Circulation of Pestiviruses in Small Ruminants from Greece: First Molecular Identification of Border Disease Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection Strategy
2.2. Serological Detection of Specific Ruminant Pestivirus Antibodies by ELISA
2.3. Real-Time RT-PCR and Antigen (Ag) ELISA
2.4. Phylogenetic Analysis
3. Results
3.1. Antibody Prevalence
3.2. Viral Detection with Real Time RT-PCR and Ag ELISA
3.3. Phylogenetic Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nettleton, P.F.; Entrican, G. Ruminant Pestiviruses. Br. Vet. J. 1995, 151, 615. [Google Scholar] [CrossRef] [PubMed]
- Ridpath, J.F. Emerging Pestiviruses Infecting Domestic and Wildlife Hosts. Anim. Health Res. Rev. 2015, 16, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Houe, H. Epidemiological Features and Economical Importance of Bovine Virus Diarrhoea Virus (BVDV) Infections. Vet. Microbiol. 1999, 64, 89–107. [Google Scholar] [CrossRef] [PubMed]
- Moennig, V.; Eicken, K.; Flebbe, U.; Frey, H.R.; Grummer, B.; Haas, L.; Greiser-Wilke, I.; Liess, B. Implementation of Two-Step Vaccination in the Control of Bovine Viral Diarrhoea (BVD). Prev. Vet. Med. 2005, 72, 109–114. [Google Scholar] [CrossRef]
- Nettleton, P.F.; Gilray, J.A.; Russo, P.; Dlissi, E. Border Disease of Sheep and Goats. Vet. Res. 1998, 29, 327–340. [Google Scholar]
- Krametter-Froetscher, R.; Duenser, M.; Preyler, B.; Theiner, A.; Benetka, V.; Moestl, K.; Baumgartner, W. Pestivirus Infection in Sheep and Goats in West Austria. Vet. J. 2010, 186, 342–346. [Google Scholar] [CrossRef]
