Transcriptional Analysis of lncRNA and Target Genes Induced by Influenza A Virus Infection in MDCK Cells
Abstract
:1. Background
2. Results
2.1. Characterization of IAV Infection in MDCKs
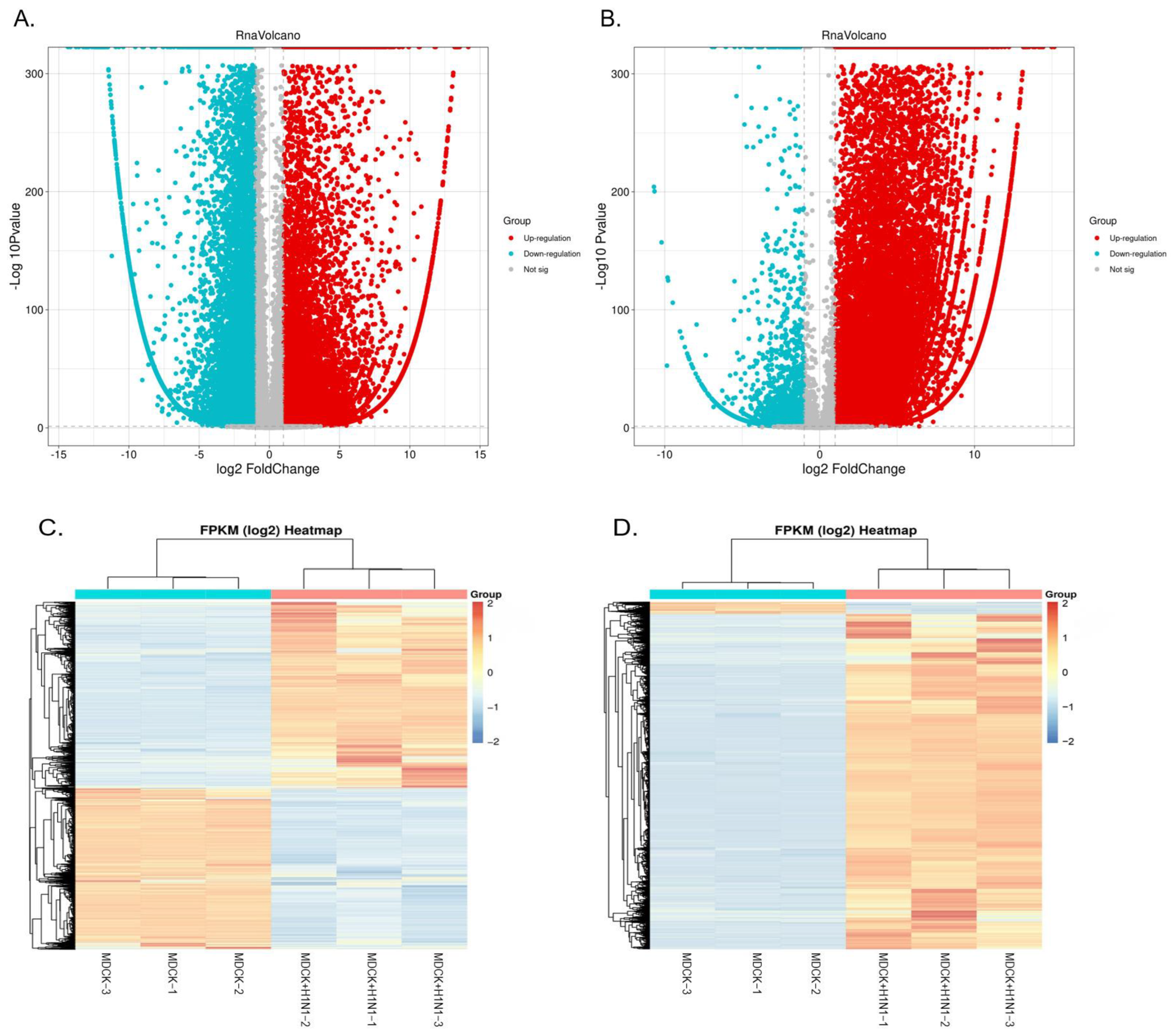
