manopwisedjaroen, S.; Purwono, P.B.; Siriwattananon, K.; Khorattanakulchai, N.; Hanittinan, O.; Boonyayothin, W.; Thitithanyanont, A.; Smith, D.R.; et al. Rapid production of SARS-CoV-2 Receptor Binding Domain (RBD) and spike specific monoclonal antibody CR3022 in Nicotiana benthamiana. Sci. Rep. 2020, 10, 17698. [Google Scholar] [CrossRef]
- Mardanova, E.S.; Kotlyarov, R.Y.; Ravin, N.V. Rapid transient expression of receptor-binding domain of SARS-CoV-2 and the conserved M2e Peptide of Influenza A virus linked to flagellin in Nicotiana benthamiana plants using self-replicating viral vector. Plants 2022, 11, 3425. [Google Scholar] [CrossRef]
- Criscuolo, E.; Caputo, V.; Diotti, R.A.; Sautto, G.A.; Kirchenbaum, G.A.; Clementi, N. Alternative methods of vaccine delivery: An overview of edible and intradermal vaccines. J. Immuno. Res. 2019, 2019, 8303648. [Google Scholar] [CrossRef]
- Czyz, M.; Pniewski, T. Thermostability of freeze-dried plant-made vlp-based vaccines. In Sustainable Drying Technologies; Del Real-Olvera, J., Ed.; InTech: London, UK, 2016; ISBN 978-953-51-2566-2. [Google Scholar]
- Pniewski, T.; Kapusta, J.; Bociąg, P.; Wojciechowicz, J.; Kostrzak, A.; Gdula, M.; Fedorowicz-Strońska, O.; Wójcik, P.; Otta, H.; Samardakiewicz, S.; et al. Low-dose oral immunization with lyophilized tissue of herbicide-resistant lettuce expressing Hepatitis B surface antigen for prototype plant-derived vaccine tablet formulation. J. Appl. Genet. 2011, 52, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Waheed, M.T.; Sameeullah, M.; Khan, F.A.; Syed, T.; Ilahi, M.; Gottschamel, J.; Lössl, A.G. Need of cost-effective vaccines in developing countries: What plant biotechnology can offer? SpringerPlus 2016, 5, 65. [Google Scholar] [CrossRef]
- Pérez Aguirreburualde, M.S.; Petruccelli, S.; Bravo Almonacid, F.; Wigdorovitz, A. Plant-based vaccine for livestock: Key points to unleash platform translation in developing countries. Curr. Mol. Biol. Rep. 2016, 2, 171–179. [Google Scholar] [CrossRef][Green Version]
- Margolin, E.; Schäfer, G.; Allen, J.D.; Gers, S.; Woodward, J.; Sutherland, A.D.; Blumenthal, M.; Meyers, A.; Shaw, M.L.; Preiser, W.; et al. A plant-produced SARS-CoV-2 spike protein elicits heterologous immunity in hamsters. Front. Plant Sci. 2023, 14, 1146234. [Google Scholar] [CrossRef]
- Hager, K.J.; Pérez Marc, G.; Gobeil, P.; Diaz, R.S.; Heizer, G.; Llapur, C.; Makarkov, A.I.; Vasconcellos, E.; Pillet, S.; Riera, F.; et al. Efficacy and Safety of a Recombinant Plant-Based Adjuvanted Covid-19 Vaccine. N. Engl. J. Med. 2022, 386, 2084–2096. [Google Scholar] [CrossRef]
- Panapitakkul, C.; Khorattanakulchai, N.; Rattanapisit, K.; Srisangsung, T.; Shanmugaraj, B.; Buranapraditkun, S.; Ketloy, C.; Prompetchara, E.; Phoolcharoen, W. Plant-produced S1 subunit protein of SARS-CoV-2 elicits immunogenic responses in mice. Vaccines 2022, 10, 1961. [Google Scholar] [CrossRef]
- O’Kennedy, M.M.; Abolnik, C.; Smith, T.; Motlou, T.; Goosen, K.; Sepotokele, K.M.; Roth, R.; du Preez, I.; Truyts, A.; Stark, H.C.; et al. Immunogenicity of adjuvanted plant-produced SARS-CoV-2 Beta spike VLP vaccine in New Zealand white rabbits. Vaccine 2023, 41, 2261–2269. [Google Scholar] [CrossRef]
- Mamedov, T.; Yuksel, D.; Ilgın, M.; Gürbüzaslan, I.; Gulec, B.; Mammadova, G.; Ozdarendeli, A.; Yetiskin, H.; Kaplan, B.; Islam Pavel, S.T.; et al. Production and characterization of Nucleocapsid and RBD cocktail antigens of SARS-CoV-2 in Nicotiana benthamiana plant as a vaccine candidate against COVID-19. Vaccines 2021, 9, 1337. [Google Scholar] [CrossRef]
