A CTB-SARS-CoV-2-ACE-2 RBD Mucosal Vaccine Protects Against Coronavirus Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Construction and Isolation of the CTB-SARS-CoV-2-ACE-2-RBD Vaccine Fusion Protein
2.2. Assessment of CTB-SARS-CoV-2-ACE-2-RBD Fusion Protein Purity by FPLC
2.3. Titration of SARS-CoV-2-ACE-2-RBD Antibody Titers
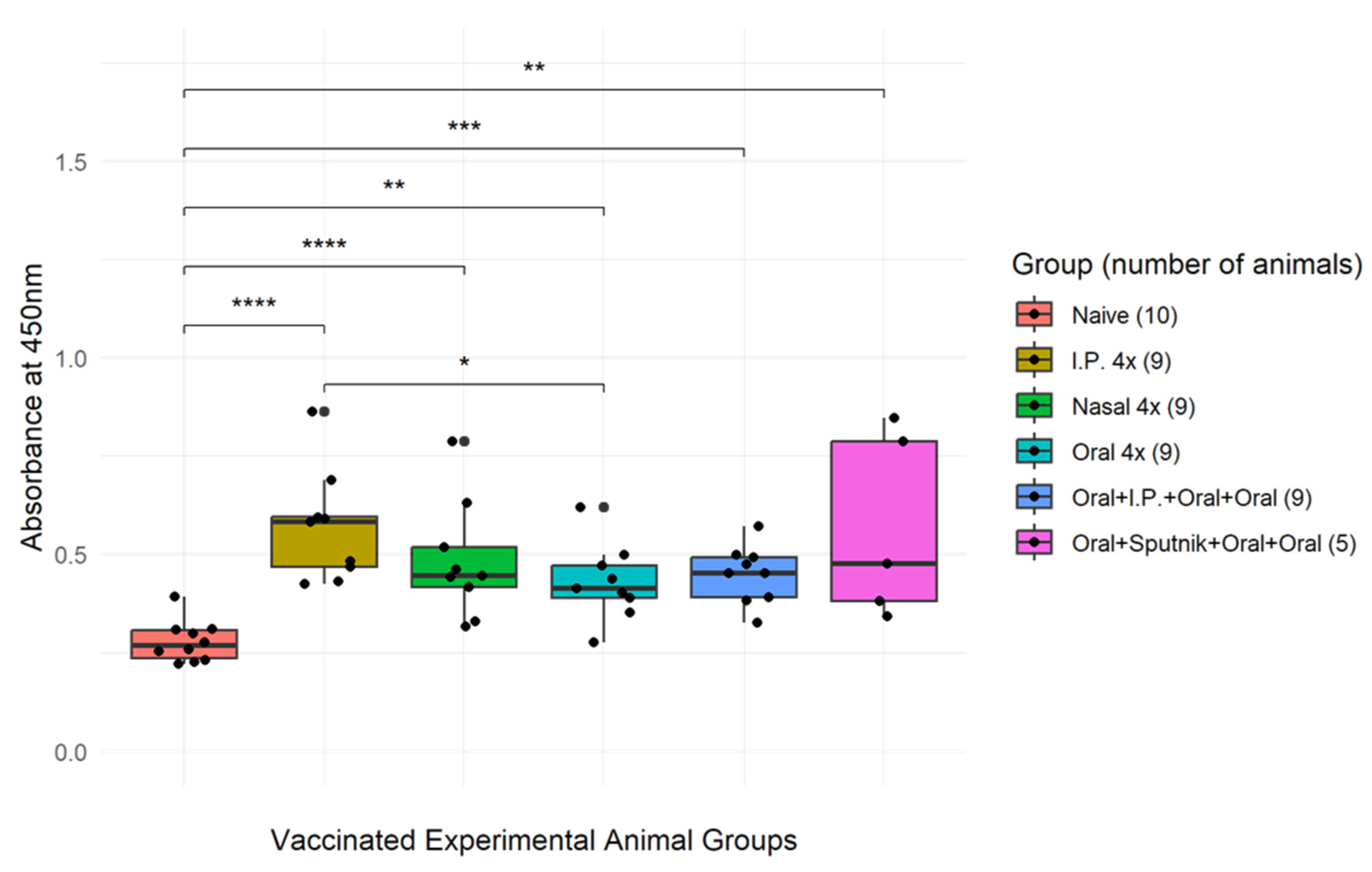
2.4. ELISA Determination of Vaccine-Induced ACE-2-RBD-Specific IgA
2.5. Mucosal Vaccine Neutralization of SARS-CoV-2 Infection In Vitro by a Virus Neutralization (VN) Assay
2.6. Detection of Anti-ACE-2-RBD Antibodies in Lung Alveolar Tissues
2.7. Statistical Analysis
3. Results
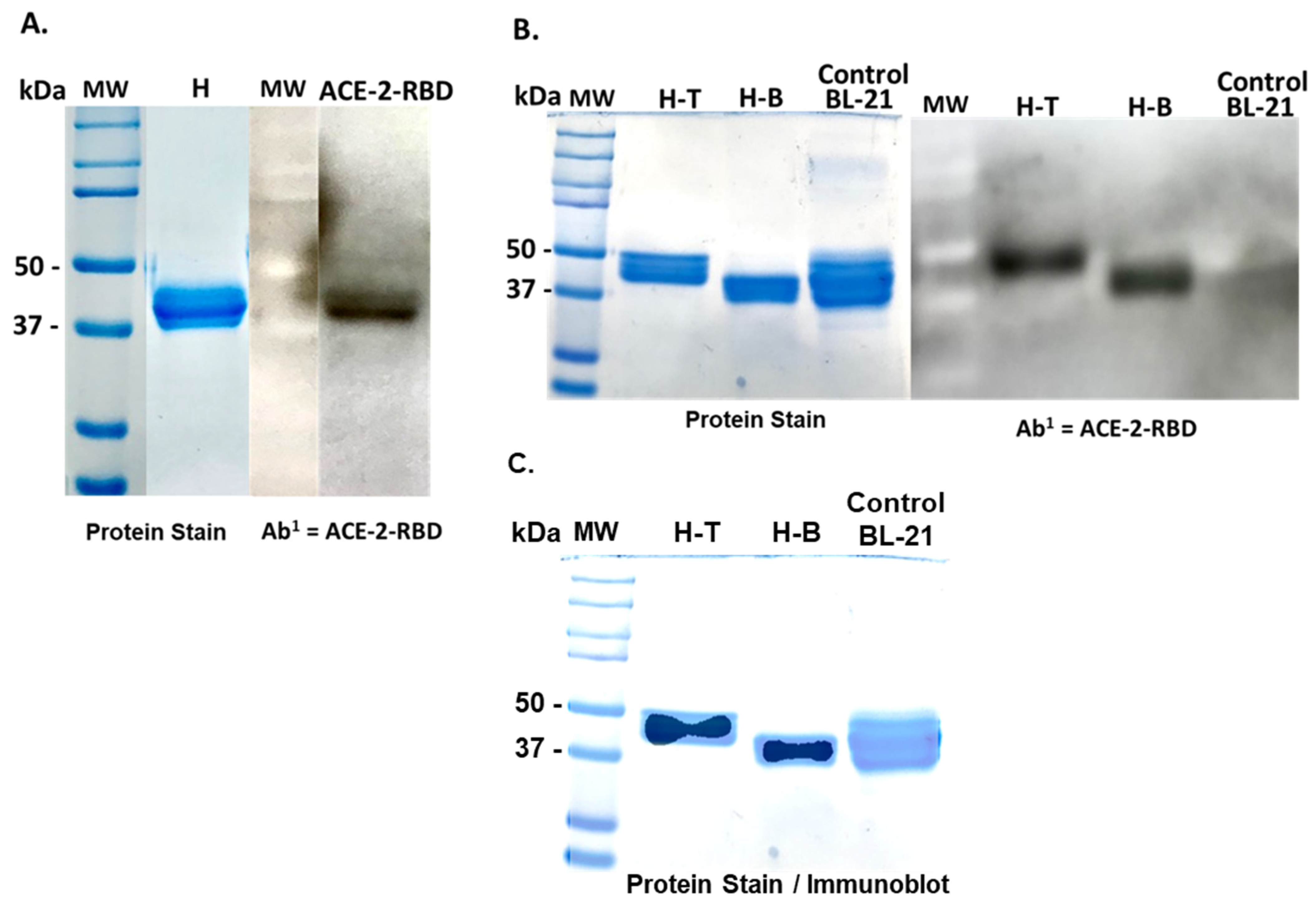
3.1. Construction of a CTB-SARS-CoV-2 Fusion Protein Subunit Mucosal Vaccine
3.2. Isolation and Purification of a CTB-SARS-CoV-2-ACE-2-RBD Fusion Protein Mucosal Vaccine
3.3. Optimization of CTB-SARS-CoV-2-ACE-2-RBD Mucosal Vaccine Immunization
3.4. Mucosal Vaccination Generates Virus Specific IgA Antibodies
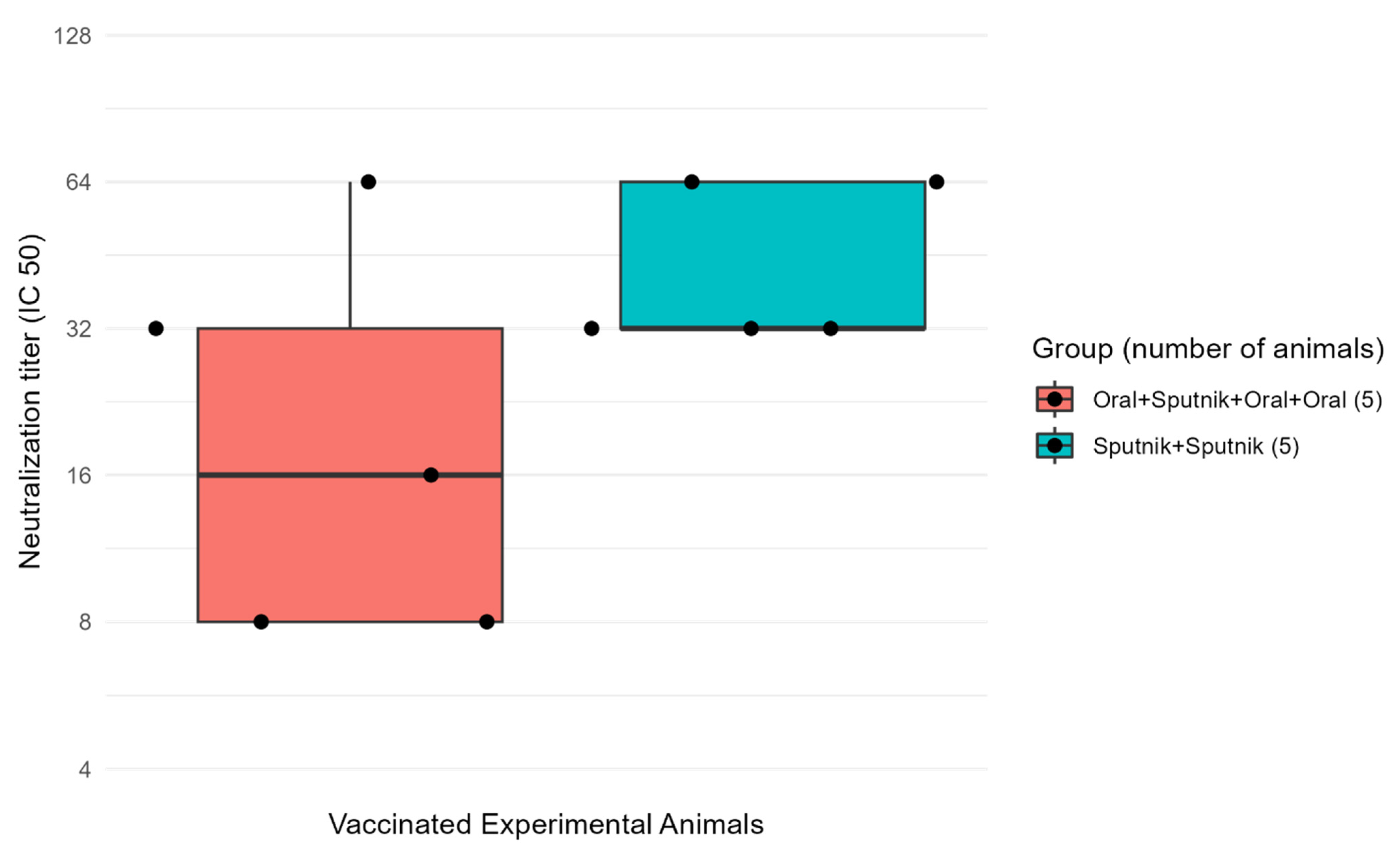
3.5. Virus Neutralization Is Enhanced by Combined Mucosal and Parenteral Immunization
3.6. Mucosal Vaccine Neutralization of SARS-CoV-2
3.7. Detection of SARS-CoV-2 Specific Antibodies in Vaccinated Mouse Lung Alveolar Tissues
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, 35. [Google Scholar] [CrossRef] [PubMed]
- de Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Xu, Y.; Bao, L.; Zhang, L.; Yu, P.; Qu, Y.; Zhu, H.; Zhao, W.; Han, Y.; Qin, C. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses 2019, 11, 59. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Zhao, G.; Li, L.; He, Y.; Zhou, Y.; Zheng, B.J.; Jiang, S. Antigenicity and immunogenicity of SARS-CoV S protein receptor-binding domain stably expressed in CHO cells. Biochem. Biophys. Res. Commun. 2009, 384, 486–490. [Google Scholar] [CrossRef]