- Løken, T.; Hyllseth, B.; Larsen, H.J. Border Disease in Norway: Serological Examination of Affected Sheep Flocks. Acta Vet. Scand. 1982, 23, 46–52. [Google Scholar] [CrossRef]
- Becher, P.; Orlich, M.; Shannon, A.D.; Horner, G.; König, M.; Thiel, H.J. Phylogenetic Analysis of Pestiviruses from Domestic and Wild Ruminants. J. Gen. Virol. 1997, 78 Pt 6, 1357–1366. [Google Scholar] [CrossRef]
- Rosamilia, A.; Grattarola, C.; Caruso, C.; Peletto, S.; Gobbi, E.; Tarello, V.; Caroggio, P.; Dondo, A.; Masoero, L.; Acutis, P.L. Detection of Border Disease Virus (BDV) Genotype 3 in Italian Goat Herds. Vet. J. 2014, 199, 446–450. [Google Scholar] [CrossRef]
- Barandika, J.F.; Aduriz, G.; Juste, R.A.; Hurtado, A. Border Disease Virus Seroprevalence Correlates to Antibodies in Bulk-Tank Milk and Reproductive Performance of Dairy Sheep Flocks. J. Dairy Sci. 2010, 93, 2444–2449. [Google Scholar] [CrossRef]
- Kittelberger, R.; Pigott, C. The Use of Pestivirus Antigen ELISA Currently Available for the Detection of Hairy Shaker Disease/Border Disease Virus in Sheep. N. Z. Vet. J. 2008, 56, 343–344. [Google Scholar] [CrossRef] [PubMed]
- Feknous, N.; Hanon, J.B.; Tignon, M.; Khaled, H.; Bouyoucef, A.; Cay, B. Seroprevalence of Border Disease Virus and Other Pestiviruses in Sheep in Algeria and Associated Risk Factors. BMC Vet. Res. 2018, 14, 339. [Google Scholar] [CrossRef] [PubMed]
- Willoughby, K.; Valdazo-González, B.; Maley, M.; Gilray, J.; Nettleton, P.F. Development of a Real Time RT-PCR to Detect and Type Ovine Pestiviruses. J. Virol. Methods 2006, 132, 187–194. [Google Scholar] [CrossRef]