- Hodgins, B.; Pillet, S.; Landry, N.; Ward, B.J. Prime-pull vaccination with a plant-derived virus-like particle influenza vaccine elicits a broad immune response and protects aged mice from death and frailty after challenge. Immun. Ageing 2019, 16, 27. [Google Scholar] [CrossRef] [PubMed]
- Pillet, S.; Couillard, J.; Trépanier, S.; Poulin, J.F.; Yassine-Diab, B.; Guy, B.; Ward, B.J.; Landry, N. Immunogenicity and safety of a quadrivalent plant-derived virus-like particle influenza vaccine candidate—Two randomized Phase II clinical trials in 18 to 49 and >50 years old adults. PLoS ONE 2019, 14, e0216533. [Google Scholar] [CrossRef]
- Hiatt, A.; Cafferkey, R.; Bowdish, K. Production of antibodies in transgenic plants. Nature 1989, 342, 76–78. [Google Scholar] [CrossRef] [PubMed]
- Mason, H.S.; Lam, D.M.K.; Arntzen, C.J. Expression of hepatitis B surface antigen in transgenic plants. Proc. Natl. Acad. Sci. USA 1992, 89, 11745–11749. [Google Scholar] [CrossRef] [PubMed]
- Mason, H.S.; Haq, T.A.; Clements, J.D.; Arntzen, C.J. Edible vaccine protects mice against Escherichia coli heat-labile enterotoxin (LT): Potatoes expressing a synthetic LT-B gene. Vaccine 1998, 16, 1336–1343. [Google Scholar] [CrossRef]
- Vermij, P.; Waltz, E. USDA approves the first plant-based vaccine. Nat. Biotechnol. 2006, 24, 233–234. [Google Scholar]
- Pujol, M.; Ramírez, N.I.; Ayala, M.; Gavilondo, J.V.; Valdés, R.; Rodríguez, M.; Brito, J.; Padilla, S.; Gómez, L.; Reyes, B.; et al. An integral approach towards a practical application for a plant-made monoclonal antibody in vaccine purification. Vaccine 2005, 23, 1833–1837. [Google Scholar] [CrossRef] [PubMed]
- Bolaños-Martínez, O.C.; Strasser, R. Plant-made poliovirus vaccines—Safe alternatives for global vaccination. Front. Plant Sci. 2022, 13, 1046346. [Google Scholar] [CrossRef]
- Naupu, P.N.; van Zyl, A.R.; Rybicki, E.P.; Hitzeroth, I.I. Immunogenicity of plant-produced Human Papillomavirus (HPV) Virus-Like Particles (VLPs). Vaccines 2020, 8, 740. [Google Scholar] [CrossRef]
- Rosales-Mendoza, S.; Nieto-Gómez, R.; Angulo, C. A Perspective on the development of plant-made vaccines in the fight against Ebola Virus. Vaccines 2017, 8, 252. [Google Scholar] [CrossRef]
- Diamos, A.G.; Pardhe, M.D.; Sun, H.; Hunter, J.G.; Mor, T.; Meador, L.; Kilbourne, J.; Chen, Q.; Mason, H.S. Codelivery of improved immune complex and virus-like particle vaccines containing zika virus envelope domain III synergistically enhances immunogenicity. Vaccine 2020, 38, 3455–3463. [Google Scholar] [CrossRef] [PubMed]
- Ponndorf, D.; Meshcheriakova, Y.; Thuenemann, E.C.; Dobon Alonso, A.; Overman, R.; Holton, N.; Dowall, S.; Kennedy, E.; Stocks, M.; Lomonossoff, G.P.; et al. Plant-made dengue virus-like particles produced by co-expression of structural and non-structural proteins induce a humoral immune response in mice. Plant Biotechnol. J. 2021, 19, 745–756. [Google Scholar] [CrossRef]
- Mardanova, E.S.; Ravin, N.V. Plant-produced recombinant Influenza A vaccines based on the M2e Peptide. Curr. Pharm. Des. 2018, 24, 1317–1324. [Google Scholar] [CrossRef] [PubMed]
- Tremouillaux-Guiller, J.; Moustafa, K.; Hefferon, K.; Gaobotse, G.; Makhzoum, A. Plant-made HIV vaccines and potential candidates. Curr. Opin. Biotechnol. 2020, 61, 209–216. [Google Scholar] [CrossRef]
- Balke, I.; Zeltins, A. Use of plant viruses and virus-like particles for the creation of novel vaccines. Adv. Drug. Deliv. Rev. 2019, 145, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. Market Analysis Report: Plant-Based Vaccines Market. The Insight Partners. 2021. Available online: https://www.theinsightpartners.com/reports/plant-based-vaccines-market (accessed on 13 July 2023).