- Roper, R.L.; Rehm, K.E. SARS vaccines: Where are we? Expert Rev. Vaccines 2009, 8, 887–898. [Google Scholar] [CrossRef]
- Jaume, M.; Yip, M.S.; Kam, Y.W.; Cheung, C.Y.; Kien, F.; Roberts, A.; Li, P.H.; Dutry, I.; Escriou, N.; Daeron, M.; et al. SARS-CoV subunit vaccine: Antibody-mediated neutralization and enhancement. Hong Kong Med. J. 2012, 18 (Suppl. S2), 31–36. [Google Scholar] [PubMed]
- Lebaeu, J. Future of mucosal immunology: Studying an integrated system-wide organ. Nat. Immunol. 2010, 11, 558–560. [Google Scholar]
- Uscher-Pines, L.; Maurer, J.; Harris, K.M. Racial and ethnic disparities in uptake and location of vaccination for 2009-H1N1 and seasonal influenza. Am. J. Public Health 2011, 101, 1252–1255. [Google Scholar] [CrossRef]
- CDC. CDC COVID-19. Hospitalization and Death by Race/Ethnicity. 2019. Available online: https://www.cdc.gov/coronavirus/2019-ncov/covid-data/investigations-discovery/hospitalization-death-by-race-ethnicity.html (accessed on 14 December 2023).
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Tan, W. Genomic characterization and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Wong, S.K.; Li, W.; Moore, M.J.; Choe, H.; Farzan, M.A. 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004, 279, 3197–3201. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Neutra, M.R.; Kozlowski, P.A. Mucosal vaccines: The promise and the challenge. Nat. Rev. Immunol. 2006, 6, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a virus attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Lu, H.; Siddiqui, P.; Zhou, Y.; Jiang, S. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation dependent epitopes that induce highly potent neutralizing antibodies. J. Immunol. 2005, 174, 4908–4915. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Zhao, G.; He, Y.; Guo, Y.; Zheng, B.J.; Jiang, S.; Zhou, Y. Receptor-binding domain of SARS-CoV spike protein induces long-term protective immunity in an animal model. Vaccine 2007, 25, 2832–2838. [Google Scholar] [CrossRef] [PubMed]