- Schweizer, M.; Peterhans, E. Pestiviruses. Annu. Rev. Anim. Biosci. 2014, 2, 141–163. [Google Scholar] [CrossRef]
- Dubois, E.; Russo, P.; Prigent, M.; Thiéry, R. Genetic characterization of ovine pestiviruses isolated in France, between 1985 and 2006. Vet. Microbiol. 2008, 130, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Yeşilbağ, K.; Alpay, G.; Becher, P. Variability and global distribution of subgenotypes of bovine viral diarrhea virus. Viruses 2017, 9, 128. [Google Scholar] [CrossRef] [PubMed]
- Righi, C.; Petrini, S.; Pierini, I.; Giammarioli, M.; de Mia, G.M. Global Distribution and Genetic Heterogeneity of Border Disease Virus. Viruses 2021, 13, 950. [Google Scholar] [CrossRef]
- Spais, A.G.; Papadopoulos, O.; Vantsis, J.T. An Extensive Outbreak of Border Disease in Greece (A Preliminary Report). In Proceedings of the 20th World Veterinary Congress, Thessaloniki, Greece, 6–12 July 1975; pp. 1461–1462. [Google Scholar]
- Vilček, S.; Herring, A.J.; Herring, J.A.; Nettleton, P.F.; Lowings, J.P.; Paton, D.J. Pestiviruses Isolated from Pigs, Cattle and Sheep Can Be Allocated into at Least Three Genogroups Using Polymerase Chain Reaction and Restriction Endonuclease Analysis. Arch. Virol. 1994, 136, 309–323. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Kaiser, V.; Nebel, L.; Schüpbach-Regula, G.; Zanoni, R.G.; Schweizer, M. Influence of Border Disease Virus (BDV) on Serological Surveillance within the Bovine Virus Diarrhea (BVD) Eradication Program in Switzerland. BMC Vet. Res. 2017, 13, 21. [Google Scholar] [CrossRef]
