The Screening of the Protective Antigens of Aeromonas hydrophila Using the Reverse Vaccinology Approach: Potential Candidates for Subunit Vaccine Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Bioinformatic Analysis of the A. hydrophila Genome
2.2.1. Data Collection
2.2.2. Signal Peptide Prediction
2.2.3. Transmembrane Helical Structure Prediction
2.2.4. Prediction of the Subcellular Localization
2.2.5. Antigenicity and Adhesion Index Analysis of the TARGET Proteins
2.2.6. Homology Analysis of the Protein Sequences
2.3. Conservative Analysis of Candidate Proteins in Different Subtypes of A. hydrophila and Aeromonas Species
2.3.1. Sample Collection
2.3.2. Molecular Characterization of the A. hydrophila Strains
2.3.3. ERIC-PCR Analysis of A. hydrophila Strains
2.3.4. Cluster Analysis of A. hydrophila Strains
2.3.5. PCR Amplification of the CDS Regions of Candidate Proteins
2.3.6. Sequence Alignment Using BLAST
2.4. Statistical Analysis
3. Results
3.1. Bioinformatic Analysis of the A. hydrophila Genome
3.1.1. Signal Peptide Prediction
3.1.2. Transmembrane Helix Prediction
3.1.3. Prediction of the Subcellular Localization
3.1.4. Antigenicity and Adhesion Index Prediction of the Candidate Proteins
3.1.5. Homology Analysis of the Candidate Proteins
3.2. Molecular Identification of Strains
3.3. ERIC-PCR
3.3.1. ERIC-PCR Typing of A. hydrophila
3.3.2. Cluster Analysis of ERIC-PCR Fingerprints
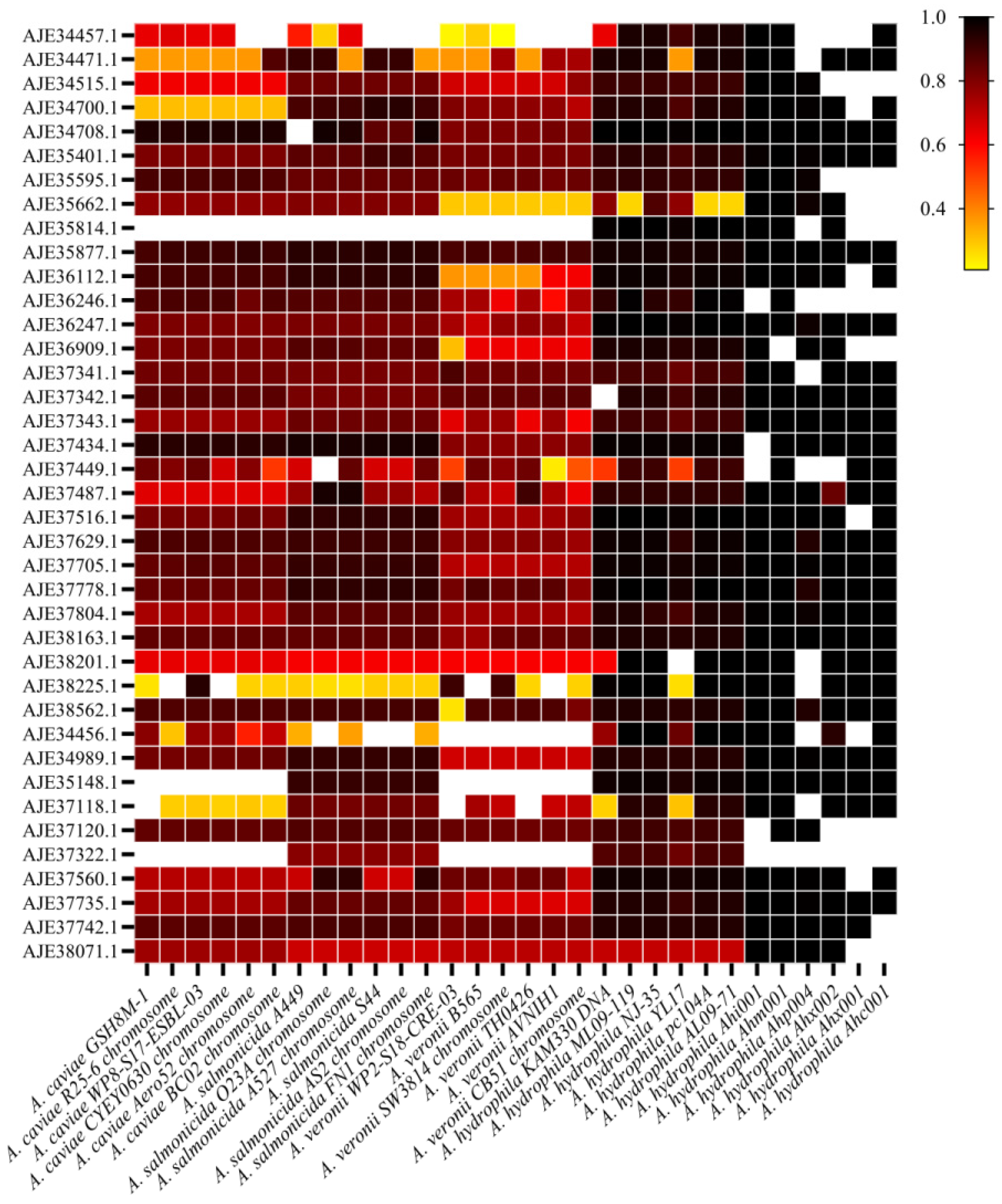
3.4. Conservative Analysis of Candidate Proteins in Aeromonas and Different Subtypes of A. hydrophila
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Citterio, B.; Francesca, B. Aeromonas hydrophila virulence. Virulence 2015, 6, 417–418. [Google Scholar] [CrossRef]
- Praveen, P.K.; Debnath, C.; Shekhar, S.; Dalai, N.; Ganguly, S. Incidence of Aeromonas spp. infection in fish and chicken meat and its related public health hazards: A review. Vet. World 2016, 9, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Zmysłowska, I.; Korzekwa, K.; Szarek, J. Aeromonas hydrophila in fish aquaculture. J. Comp. Pathol. 2009, 141, 313. [Google Scholar] [CrossRef]
- Davin-Regli, A.; Bollet, C.; Chamorey, E.; Colonna D’istria, V.; Cremieux, A. A cluster of cases of infections due to Aeromonas hydrophila revealed by combined RAPD and ERIC-PCR. J. Med. Microbiol. 1998, 47, 499–504. [Google Scholar] [CrossRef]
- Aguilera-Arreola, M.G.; Hernández-Rodríguez, C.; Zúñiga, G.; Figueras, M.J.; Castro-Escarpulli, G. Aeromonas hydrophila clinical and environmental ecotypes as revealed by genetic diversity and virulence genes. FEMS Microbiol. Lett. 2005, 242, 231–240. [Google Scholar] [CrossRef]
- Awan, F.; Dong, Y.; Wang, N.; Liu, J.; Ma, K.; Liu, Y. The fight for invincibility: Environmental stress response mechanisms and Aeromonas hydrophila. Microb. Pathog. 2018, 116, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Khushiramani, R.; Girisha, S.K.; Karunasagar, I.; Karunasagar, I. Cloning and expression of an outer membrane protein ompTS of Aeromonas hydrophila and study of immunogenicity in fish. Protein Expr. Purif. 2007, 51, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Meena, J.K.; Sharma, M.; Dixit, A. Recombinant outer membrane protein C of Aeromonas hydrophila elicits mixed immune response and generates agglutinating antibodies. Immunol. Res. 2016, 64, 1087–1099. [Google Scholar] [CrossRef]