- Arntzen, C. Plant-made pharmaceuticals: From ‘Edible Vaccines’ to Ebola therapeutics. Plant Biotechnol. J. 2015, 13, 1013–1016. [Google Scholar] [CrossRef]
- Chen, Q.; Davis, K.R. The potential of plants as a system for the development and production of human biologics. F1000Res 2016, 5, F1000 Faculty Rev-912. [Google Scholar] [CrossRef]
- Chen, Q.; He, J.; Phoolcharoen, W.; Mason, H.S. Geminiviral vectors based on bean yellow dwarf virus for production of vaccine antigens and monoclonal antibodies in plants. Hum. Vaccines 2011, 7, 331–338. [Google Scholar] [CrossRef]
- Klimyuk, V.; Pogue, G.; Herz, S.; Butler, J.; Haydon, H. Production of recombinant antigens and antibodies in Nicotiana benthamiana using ‘magnifection’ technology: GMP-compliant facilities for small- and large-scale manufacturing. Curr. Top. Microbiol. Immunol. 2014, 375, 127–154. [Google Scholar] [PubMed]
- Peyret, H.; Lomonossoff, G.P. When plant virology met Agrobacterium: The rise of the deconstructed clones. Plant Biotechnol. J. 2015, 13, 1121–1135. [Google Scholar] [CrossRef]
- Rybicki, E.P. Plant-made vaccines and reagents for the one health initiative. Hum. Vaccin. Immunother. 2017, 13, 2912–2917. [Google Scholar] [CrossRef]
- Babcock, G.J.; Esshaki, D.J.; Thomas, W.D., Jr.; Ambrosino, D.M. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 2004, 78, 4552–4560. [Google Scholar] [CrossRef]
- Gómez, N.; Carrillo, C.; Salinas, J.; Parra, F.; Borca, M.V.; Escribano, J.M. Expression of immunogenic glycoprotein S polypeptides from transmissible gastroenteritis coronavirus in transgenic plants. Virology 1998, 249, 352–358. [Google Scholar] [CrossRef]
- Gomez, N.; Wigdorovitz, A.; Castanon, S.; Gil, F.; Ordas, R.; Borca, M.V.; Escribano, J.M. Oral immunogenicity of the plant-derived spike protein from swine-transmissible gastroenteritis coronavirus. Arch. Virol. 2000, 145, 1725–1732. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.Y.; Cheng, L.Q.; Zheng, X.J.; Wu, J.-X.; Shang, S.-B.; Wang, J.-Y.; Chen, J.-G. Generation of the transgenic potato expressing full-length spike protein of infectious bronchitis virus. J. Biotechnol. 2004, 111, 121–130. [Google Scholar] [CrossRef]
- Zhou, J.Y.; Wu, J.X.; Cheng, L.Q.; Zheng, X.-J.; Gong, H.; Shang, S.-B.; Zhou, E.-M. Expression of immunogenic S1 glycoprotein of infectious bronchitis virus in transgenic potatoes. J. Virol. 2003, 77, 9090–9093. [Google Scholar] [CrossRef][Green Version]
- Lamphear, B.J.; Jilka, J.M.; Kesl, L.; Welter, M.; Howard, J.A.; Streatfield, S.J. A corn-based delivery system for animal vaccines: An oral transmissible gastroenteritis virus vaccine boosts lactogenic immunity in swine. Vaccine 2004, 22, 2420–2424. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Ramalingam, S.; Chye, M.L. Accumulation of recombinant SARS-CoV spike protein in plant cytosol and chloroplasts indicate potential for development of plant-derived oral vaccines. Exp. Biol. Med. 2006, 231, 1346–1352. [Google Scholar] [CrossRef]