- Arnal, M.C.; Fernández-de-Luco, D.; Riba, L.; Maley, M.; Gilray, J.; Willoughby, K.; Vilcek, S.; Nettleton, P.F. A Novel Pestivirus Associated with Deaths in Pyrenean Chamois (Rupicapra Pyrenaica Pyrenaica). J. Gen. Virol. 2004, 85, 3653–3657. [Google Scholar] [CrossRef] [PubMed]
- Valdazo-González, B.; Álvarez-Martínez, M.; Sandvik, T. Genetic and Antigenic Typing of Border Disease Virus Isolates in Sheep from the Iberian Peninsula. Vet. J. 2007, 174, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Luzzago, C.; Ebranati, E.; Cabezon, O.; Fernandez-Sirera, L.; Lavin, S.; Rosell, R.; Veo, C.; Rossi, L.; Cavallero, S.; Lanfranchi, P.; et al. Spatial and Temporal Phylogeny of Border Disease Virus in Pyrenean Chamois (Rupicapra p. Pyrenaica). PLoS ONE 2016, 11, e0168232. [Google Scholar] [CrossRef]
- García-Pérez, A.L.; Minguijón, E.; Barandika, J.F.; Aduriz, G.; Povedano, I.; Juste, R.A.; Hurtado, A. Detection of Border Disease Virus in Fetuses, Stillbirths, and Newborn Lambs from Natural and Experimental Infections. J. Vet. Diagn. Investig. 2009, 21, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Van Rijn, P.A.; Van Gennip, H.G.P.; Leendertse, C.H.; Bruschke, C.J.M.; Paton, D.J.; Moormann, R.J.M.; van Oirschot, J.T. Subdivision of the Pestivirus genus based on envelope glycoprotein E2. Virology 1997, 237, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Becher, P.; Ramirez, R.A.; Orlich, M.; Rosales, S.C.; König, M.; Schweizer, M.; Stalder, H.; Schirrmeier, H.; Thiel, H.-J. Genetic and antigenic characterization of novel pestivirus genotypes: Implications for classification. Virology 2003, 311, 96–104. [Google Scholar] [CrossRef]
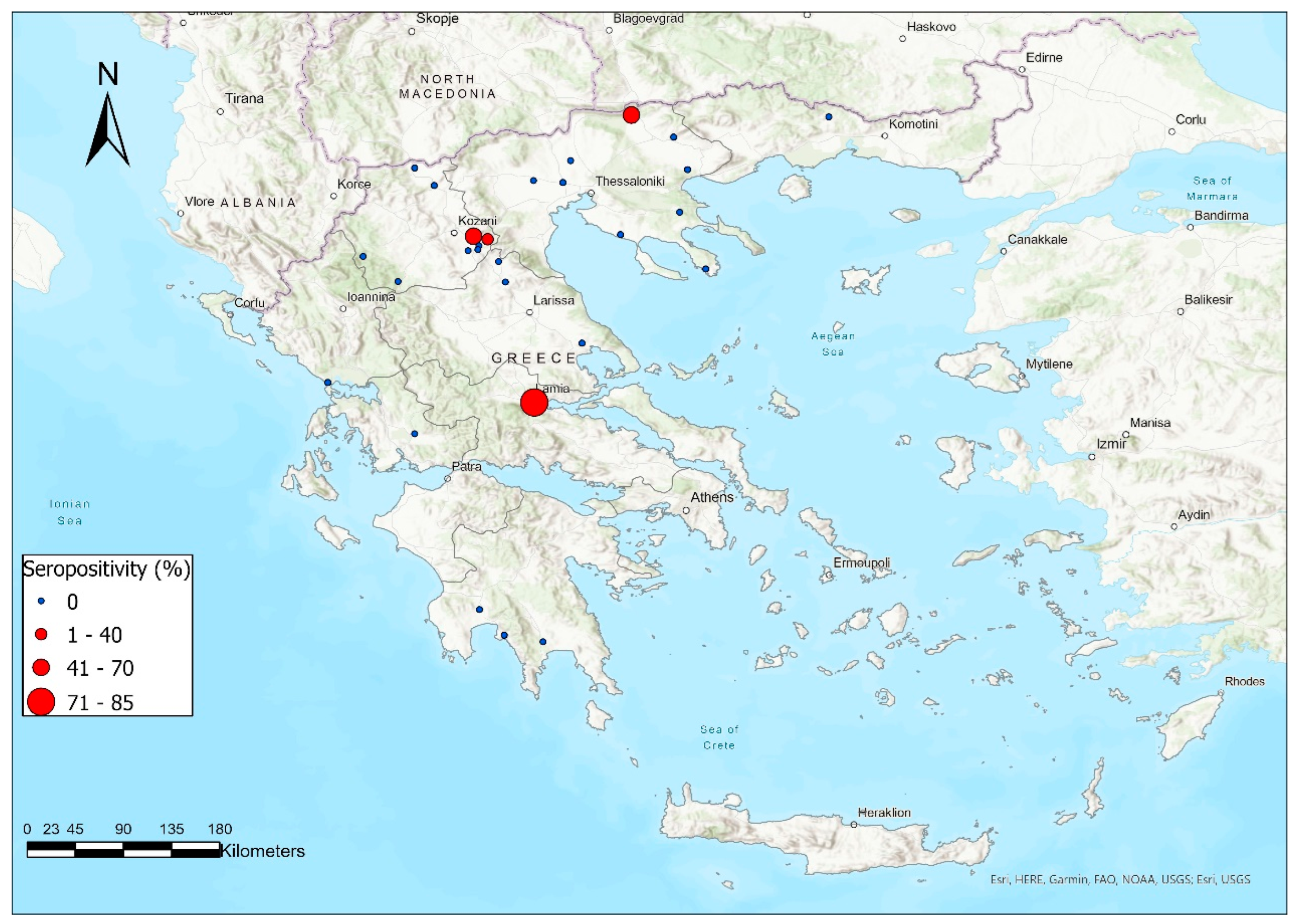