- Maiti, B.; Dubey, S.; Munang’andu, H.M.; Karunasagar, I.; Karunasagar, I.; Evensen, Ø. Application of Outer Membrane Protein-Based Vaccines Against Major Bacterial Fish Pathogens in India. Front. Immunol. 2020, 11, 1362. [Google Scholar] [CrossRef]
- Mzula, A.; Wambura, P.N.; Mdegela, R.H.; Shirima, G.M. Current State of Modern Biotechnological-Based Aeromonas hydrophila Vaccines for Aquaculture: A Systematic Review. BioMed Res. Int. 2019, 2019, 3768948. [Google Scholar] [CrossRef]
- Wang, N.; Yang, Z.; Zang, M.; Liu, Y.; Lu, C. Identification of Omp38 by immunoproteomic analysis and evaluation as a potential vaccine antigen against Aeromonas hydrophila in Chinese breams. Fish Shellfish Immunol. 2013, 34, 74–81. [Google Scholar] [CrossRef]
- Abdelhamed, H.; Ibrahim, I.; Nho, S.W.; Banes, M.M.; Wills, R.W.; Karsi, A.; Lawrence, M.L. Evaluation of three recombinant outer membrane proteins, OmpA1, Tdr, and TbpA, as potential vaccine antigens against virulent Aeromonas hydrophila infection in channel catfish (Ictalurus punctatus). Fish Shellfish Immunol. 2017, 66, 480–486. [Google Scholar] [CrossRef]
- Dubey, S.; Avadhani, K.; Mutalik, S.; Sivadasan, S.M.; Maiti, B.; Paul, J.; Girisha, S.K.; Venugopal, M.N.; Mutoloki, S.; Evensen, Ø.; et al. Aeromonas hydrophila OmpW PLGA nanoparticle oral vaccine shows a dose-dependent protective immunity in rohu (Labeo rohita). Vaccines 2016, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Kozińska, A.; Pękala, A. Characteristics of disease spectrum in relation to species, serogroups, and adhesion ability of motile Aeromonads in fish. Sci. World J. 2012, 2012, 949358. [Google Scholar] [CrossRef] [PubMed]
- Seib, K.L.; Zhao, X.; Rappuoli, R. Developing vaccines in the era of genomics: A decade of reverse vaccinology. Clin. Microbiol. Infect. 2012, 18 (Suppl. S5), 109–116. [Google Scholar] [CrossRef] [PubMed]
- Kaur, B.; Naveen Kumar, B.T.; Tyagi, A.; Admane Holeyappa, S.; Singh, N.K. Identification of novel vaccine candidates in the whole-cell Aeromonas hydrophila biofilm vaccine through reverse vaccinology approach. Fish Shellfish Immunol. 2021, 114, 132–141. [Google Scholar] [CrossRef]
- Bi, Z.X.; Liu, Y.J.; Lu, C.P. Contribution of AhyR to virulence of Aeromonas hydrophila J-1. Res. Vet. Sci. 2007, 83, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Enwall, K.; Philippot, L.; Hallin, S. Activity and composition of the denitrifying bacterial community respond differently to long-term fertilization. Appl. Environ. Microbiol. 2005, 71, 8335–8343. [Google Scholar] [CrossRef]
- He, P.; Wang, H.; Luo, J.; Yan, Y.; Chen, Z. A Real-Time PCR with Melting Curve Analysis for Molecular Typing of Vibrio parahaemolyticus. Curr. Microbiol. 2018, 75, 1206–1213. [Google Scholar] [CrossRef]