- Siegrist, C.A. Mechanisms underlying adverse reactions to vaccines. J. Comp. Pathol. 2007, 137 (Suppl. S1), S46–S50, Erratum in J. Comp. Pathol. 2023, 138, 169. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fett, C.; Zhao, J.; Meyerholz, D.K.; Perlman, S. Virus-Specific Memory CD8 T Cells Provide Substantial Protection from Lethal Severe Acute Respiratory Syndrome Coronavirus Infection. J. Virol. 2014, 88, 11034–11044. [Google Scholar] [CrossRef] [PubMed]
- Kiyono, H.; Fukuyama, S. NALT- versus Peyer’s-patch-mediated mucosal immunity. Nat. Rev. Immunol. 2004, 4, 699–710. [Google Scholar] [CrossRef]
- Kunkel, E.J.; Kim, C.H.; Lazarus, N.H.; Vierra, M.A.; Soler, D.; Bowman, E.P.; Butcher, E.C. CCR10 expression is a common feature of circulating and mucosal epithelial tissue IgA Ab-secreting 3. cells. J. Clin. Investig. 2003, 111, 1001–1010. [Google Scholar] [CrossRef]
- Lazarus, N.H.; Kunkel, E.J.; Johnston, B.; Wilson, E.; Youngman, K.R.; Butcher, E.C. A common mucosal chemokine (mucosae-associated epithelial chemokine/CCL28) selectively attracts IgA plasmablasts. J. Immunol. 2003, 170, 3799–3805. [Google Scholar] [CrossRef]
- Silva-Sanchez, A.; Randall, T.D. Anatomical Uniqueness of the Mucosal Immune System (GALT, NALT, iBALT) for the Induction and Regulation of Mucosal Immunity and Tolerance. Mucosal Vaccines 2020, 21, 7149644. [Google Scholar]
- Chen, K.; Cerutti, A. Vaccination strategies to promote mucosal antibody responses. Immunity 2010, 33, 479–491. [Google Scholar] [CrossRef]
- Cerutti, A.; Chen, K.; Chorny, A. Immunoglobulin responses at the mucosal interface. Annu. Rev. Immunol. 2011, 29, 273–293. [Google Scholar] [CrossRef]
- van Ginkel, F.W.; Nguyen, H.H.; McGhee, J.R. Vaccines for mucosal immunity to combat emerging infectious diseases. Emerg. Infect. Dis. 2000, 6, 123–132. [Google Scholar] [CrossRef]