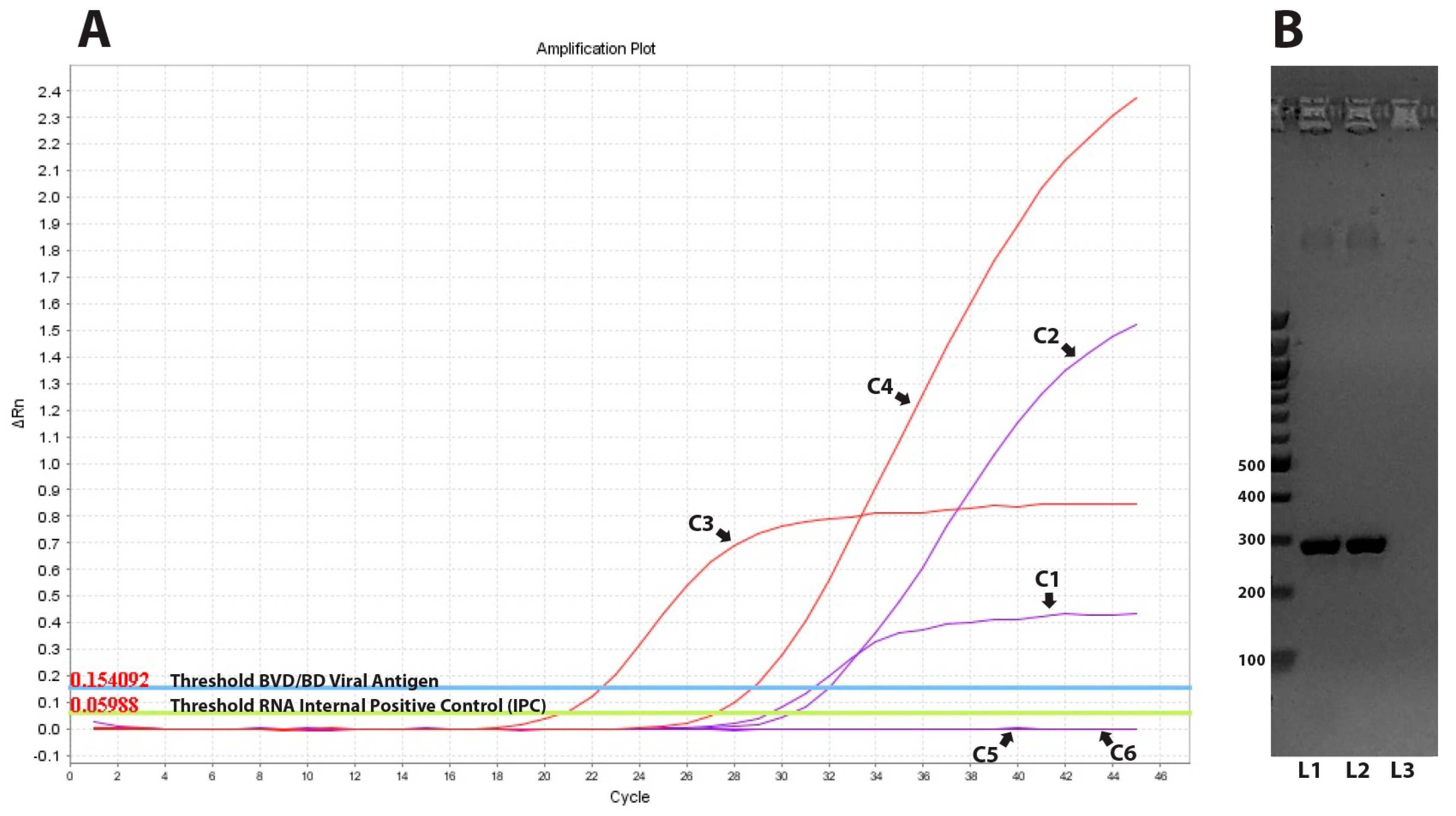
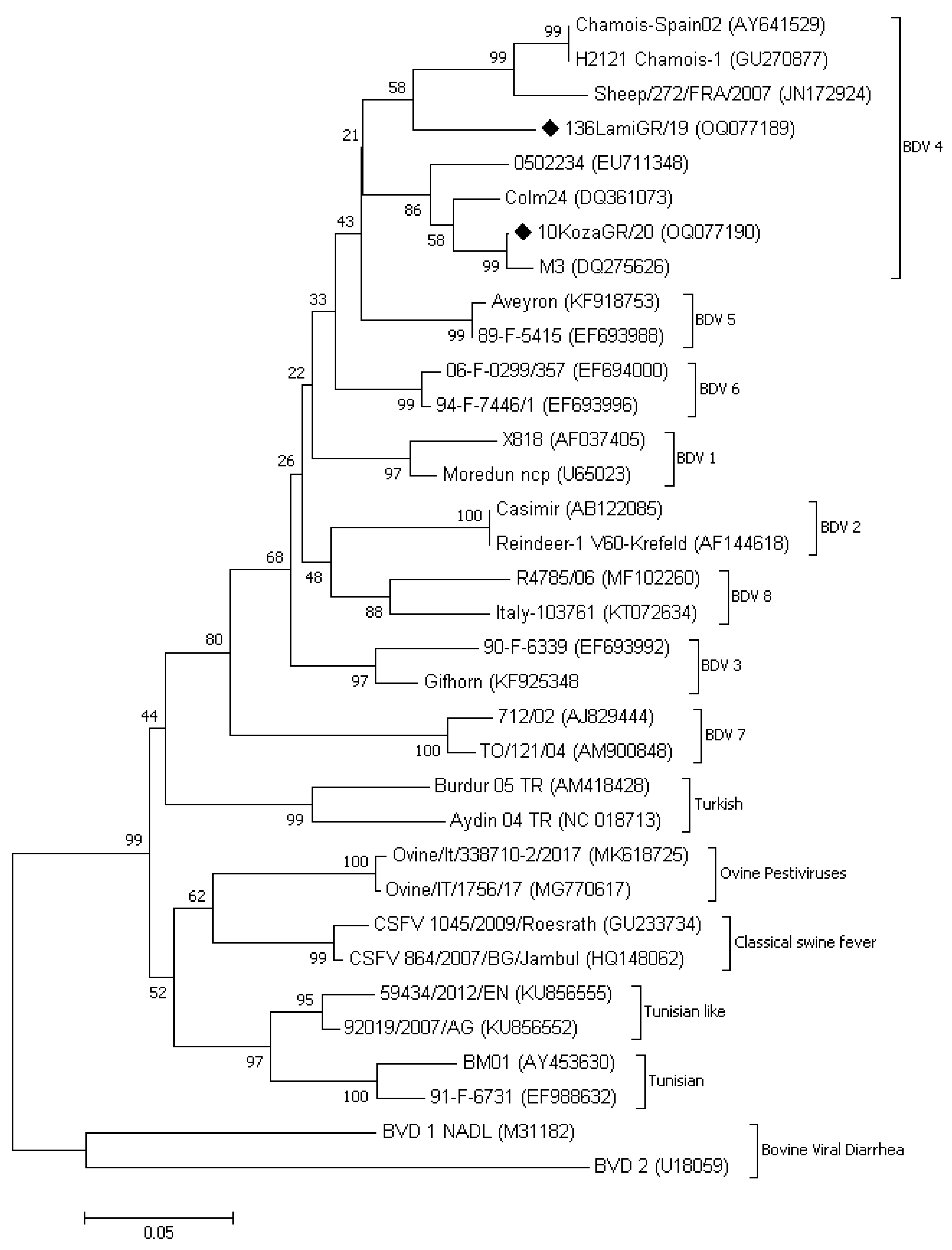
| Flock/ Herd ID | Area | Species | Breed * | ELISA Positive/Tested Animals (Seropositivity%) | PCR Positive/Tested Animals (Genotype) |
|---|---|---|---|---|---|
| 1 | Kozani | Sheep | Lacaune | 7/10 (70%) | 0/3 |
| 2 | Kozani | Sheep | Lacaune | 0/10 (0%) | 0/0 |
| 3 | Kozani | Sheep | Chios cross | 0/10 (0%) | 0/0 |
| 4 | Xanthi | Sheep | Assaf cross | 0/20 (0%) | 0/0 |
| 5 | Lakonia | Sheep | Lacaune cross | 0/20 (0%) | 0/0 |
| 6 | Serres | Sheep | Assaf cross | 0/19 (0%) | 0/0 |
| 7 | Messinia | Sheep | Chios cross | 0/20 (0%) | 0/0 |
| 8 | Thessaloniki | Sheep | Chios | 0/20 (0%) | 0/0 |
| 9 | Kozani | Goat | Alpine cross | 0/20 (0%) | 0/0 |
| 10 | Florina | Goat | Saanen cross | 0/7 (0%) | 0/0 |
| 11 | Magnisia | Sheep | Chios | 0/20 (0%) | 0/0 |
| 12 | Serres | Sheep | Lacaune | 12/20 (60%) | 0/8 |
| 13 | Fthiotida | Sheep | Lacaune | 17/20 (85%) | 1/3 (BDV-4) ** |
| 14 | Florina | Goat | Alpine cross | 0/16 (0%) | 0/0 |
| 15 | Aitolokarnania | Sheep | Chios | 0/20 (0%) | 0/0 |
| 16 | Chalkidiki | Sheep | Chios | 0/20 (0%) | 0/0 |
| 17 | Kilkis | Sheep | Chios | 0/20 (0%) | 0/0 |
| 18 | Preveza | Sheep | Chios | 0/20 (0%) | 0/0 |
| 19 | Pella | Sheep | Chios | 0/20 (0%) | 0/0 |
| 20 | Grevena | Sheep | Lacaune cross | 0/20 (0%) | 0/0 |
| 21 | Grevena | Sheep | Assaf cross | 0/10 (0%) | 0/0 |
| 22 | Chalkidiki | Goat | Alpine cross | 0/10 (0%) | 0/0 |
| 23 | Messinia | Sheep | Lacaune cross | 0/20 (0%) | 0/0 |
| 24 | Larissa | Sheep | Chios cross | 0/20 (0%) | 0/0 |
| 25 | Larissa | Sheep | Lacaune cross | 0/20 (0%) | 0/0 |