- Szczuka, E.; Kaznowski, A. Typing of clinical and environmental Aeromonas sp. strains by random amplified polymorphic DNA PCR, repetitive extragenic palindromic PCR, and enterobacterial repetitive intergenic consensus sequence PCR. J. Clin. Microbiol. 2004, 42, 220–228. [Google Scholar] [CrossRef]
- Pizza, M.; Scarlato, V.; Masignani, V.; Giuliani, M.M.; Aricò, B.; Comanducci, M.; Jennings, G.T.; Baldi, L.; Bartolini, E.; Capecchi, B.; et al. Identification of vaccine candidates against serogroup B Meningococcus by whole-genome sequencing. Science 2000, 287, 1816–1820. [Google Scholar] [CrossRef] [PubMed]
- Leow, C.Y.; Kazi, A.; Hisyam Ismail CM, K.; Chuah, C.; Lim, B.H.; Leow, C.H.; Banga Singh, K.K. Reverse vaccinology approach for the identification and characterization of outer membrane proteins of Shigella flexneri as potential cellular- and antibody-dependent vaccine candidates. Clin. Exp. Vaccine Res. 2020, 9, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, J.; Guo, S.; Liu, L.; Yuan, Q.; Guo, L.; Pan, S. Identification of Vibrio parahaemolyticus and Vibrio spp. Specific Outer Membrane Proteins by Reverse Vaccinology and Surface Proteome. Front. Microbiol. 2021, 11, 625315. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Yao, S.; Hart, D.J.; An, Y. Signal peptide-dependent protein translocation pathway is crucial for the sucrose sensitivity of SacB-expressing Escherichia coli. Biochem. Eng. J. 2017, 122, 71–74. [Google Scholar] [CrossRef]
- Claros, M.G.; Brunak, S.; von Heijne, G. Prediction of N-terminal protein sorting signals. Curr. Opin. Struct. Biol. 1997, 7, 394–398. [Google Scholar] [CrossRef]
- Nielsen, H. Predicting Secretory Proteins with SignalP. Methods Mol. Biol. 2017, 1611, 59–73. [Google Scholar] [CrossRef]
- Crane, J.M.; Randall, L.L. The Sec System: Protein Export in Escherichia coli. EcoSal Plus 2017, 7, 10–1128. [Google Scholar] [CrossRef]
- Palmer, T.; Berks, B.C.; Sargent, F. Analysis of Tat targeting function and twin-arginine signal peptide activity in Escherichia coli. Methods Mol. Biol. 2010, 619, 191–216. [Google Scholar] [CrossRef]
- Kitamura, S.; Wolan, D.W. Probing substrate recognition of bacterial lipoprotein signal peptidase using FRET reporters. FEBS Lett. 2018, 592, 2289–2296. [Google Scholar] [CrossRef]
- Tusnády, G.E.; Simon, I. The HMMTOP transmembrane topology prediction server. Bioinformatics 2001, 17, 849–850. [Google Scholar] [CrossRef]
- Gardy, J.L.; Laird, M.R.; Chen, F.; Rey, S.; Walsh, C.J.; Ester, M.; Brinkman, F.S. PSORTb v.2.0: Expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 2005, 21, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.B.; Chou, K.C. Gneg-mPLoc: A top-down strategy to enhance the quality of predicting subcellular localization of Gram-negative bacterial proteins. J. Theor. Biol. 2010, 264, 326–333. [Google Scholar] [CrossRef]