- Mestecky, J. The common mucosal immune system and current strategies for induction of immune responses in external secretions. J. Clin. Immunol. 1987, 7, 265–276. [Google Scholar] [CrossRef]
- Arakawa, T.; Chong, D.K.X.; Langridge, W.H.R. Efficacy of a Food Plant-Based Oral Cholera Toxin B Subunit Vaccine. Nat. Biotechnol. 1998, 16, 292–297. [Google Scholar] [CrossRef]
- Kim, P.H.; Eckmann, L.; Lee, W.J.; Han, W.; Kagnoff, M.F. Cholera toxin and cholera toxin B subunit induce IgA switching through the action of TGF-beta 1. J. Immunol. 1998, 160, 1198–1203. [Google Scholar] [CrossRef]
- Leach, R. Why a COVID-19 vaccine might not be the answer we’re hoping for. Infect. Control Today 2020, 24, 22–23. [Google Scholar]
- Arakawa, T.; Daniel, K.X.; Chong, J.; Merritt, L.; Langridge, W.H.R. Expression of cholera toxin B subunit oligomers in transgenic potato plants. Transgenic Res. 1997, 6, 403–413. [Google Scholar] [CrossRef]
- Yu, J.; Arakawa, T.; Chong, D.; Langridge, W. Assembly of cholera toxin–antigen fusion proteins in transgenic potato. Transgenics 2001, 3, 153–162. [Google Scholar]
- Kim, T.-G.; Huy, N.; Kim, M.; Jeong, D.; Jang, Y.; Yang, M.; Langridge, W.; Lee, J. Immunogenicity of a cholera toxin B subunit Porphyromonas gingivalis fimbrial antigen fusion protein expressed in E. coli. Mol. Biotechnol. 2009, 41, 157–164. [Google Scholar] [CrossRef]
- Kim, T.-G.; Gruber, A.; Langridge, W. HIV-1 gp120 V3 cholera toxin B subunit fusion gene expression in transgenic potato. Protein Expr. Purif. 2004, 37, 196–202. [Google Scholar] [CrossRef]
- Hou, J.; Ying, L.; Jenny, H.; Hongzhi, W.; Ran, T.; Yiming, S. Cholera toxin B subunit acts as a potent systemic adjuvant for HIV-1 DNA vaccination intramuscularly in mice. Hum. Vaccin. Immunother. 2014, 10, 1274–1283. [Google Scholar] [CrossRef]