- Pogrebnyak, N.; Golovkin, M.; Andrianov, V.; Spitsin, S.; Smirnov, Y.; Egolf, R.; Koprowski, H. Severe acute respiratory syndrome (SARS) S protein production in plants: Development of recombinant vaccine. Proc. Natl. Acad. Sci. USA 2005, 102, 9062–9067. [Google Scholar] [CrossRef]
- Zheng, N.; Xia, R.; Yang, C.; Yin, B.; Li, Y.; Duan, C.; Liang, L.; Guo, H.; Xie, Q. Boosted expression of the SARS-CoV nucleocapsid protein in tobacco and its immunogenicity in mice. Vaccine 2009, 27, 5001–5007. [Google Scholar] [CrossRef]
- Wirz, H.; Sauer-Budge, A.F.; Briggs, J.; Sharpe, A.; Shu, S.; Sharon, A. Automated production of plant-based vaccines and pharmaceuticals. SLAS Technol. 2012, 17, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Huebbers, J.W.; Buyel, J.F. On the verge of the market—Plant factories for the automated and standardized production of biopharmaceuticals. Biotechnol. Adv. 2021, 46, 107681. [Google Scholar] [CrossRef] [PubMed]
- Laughlin, R.C.; Madera, R.; Peres, Y.; Berquist, B.R.; Wang, L.; Buist, S.; Burakova, Y.; Palle, S.; Chung, C.J.; Rasmussen, M.V.; et al. Plant-made E2 glycoprotein single-dose vaccine protects pigs against classical swine fever. Plant Biotechnol. J. 2019, 17, 410–420. [Google Scholar] [CrossRef]
- Zahmanova, G.; Aljabali, A.A.; Takova, K.; Toneva, V.; Tambuwala, M.M.; Andonov, A.P.; Lukov, G.L.; Minkov, I. The Plantviruses and molecular farming: How beneficial they might be for human and animal health? Int. J. Mol. Sci. 2023, 24, 1533. [Google Scholar] [CrossRef]
- Medicago. Medicago Announces Production of a Viable Vaccine Candidate for COVID-19; Medicago: Québec City, QC, Canada, 2020. [Google Scholar]
- DARPA Makes 10 Million Strides in the Race to Contain a Hypothetical Pandemic. 2012. Available online: http://www.darpa.mil/NewsEvents/Releases/2012/07/25.aspx (accessed on 1 April 2020).
- Medicago. Medicago Announces Positive Results in Animal Trials for Its Vaccine Candidate against COVID-19; Medicago: Québec City, QC, Canada, 2020. [Google Scholar]
- Ward, B.J.; Gobeil, P.; Séguin, A.; Atkins, J.; Boulay, I.; Charbonneau, P.-Y.; Couture, M.; D’Aoust, M.-A.; Dhaliwall, J.; Finkle, C.; et al. Phase 1 randomized trial of a plant-derived virus-like particle vaccine for COVID-19. Nat. Med. 2021, 27, 1071–1078. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, Q.; Du, L.; Lu, L.; Jiang, S. Receptor-binding domain as a target for developing SARS vaccines. J. Thorac. Dis. 2013, 5, S142–S148. [Google Scholar]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef]
- Zlotnick, A.; Venkatakrishnan, B.; Tan, Z.; Lewellyn, E.; Turner, W.; Francis, S. Core protein: A pleiotropic keystone in the HBV lifecycle. Antivir. Res. 2015, 121, 82–93. [Google Scholar] [CrossRef] [PubMed]
- iBio. iBio Announces Development of Proprietary COVID-19 Vaccine Candidates. 2020. Available online: www.ibioinc.com (accessed on 1 June 2020).