- Bert van den Berg, B. The FadL family: Unusual transporters for unusual substrates. Curr. Opin. Struct. Biol. 2005, 15, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Koronakis, V.; Eswaran, J.; Hughes, C. Structure and function of TolC: The bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 2004, 73, 467–489. [Google Scholar] [CrossRef] [PubMed]
- Kuehl, C.J.; Crosa, J.H. The TonB energy transduction systems in Vibrio species. Future Microbiol. 2010, 5, 1403–1412. [Google Scholar] [CrossRef]
- Funahashi, T.; Tanabe, T.; Miyamoto, K.; Tsujibo, H.; Maki, J.; Yamamoto, S. Characterization of a gene encoding the outer membrane receptor for ferric enterobactin in Aeromonas hydrophila ATCC 7966(T). Biosci. Biotechnol. Biochem. 2013, 77, 353–360. [Google Scholar] [CrossRef]
- Guan, Q.; Bhowmick, B.; Upadhyay, A.; Han, Q. Structure and functions of bacterial outer membrane protein A, a potential therapeutic target for bacterial infection. Curr. Top. Med. Chem. 2021, 21, 1129–1138. [Google Scholar] [CrossRef]
- Nie, D.; Hu, Y.; Chen, Z.; Li, M.; Hou, Z.; Luo, X.; Mao, X.; Xue, X. Outer membrane protein A (OmpA) as a potential therapeutic target for Acinetobacter baumannii infection. J. Biomed. Sci. 2020, 27, 26. [Google Scholar] [CrossRef]
- Chaturvedi, D.; Mahalakshmi, R. Transmembrane β-barrels: Evolution, folding and energetics. Biochimica et biophysica acta. Biomembranes 2017, 1859, 2467–2482. [Google Scholar] [CrossRef]
- Noinaj, N.; Rollauer, S.E.; Buchanan, S.K. The β-barrel membrane protein insertase machinery from Gram-negative bacteria. Curr. Opin. Struct. Biol. 2015, 31, 35–42. [Google Scholar] [CrossRef]
- Bakelar, J.; Buchanan, S.K.; Noinaj, N. The structure of the β-barrel assembly machinery complex. Science 2016, 351, 180–186. [Google Scholar] [CrossRef]
- Namdari, F.; Hurtado-Escobar, G.A.; Abed, N.; Trotereau, J.; Fardini, Y.; Giraud, E.; Velge, P.; Virlogeux-Payant, I. Deciphering the roles of BamB and its interaction with BamA in outer membrane biogenesis, T3SS expression and virulence in Salmonella. PLoS ONE 2012, 7, e46050. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Kang, H.S.; Okon, M.; Escobar-Cabrera, E.; McIntosh, L.P.; Paetzel, M. Structural characterization of Escherichia coli BamE, a lipoprotein component of the β-barrel assembly machinery complex. Biochemistry 2011, 50, 1081–1090. [Google Scholar] [CrossRef]
- Nivaskumar, M.; Francetic, O. Type II secretion system: A magic beanstalk or a protein escalator. Biochim. Biophys. Acta 2014, 1843, 1568–1577. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.L.; Gurung, I.; Anonsen, J.H.; Spielman, I.; Harper, E.; Hall AM, J.; Goosens, V.J.; Raynaud, C.; Koomey, M.; Biais, N.; et al. Global biochemical and structural analysis of the type IV pilus from the Gram-positive bacterium Streptococcus sanguinis. J. Biol. Chem. 2019, 294, 6796–6808. [Google Scholar] [CrossRef]
- Yang, T.C.; Ma, X.C.; Liu, F.; Lin, L.R.; Liu, L.L.; Liu, G.L.; Tong, M.L.; Fu, Z.G.; Zhou, L. Screening of the Salmonella paratyphi ACMCC 50973 strain outer membrane proteins for the identification of potential vaccine targets. Mol. Med. Rep. 2012, 5, 78–83. [Google Scholar] [CrossRef]