- Wu, H.-Y. Nasal lymphoid tissue intranasal immunization and compartmentalization of the common mucosal immune system. Immunol. Res. 1997, 16, 187–201. [Google Scholar] [CrossRef]
- Gloudemans, A.K.; Plantinga, M.; Guilliams, M.; Willart, M.A.; Ozir-Fazalalikhan, A.; Van Der Ham, A.; Boon, L.; Harris, N.L.; Hammad, H.; Hoogsteden, H.C. The Mucosal Adjuvant Cholera Toxin B Instructs Non-Mucosal Dendritic Cells to Promote IgA Production via Retinoic Acid and TGF-beta. PLoS ONE 2013, 8, e59822. [Google Scholar] [CrossRef]
- Paudel, P.C.; Brochu, H.; Popowski, K.D.; Gracieux, M.C.; Cores, J.; Huang, K.; Hensley, M.T.; Harrell, E.; Vandergriff, A.C. Inhalation of lung spheroid cell secretome and exosomes promotes lung repair in pulmonary fibrosis. Nat. Commun. 2020, 11, 1064. [Google Scholar]
- Gierer, S.; Bertram, S.; Kaup, F.; Wrensch, F.; Heurich, A.; Krämer-Kühl, A.; Welsch, K.; Winkler, M.; Meyer, B.; Drosten, C.; et al. The spike protein of the emerging beta coronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 2013, 87, 5502–5511. [Google Scholar] [CrossRef]
- Brandtzaeg, P. Mucosal immunity: Induction, dissemination, and effector functions. Scand. J. Immunol. 2009, 70, 505–515. [Google Scholar] [CrossRef]
- Renz, H.; Brandtzaeg, P.; Hornef, M. the impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat. Rev. Immunol. 2012, 12, 9–23. [Google Scholar] [CrossRef]
- Ahlawat, S.; Krishna, A.; Sharma, K. Immunological co-ordination between gut and lungs in SARS-CoV-2. Virus Res. 2020, 286, 198103. [Google Scholar] [CrossRef]
- Sencio, V.; Machado, M.G.; Trottein, F. The lung-gut axis during viral respiratory infections: The impact of gut dysbiosis on secondary disease outcomes. Mucosal Immunol. 2021, 14, 296–304. [Google Scholar] [CrossRef] [PubMed]




| Group | Delivery | Vaccine Treatment (Total of 4 Doses at 4-Week Intervals) | Dose |
|---|---|---|---|
| 1 (n = 10) | Oral | (4×) Bicarbonate buffer (negative control) | 0.5 mL |