- Rybicki, E.P. Plant molecular farming of virus-like nanoparticles as vaccines and reagents. WIREs Nanomed. Nanobiotechnol. 2019, 12, e1587. [Google Scholar] [CrossRef] [PubMed]
- iBio. iBio-201 Demonstrates Ability to Elicit anti-SARS-CoV-2 Immune Response in Preclinical Studies. 2020. Available online: www.ibioinc.com (accessed on 1 June 2020).
- iBio. iBio Reports Successful COVID-19 Vaccine Toxicology Study Results and Announces Next-Gen COVID-19 Vaccine Program. 2021. Available online: www.ibioinc.com (accessed on 1 June 2021).
- BaiyaPhytopharm. COVID-19 Vaccine Development. Available online: https://baiyaphytopharm.com/covid-19/ (accessed on 12 August 2021).
- Kang, H.; Kim, D.; Min, K.; Park, M.; Kim, S.H.; Sohn, E.J.; Choi, B.H.; Hwang, I. Recombinant proteins of spike protein of SARS-CoV-2 with the Omicron receptor-binding domain induce production of highly Omicron-specific neutralizing antibodies. Clin. Exp. Vaccine Res. 2022, 11, 285–289. [Google Scholar] [CrossRef]
- Royal, J.M.; Simpson, C.A.; McCormick, A.A.; Phillips, A.; Hume, S.; Morton, J.; Shepherd, J.; Oh, Y.; Swope, K.; DeBeauchamp, J.L.; et al. Development of a SARS-CoV-2 vaccine candidate using plant-based manufacturing and a Tobacco Mosaic Virus-like nano-particle. Vaccines 2021, 9, 1347. [Google Scholar] [CrossRef] [PubMed]
- Takeyama, N.; Kiyono, H.; Yuki, Y. Plant-based vaccines for animals and humans: Recent advances in technology and clinical trials. Ther. Adv. Vaccines 2015, 3, 139–154. [Google Scholar] [CrossRef]
- Covarrubias, C.E.; Rivera, T.A.; Soto, C.A.; Deeks, T.; Kalergis, A.M. Current GMP standards for the production of vaccines and antibodies: An overview. Front. Public Health 2022, 10, 1021905. [Google Scholar] [CrossRef]
- Daniell, H.; Kulis, M.; Herzog, R.W. Plant cell-made protein antigens for induction of oral tolerance. Biotechnol. Adv. 2019, 37, 107413. [Google Scholar] [CrossRef] [PubMed]
- Schwestka, J.; König-Beihammer, J.; Shin, Y.-J.; Vavra, U.; Kienzl, N.F.; Grünwald-Gruber, C.; Maresch, D.; Klausberger, M.; Laurent, E.; Stadler, M.; et al. Impact of specific N-Glycan modifications on the use of plant-produced SARS-CoV-2 antigens in serological assays. Front. Plant Sci. 2021, 12, 747500. [Google Scholar] [CrossRef]
- Strasser, R. Plant Glycoengineering for designing next-generation vaccines and therapeutic proteins. Biotechnol. Adv. 2023, 67, 108197. [Google Scholar] [CrossRef]
- Jailani, A.A.K.; Shilpi, S.; Mandal, B. Rapid demonstration of infectivity of a hybrid strain of potato virus Y occurring in India through overlapping extension PCR. Physiol. Mol. Plant Pathol. 2017, 98, 62–68. [Google Scholar] [CrossRef]
- Jailani, A.A.K.; Solanki, V.; Roy, A.; Sivasudha, T.; Mandal, B. A CGMMV genome-replicon vector with partial sequences of coat protein gene efficiently expresses GFP in Nicotiana benthamiana. Virus Res. 2017, 233, 77–85. [Google Scholar] [CrossRef]
- Kumar, A.; Abdul Kader Jailani, A.; Roy, A.; Mandal, B. The occurrence, biology and genomic properties of tobamoviruses infecting crop plants in India. In A Century of Plant Virology in India; Mandal, B., Rao, G.P., Baranwal, V.K., Jain, R.K., Eds.; Springer: Singapore, 2017; pp. 429–444. [Google Scholar]
- Sandra, N.; Jailani, A.A.K.; Jain, R.K.; Mandal, B. Development of Soybean yellow mottle mosaic virus-based expression vector for heterologous protein expression in French bean. Mol. Biotechnol. 2019, 61, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Rybicki, E.P. Plant-made vaccines for humans and animals: Plant-made vaccines. Plant Biotechnol. J. 2010, 8, 620–637. [Google Scholar] [CrossRef] [PubMed]