- Baarda, B.I.; Zielke, R.A.; Holm, A.K.; Sikora, A.E. Comprehensive bioinformatic assessments of the variability of Neisseria gonorrhoeae vaccine candidates. mSphere 2021, 6, e00977-20. [Google Scholar] [CrossRef] [PubMed]
- Ahmadbeigi, Y.; Chirani, A.S.; Soleimani, N.; Mahdavi, M.; Goudarzi, M. Immunopotentiation of the engineered low-molecular-weight pilin targeting Pseudomonas aeruginosa: A combination of immunoinformatics investigation and active immunization. Mol. Immunol. 2020, 124, 70–82. [Google Scholar] [CrossRef]
- Gholami, M.; Chirani, A.S.; Razavi, S.; Falak, R.; Irajian, G. Immunogenicity of a fusion protein containing PilQ and disulphide turn region of PilA from Pseudomonas aeruginosa in mice. Lett. Appl. Microbiol. 2017, 65, 439–445. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, Z.; Dong, Y.; Chen, Y. Recombinant PAL/PilE/FlaA DNA vaccine provides protective immunity against Legionella pneumophila in BALB/c mice. BMC Biotechnol. 2020, 20, 28. [Google Scholar] [CrossRef]
- Du, Y.; Hu, X.; Miao, L.; Chen, J. Current status and development prospects of aquatic vaccines. Front. Immunol. 2022, 13, 1040336. [Google Scholar] [CrossRef] [PubMed]
- Assefa, A.; Abunna, F. Maintenance of Fish Health in Aquaculture: Review of Epidemiological Approaches for Prevention and Control of Infectious Disease of Fish. Vet. Med. Int. 2018, 2018, 5432497. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.B.; Xu, J. Better Adjuvants for Better Vaccines: Progress in Adjuvant Delivery Systems, Modifications, and Adjuvant-Antigen Codelivery. Vaccines 2020, 8, 128. [Google Scholar] [CrossRef]
- Tafalla, C.; Bøgwald, J.; Dalmo, R.A. Adjuvants and immunostimulants in fish vaccines: Current knowledge and future perspectives. Fish Shellfish Immunol. 2013, 35, 1740–1750. [Google Scholar] [CrossRef]
- Moran, H.B.T.; Turley, J.L.; Andersson, M.; Lavelle, E.C. Immunomodulatory properties of chitosan polymers. Biomaterials. 2018, 184, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Rivera-Hernandez, T.; Chatterjee, O.; Walker, M.J.; Moyle, P.M. Semisynthetic, self-adjuvanting vaccine development: Efficient, site-specific sortase A-mediated conjugation of Toll-like receptor 2 ligand FSL-1 to recombinant protein antigens under native conditions and application to a model group A streptococcal vaccine. J. Control. Release 2020, 317, 96–108. [Google Scholar] [CrossRef]
- Dash, P.; Sahoo, P.K.; Gupta, P.K.; Garg, L.C.; Dixit, A. Immune responses and protective efficacy of recombinant outer membrane protein R (rOmpR)-based vaccine of Aeromonas hydrophila with a modified adjuvant formulation in rohu (Labeo rohita). Fish Shellfish Immunol. 2014, 39, 512–523. [Google Scholar] [CrossRef]
- Rauta, P.R.; Nayak, B. Parenteral immunization of PLA/PLGA nanoparticle encapsulating outer membrane protein (Omp) from Aeromonas hydrophila: Evaluation of immunostimulatory action in Labeo rohita (rohu). Fish Shellfish Immunol. 2015, 44, 287–294. [Google Scholar] [CrossRef]