| 2 (n = 10) | Oral | (4×) 15 μg of CTB-SARS-CoV-2–ACE2-RBD in Bicarbonate Buffer | 0.5 mL |
| 3 (n = 10) | Parenteral (i.p.) | (4×) 15 μg of CTB-SARS-CoV-2–ACE2-RBD in PBS | 0.3 mL |
| 4 (n = 10) | Oral + Parenteral + Oral + Oral | (Oral 3×) 15 μg of CTB-SARS-CoV-2–ACE2-RBD in Bicarbonate Buffer (i.p. 1×) 15 μg of CTB-SARS-CoV-2–ACE2-RBD in PBS | 0.5 mL 0.3 mL |
| 5 (n = 10) | Nasal | (4×) 15 μg of CTB-SARS-CoV-2–ACE2-RBD in Bicarbonate Buffer | 0.05 mL |
| 6 (n = 10) | Oral + Parenteral + Oral + Oral | (Oral 3×) 15 μg of CTB-SARS-CoV-2–ACE2-RBD in Bicarbonate Buffer (i.p. 1×) Sputnik V (full human dose) | 0.5 mL 0.5 mL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dénes, B.; Fuller, R.N.; Kelin, W.; Levin, T.R.; Gil, J.; Harewood, A.; Lőrincz, M.; Wall, N.R.; Firek, A.F.; Langridge, W.H.R. A CTB-SARS-CoV-2-ACE-2 RBD Mucosal Vaccine Protects Against Coronavirus Infection. Vaccines 2023, 11, 1865. https://doi.org/10.3390/vaccines11121865
Dénes B, Fuller RN, Kelin W, Levin TR, Gil J, Harewood A, Lőrincz M, Wall NR, Firek AF, Langridge WHR. A CTB-SARS-CoV-2-ACE-2 RBD Mucosal Vaccine Protects Against Coronavirus Infection. Vaccines. 2023; 11(12):1865. https://doi.org/10.3390/vaccines11121865
Chicago/Turabian StyleDénes, Béla, Ryan N. Fuller, Wayne Kelin, Tessa R. Levin, Jaipuneet Gil, Aaren Harewood, Márta Lőrincz, Nathan R. Wall, Anthony F. Firek, and William H. R. Langridge. 2023. "A CTB-SARS-CoV-2-ACE-2 RBD Mucosal Vaccine Protects Against Coronavirus Infection" Vaccines 11, no. 12: 1865. https://doi.org/10.3390/vaccines11121865
APA StyleDénes, B., Fuller, R. N., Kelin, W., Levin, T. R., Gil, J., Harewood, A., Lőrincz, M., Wall, N. R., Firek, A. F., & Langridge, W. H. R. (2023). A CTB-SARS-CoV-2-ACE-2 RBD Mucosal Vaccine Protects Against Coronavirus Infection. Vaccines, 11(12), 1865. https://doi.org/10.3390/vaccines11121865