- Rosales-Mendoza, S.; Márquez-Escobar, V.A.; González-Ortega, O.; Nieto-Gómez, R.; Arévalo-Villalobos, J.I. What does plant-based vaccine technology offer to the fight against COVID-19? Vaccines 2020, 8, 183. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Van Eerde, A.; Rimstad, E.; Bock, R.; Branza-Nichita, N.; Yakovlev, I.A.; Clarke, J.L. Plant-made vaccines against viral diseases in humans and farm animals. Front. Plant Sci. 2023, 14, 1170815. [Google Scholar] [CrossRef]

| Sr No. | Candidate Vaccine | Content | Approved in Countries | Administration | Developer/Manufacturer |
|---|---|---|---|---|---|
| 1 | CoronaVac | Inactivated virus (Vero cell) | 56 | Intramuscular injection of 2 doses | Sinovac Life Sciences Co., Ltd. (Beijing, China) |
| 2 | Covilo | Inactivated virus (Vero cell) | 93 | Intramuscular injection of 1, 2 or 5 doses | Beijing Institute of Biological Products Co., Ltd. (BIBP) (Sinopharm, Beijing, China) |
| 3 | COVAXIN | Inactivated virus (Whole Virion) | 14 | Intramuscular injection of 1, 2, 5 or 10 doses | Bharat Biotech International Ltd. (Hyderabad, India) |
| 4 | Covishield | Non Replicating Viral Vector ChAdOx1-S (recombinant) | 49 | Intramuscular injection of 2 or 10 doses | Serum Institute of India Pvt. Ltd. (Pune, India) |
| 5 | VAXZEVRIA | Non Replicating Viral Vector ChAdOx1-S (recombinant) | 149 | Intramuscular injection of 2 or 10 doses | Oxford/AstraZeneca with SK Bioscience Co. Ltd. (Gyeonggi-do, Republic of Korea) |
| 6 | Convidecia | Non Replicating Viral Vector Ad5-nCoV-S [Recombinant]) | 10 | Intramuscular injection of 1 and 3 doses (0.5 mL) | CanSino Biologics Inc. (Tianjin, China) |
| 7 | Jcovden | Non Replicating Viral Vector Ad26.COV2-S (recombinant) | 113 | Intramuscular injection of 5 doses | Janssen (Johnson & Johnson) (Beerse, Belgium) |
| 8 | Comirnaty | mRNA vaccine (nucleoside modified) | 149 | Intramuscular injection of 2 or 6 doses (30 µg, 0.3 mL each) | Pfizer/BioNTech (Goldgrubae, Germany) |
| 9 | SPIKEVAX | mRNA(1273)Vaccine (nucleoside modified) | 88 | Intramuscular injection of Intramuscular injection of 10 doses (0.5 mL per dose) | Moderna Biotech (Madrid, Spain) |
| 10 | Covovax | SARS-CoV-2 rS Protein Nanoparticle (Recombinant) | 6 | Intramuscular injection of 1 dose and 10 doses (0.5 mL per dose) | Serum Institute of India Pvt. Ltd. (Pune, India) |
| 11 | Nuvaxovid | (SARS-CoV-2 rS [Recombinant, adjuvanted]) | 40 | 10 doses (0.5 mL per dose) | Novavax CZ a.s. (Jevany, Czech Republic) |
| Class | Types | Description | Application | Pros | Cons |
|---|---|---|---|---|---|
| Pathogen-based vaccine | Inactivated pathogen | Whole celled pathogens killed by chemical, heat or radiationtreatment | Difficulty in epitope designing for highly mutating pathogens like influenza, polio, etc | 1. Easy to develop 2. Immune response induced by original pathogen | With emergence of new strains, the immunogenicity of vaccines reduced |
| Live-attenuated pathogen | Genetically engineered, weakened or attenuated strains of pathogen with reduced virulence | Established platform for multiple human pathogens, viz., measles, mumps, rubella, chicken-pox, etc | Induce strong cellular and humoralimmune responses | 1. Lengthy andtime-consuming development process 2. Risk of virulence reversion of virus strain via recombination and mutation | |
| Recombinant vector vaccine | Replicating Virus Vector | Efficient expressionof antigen using replicating virus vector-based expression system | Efficient delivery of antigen into human cells and tissues | Induction of cellular and humoral immune response at low dose | 1. Pre-existing immunity against the virusvector reduced replicability of the construct, 2. Safety and immunogenicity issues |