- Unajak, S.; Pumchan, A.; Roytrakul, S.; Sawatdichaikul, O.; Areechon, N. Novel Vaccine Development for Fish Culture Based on the Multiepitope Concept. Methods Mol. Biol. 2022, 2411, 219–240. [Google Scholar] [CrossRef]
- Zhu, F.; Tan, C.; Li, C.; Ma, S.; Wen, H.; Yang, H.; Rao, M.; Zhang, P.; Peng, W.; Cui, Y.; et al. Design of a multi-epitope vaccine against six Nocardia species based on reverse vaccinology combined with immunoinformatics. Front. Immunol. 2023, 14, 1100188. [Google Scholar] [CrossRef]
- Li, M.; Zhu, Y.; Niu, C.; Xie, X.; Haimiti, G.; Guo, W.; Yu, M.; Chen, Z.; Ding, J.; Zhang, F. Design of a multi-epitope vaccine candidate against Brucella melitensis. Sci. Rep. 2022, 12, 10146. [Google Scholar] [CrossRef]
- Sheng, X.; Zhang, H.; Liu, M.; Tang, X.; Xing, J.; Chi, H.; Zhan, W. Development and Evaluation of Recombinant B-Cell Multi-Epitopes of PDHA1 and GAPDH as Subunit Vaccines against Streptococcus iniae Infection in Flounder (Paralichthys olivaceus). Vaccines 2023, 11, 624. [Google Scholar] [CrossRef]
- Nawaz, M.; Ullah, A.; Al-Harbi, A.I.; Haq, M.U.; Hameed, A.R.; Ahmad, S.; Aziz, A.; Raziq, K.; Khan, S.; Irfan, M.; et al. Genome-Based Multi-Antigenic Epitopes Vaccine Construct Designing against Staphylococcus hominis Using Reverse Vaccinology and Biophysical Approaches. Vaccines 2022, 10, 1729. [Google Scholar] [CrossRef] [PubMed]
- Shafaghi, M.; Bahadori, Z.; Madanchi, H.; Ranjbar, M.M.; Shabani, A.A.; Mousavi, S.F. Immunoinformatics-aided design of a new multi-epitope vaccine adjuvanted with domain 4 of pneumolysin against Streptococcus pneumoniae strains. BMC Bioinform. 2023, 24, 67. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.I.; Mou, M.J.; Sanjida, S. Application of reverse vaccinology to design a multi-epitope subunit vaccine against a new strain of Aeromonas veronii. J. Genet. Eng. Biotechnol. 2022, 20, 118. [Google Scholar] [CrossRef] [PubMed]



| Strain | Host Fish Species | Isolated Tissues | Location |
|---|---|---|---|
| A. hydrophila Ahp001 | Parabramis pekinensis | kidney | Nanjing |
| A. hydrophila Ahp002 | Parabramis pekinensis | liver | Nanjing |
| A. hydrophila Ahp003 | Parabramis pekinensis | gill | Huai’an |
| A. hydrophila Ahp004 | Parabramis pekinensis | liver | Huai’an |
| A. hydrophila Ahm001 | Megalobrama amblycephala | liver | Wuhan |
| A. hydrophila Ahc001 | Carassius auratus gibelio | body surface | Hubei |
| A. hydrophila Ahi001 | Ictalurus punctatus | kidney | Sichuan |
| A. hydrophila Ahx001 | Ictalurus punctatus | body surface | Sichuan |
| A. hydrophila Ahx002 | Ictalurus punctatus | body surface | Sichuan |
| A. veronii Avc001 | Carassius auratus | gill | Lianyungang |
| Protein Number | Protein Description | α-Helix | Adhesion | Antigenicity | Percentage of Occurrence (%) |
|---|---|---|---|---|---|
| AJE34457.1 | fimbrial biogenesis outer membrane usher protein | 0 | 0.715 | 0.6061 | 8 |
| AJE34471.1 | TonB-dependent receptor | 0 | 0.555 | 0.6085 | 38 |
| AJE34515.1 | porin OmpA | 0 | 0.715 | 0.6038 | 16 |