| 26 | Chalkidiki | Sheep | Assaf | 0/19 (0%) | 0/0 |
| 27 | Chalkidiki | Sheep | Lacaune cross | 0/9 (0%) | 0/0 |
| 28 | Kozani | Sheep | Lacaune cross | 4/10 (40%) | 1/6 (BDV-4) *** |
| TOTAL | 40/470 (8.51%) | 2/20 (BDV-4) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouzalas, I.G.; Gelasakis, A.I.; Chassalevris, T.; Apostolidi, E.D.; Pappas, F.; Ekateriniadou, L.; Boukouvala, E.; Zdragas, A. Circulation of Pestiviruses in Small Ruminants from Greece: First Molecular Identification of Border Disease Virus. Vaccines 2023, 11, 918. https://doi.org/10.3390/vaccines11050918
Bouzalas IG, Gelasakis AI, Chassalevris T, Apostolidi ED, Pappas F, Ekateriniadou L, Boukouvala E, Zdragas A. Circulation of Pestiviruses in Small Ruminants from Greece: First Molecular Identification of Border Disease Virus. Vaccines. 2023; 11(5):918. https://doi.org/10.3390/vaccines11050918
Chicago/Turabian StyleBouzalas, Ilias G., Athanasios I. Gelasakis, Taxiarchis Chassalevris, Evangelia D. Apostolidi, Fotis Pappas, Loukia Ekateriniadou, Evridiki Boukouvala, and Antonios Zdragas. 2023. "Circulation of Pestiviruses in Small Ruminants from Greece: First Molecular Identification of Border Disease Virus" Vaccines 11, no. 5: 918. https://doi.org/10.3390/vaccines11050918
APA StyleBouzalas, I. G., Gelasakis, A. I., Chassalevris, T., Apostolidi, E. D., Pappas, F., Ekateriniadou, L., Boukouvala, E., & Zdragas, A. (2023). Circulation of Pestiviruses in Small Ruminants from Greece: First Molecular Identification of Border Disease Virus. Vaccines, 11(5), 918. https://doi.org/10.3390/vaccines11050918