| Non-replicating virus vectors | Efficient expressionof antigen using non-replicating virus vector-based expression system | Efficient delivery of antigen into human cells and tissues | Induction of cellular and humoral immune response at low dose | 1. Risk of reversion of pathogenicity is very minimal 2. very safe to use | |
| Nucleic acid-based vaccine | DNA | Antigen encoding DNA (gene sequence) is delivered to human cell using bacterial plasmid | Efficient delivery of antigen into human cells and tissues | Easy to design, Rapid manufacture, Noninfectious nature | 1. Very difficult to deliver into human cell, may require some special care; 2. Low transfection and lesser protein expression |
| RNA | Antigen expression through through self-replicating or non-replicating mRNA | Efficient administration of mRNA via lipid-based delivery systems like lipoplexes and polyplexes | Noninfectious molecules induce humoral and cellular immune responses | Risk of side effects like cardiac arrest | |
| Protein-based vaccine | Protein subunit | Heterologous expression of certain part of the pathogenshowing immunogenicity used as protein subunit vaccine | Preparation of Subunit vaccine formulations by mixing purified antigens with potent adjuvants | Low risk, Safe and stable, fast manufacturing, | Lower immunogenicity; Requirement of adjuvant or conjugate to increase immunogenicity |
| VLP | Aggregation of protein forming a virus like configuration without anyvirus genome | Self-assembly of virus capsid protein forming a nanoparticle like structure with potential antigenic response; applicable against different diseases | Noninfectious, Strong humoral response, safe for immune compromisedindividuals, More stable than subunit vaccine | Noninfectious, Difficulty in scaling up of production |
| Parameters | Bacteria (E. coli) | Fungi (Yeast) | Insect Cell | Mammalian Cell | Plant |
|---|---|---|---|---|---|
| Amount of Protein expression | High | Low | Low | Low | High |
| Speed of Protein expression | Very high | High | High | High | High |
| Cost | Very low | Low | Low | High | Medium |
| Scalability | High | High | Medium | Medium | Medium |
| Yield | High | Medium | Medium | Medium | Medium |
| Post-translational modification (N-glycosylation) | No | Yes | Yes | Yes | Yes |
| Protein purification | Difficult | Difficult | Difficult | Difficult | Easy * |
| Chance of contamination | Yes | Yes | Yes | Yes | No |
| Immunogenicity | Low | Medium | Medium | Very high | High |
| Oral delivery | No | No | No | No | Yes |
| Cool chain transportation | Require | Require | Require | Require | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chattopadhyay, A.; Jailani, A.A.K.; Mandal, B. Exigency of Plant-Based Vaccine against COVID-19 Emergence as Pandemic Preparedness. Vaccines 2023, 11, 1347. https://doi.org/10.3390/vaccines11081347
Chattopadhyay A, Jailani AAK, Mandal B. Exigency of Plant-Based Vaccine against COVID-19 Emergence as Pandemic Preparedness. Vaccines. 2023; 11(8):1347. https://doi.org/10.3390/vaccines11081347
Chicago/Turabian StyleChattopadhyay, Anirudha, A. Abdul Kader Jailani, and Bikash Mandal. 2023. "Exigency of Plant-Based Vaccine against COVID-19 Emergence as Pandemic Preparedness" Vaccines 11, no. 8: 1347. https://doi.org/10.3390/vaccines11081347
APA StyleChattopadhyay, A., Jailani, A. A. K., & Mandal, B. (2023). Exigency of Plant-Based Vaccine against COVID-19 Emergence as Pandemic Preparedness. Vaccines, 11(8), 1347. https://doi.org/10.3390/vaccines11081347