| AJE34700.1 | ligand-gated channel protein | 0 | 0.616 | 0.6769 | 30 |
| AJE34708.1 | outer membrane beta-barrel protein | 0 | 0.838 | 0.7068 | 30 |
| AJE35401.1 | type IV pilus secretin PilQ | 0 | 0.565 | 0.6985 | 57 |
| AJE35595.1 | carbohydrate porin | 0 | 0.663 | 0.7585 | 51 |
| AJE35662.1 | efflux transporter outer membrane subunit | 0 | 0.539 | 0.6502 | 32 |
| AJE35814.1 | MtrB/PioB family decaheme-associated outer membrane protein | 0 | 0.723 | 0.5953 | 11 |
| AJE35877.1 | TolC family outer membrane protein | 0 | 0.535 | 0.6247 | 70 |
| AJE36112.1 | siderophore amonabactin TonB-dependent receptor | 0 | 0.657 | 0.6506 | 41 |
| AJE36246.1 | OmpP1/FadL family transporter | 0 | 0.71 | 0.5389 | 22 |
| AJE36247.1 | outer membrane protein transport protein | 0 | 0.825 | 0.6604 | 11 |
| AJE36909.1 | TonB-dependent hemoglobin/transferrin/lactoferrin family receptor | 0 | 0.525 | 0.7156 | 14 |
| AJE37341.1 | OmpA family protein | 0 | 0.612 | 0.725 | 19 |
| AJE37342.1 | porin OmpA | 0 | 0.625 | 0.6875 | 54 |
| AJE37343.1 | OmpA family protein | 0 | 0.559 | 0.6993 | 24 |
| AJE37434.1 | outer membrane protein assembly factor BamA | 0 | 0.637 | 0.6605 | 49 |
| AJE37449.1 | maltoporin LamB | 0 | 0.774 | 0.6319 | 43 |
| AJE37487.1 | outer membrane protein OmpK | 0 | 0.726 | 0.5222 | 41 |
| AJE37516.1 | outer membrane beta-barrel protein | 0 | 0.764 | 0.6778 | 11 |
| AJE37629.1 | TonB-dependent hemoglobin/transferrin/lactoferrin family receptor | 1 | 0.624 | 0.665 | 43 |
| AJE37705.1 | LPS assembly protein LptD | 1 | 0.527 | 0.7443 | 32 |
| AJE37778.1 | porin | 0 | 0.704 | 0.7588 | 19 |
| AJE37804.1 | peptidoglycan DD-metalloendopeptidase family protein | 0 | 0.787 | 0.7381 | 14 |
| AJE38163.1 | porin | 0 | 0.767 | 0.7306 | 54 |
| AJE38201.1 | OmpA family protein | 0 | 0.611 | 0.7071 | 3 |
| AJE38225.1 | porin | 0 | 0.811 | 0.748 | 16 |
| AJE38562.1 | TonB-dependent siderophore receptor | 0 | 0.591 | 0.5896 | 62 |
| AJE34456.1 | hypothetical protein | 1 | 0.667 | 0.5206 | 5 |
| AJE34989.1 | TIGR04219 family outer membrane beta-barrel protein | 0 | 0.691 | 0.7105 | 24 |
| AJE35148.1 | immune inhibitor A | 0 | 0.572 | 0.5513 | 41 |
| AJE37118.1 | DUF1566 domain-containing protein | 0 | 0.605 | 0.6726 | 5 |
| AJE37120.1 | outer membrane protein assembly factor BamE | 0 | 0.608 | 0.6881 | 46 |
| AJE37322.1 | M23 family metallopeptidase | 0 | 0.74 | 0.5346 | 11 |
| AJE37560.1 | OprD family outer membrane porin | 0 | 0.644 | 0.6276 | 19 |
| AJE37735.1 | DUF2860 family protein | 0 | 0.673 | 0.5925 | 16 |
| AJE37742.1 | outer membrane protein assembly factor BamC | 0 | 0.643 | 0.6185 | 57 |
| AJE38071.1 | glycine zipper 2TM domain-containing protein | 0 | 0.815 | 0.8437 | 57 |
| No. | Protein Number | NCBI NR Data Base | Sample Strains | |||
|---|---|---|---|---|---|---|
| A. caviae | A. salmonicida | A. veronii | A. hydrophila | |||
| 1 | AJE34457.1 | 67 | 50 | 50 | 100 | 50 |
| 2 | AJE34471.1 | 100 | 100 | 100 | 100 | 83 |
| 3 | AJE34515.1 | 100 | 100 | 100 | 100 | 50 |
| 4 | AJE34700.1 | 100 | 100 | 100 | 100 | 83 |
| 5 | AJE34708.1 | 100 | 83 | 100 | 100 | 100 |
| 6 | AJE35401.1 | 100 | 100 | 100 | 100 | 100 |
| 7 | AJE35595.1 | 100 | 100 | 100 | 100 | 50 |
| 8 | AJE35662.1 | 100 | 100 | 100 | 100 | 67 |
| 9 | AJE35814.1 | 0 | 0 | 0 | 100 | 50 |
| 10 | AJE35877.1 | 100 | 100 | 100 | 100 | 100 |
| 11 | AJE36112.1 | 100 | 100 | 100 | 100 | 83 |
| 12 | AJE36246.1 | 100 | 100 | 100 | 100 | 17 |
| 13 | AJE36247.1 | 100 | 100 | 100 | 100 | 100 |
| 14 | AJE36909.1 | 100 | 100 | 100 | 100 | 50 |
| 15 | AJE37341.1 | 100 | 100 | 100 | 100 | 83 |
| 16 | AJE37342.1 | 100 | 100 | 100 | 83 | 100 |
| 17 | AJE37343.1 | 100 | 100 | 100 | 100 | 100 |
| 18 | AJE37434.1 | 100 | 100 | 100 | 100 | 83 |
| 19 | AJE37449.1 | 100 | 83 | 100 | 100 | 50 |
| 20 | AJE37487.1 | 100 | 100 | 100 | 100 | 100 |
| 21 | AJE37516.1 | 100 | 100 | 100 | 100 | 83 |
| 22 | AJE37629.1 | 100 | 100 | 100 | 100 | 100 |
| 23 | AJE37705.1 | 100 | 100 | 100 | 100 | 100 |
| 24 | AJE37778.1 | 100 | 100 | 100 | 100 | 100 |
| 25 | AJE37804.1 | 100 | 100 | 100 | 100 | 100 |
| 26 | AJE38163.1 | 100 | 100 | 100 | 100 | 100 |
| 27 | AJE38201.1 | 100 | 100 | 100 | 83 | 83 |
| 28 | AJE38225.1 | 67 | 100 | 67 | 100 | 83 |
| 29 | AJE38562.1 | 100 | 100 | 100 | 100 | 100 |
| 30 | AJE34456.1 | 100 | 50 | 0 | 100 | 67 |
| 31 | AJE34989.1 | 100 | 100 | 100 | 100 | 100 |
| 32 | AJE35148.1 | 0 | 100 | 0 | 100 | 100 |
| 33 | AJE37118.1 | 83 | 100 | 67 | 100 | 83 |
| 34 | AJE37120.1 | 100 | 100 | 100 | 100 | 33 |
| 35 | AJE37322.1 | 0 | 100 | 0 | 100 | 0 |
| 36 | AJE37560.1 | 100 | 100 | 100 | 100 | 83 |
| 37 | AJE37735.1 | 100 | 100 | 100 | 100 | 100 |
| 38 | AJE37742.1 | 100 | 100 | 100 | 100 | 83 |
| 39 | AJE38071.1 | 100 | 100 | 100 | 100 | 67 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Zhang, M.; Xu, Z.; He, Y.; Zhao, X.; Cheng, H.; Chen, X.; Xu, J.; Ding, Z. The Screening of the Protective Antigens of Aeromonas hydrophila Using the Reverse Vaccinology Approach: Potential Candidates for Subunit Vaccine Development. Vaccines 2023, 11, 1266. https://doi.org/10.3390/vaccines11071266
Zhang T, Zhang M, Xu Z, He Y, Zhao X, Cheng H, Chen X, Xu J, Ding Z. The Screening of the Protective Antigens of Aeromonas hydrophila Using the Reverse Vaccinology Approach: Potential Candidates for Subunit Vaccine Development. Vaccines. 2023; 11(7):1266. https://doi.org/10.3390/vaccines11071266
Chicago/Turabian StyleZhang, Ting, Minying Zhang, Zehua Xu, Yang He, Xiaoheng Zhao, Hanliang Cheng, Xiangning Chen, Jianhe Xu, and Zhujin Ding. 2023. "The Screening of the Protective Antigens of Aeromonas hydrophila Using the Reverse Vaccinology Approach: Potential Candidates for Subunit Vaccine Development" Vaccines 11, no. 7: 1266. https://doi.org/10.3390/vaccines11071266
APA StyleZhang, T., Zhang, M., Xu, Z., He, Y., Zhao, X., Cheng, H., Chen, X., Xu, J., & Ding, Z. (2023). The Screening of the Protective Antigens of Aeromonas hydrophila Using the Reverse Vaccinology Approach: Potential Candidates for Subunit Vaccine Development. Vaccines, 11(7), 1266. https://doi.org/10.3390/vaccines11071266