2.2. Overview of Sequencing Data
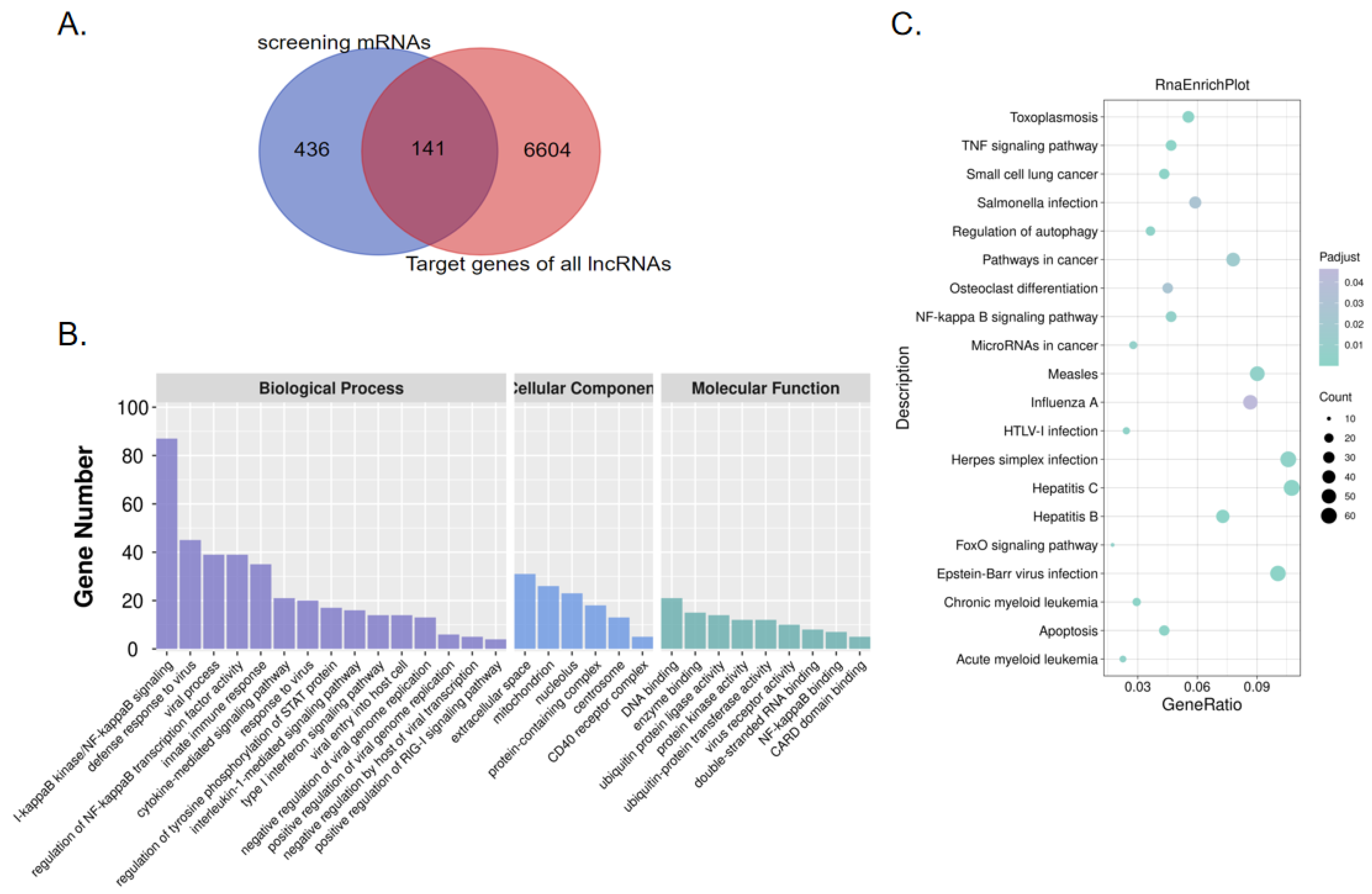
2.3. Screening of Virus-Related DEGs Revealed Enrichment Changes in Several Signaling Pathways in MDCK Cells after Infection
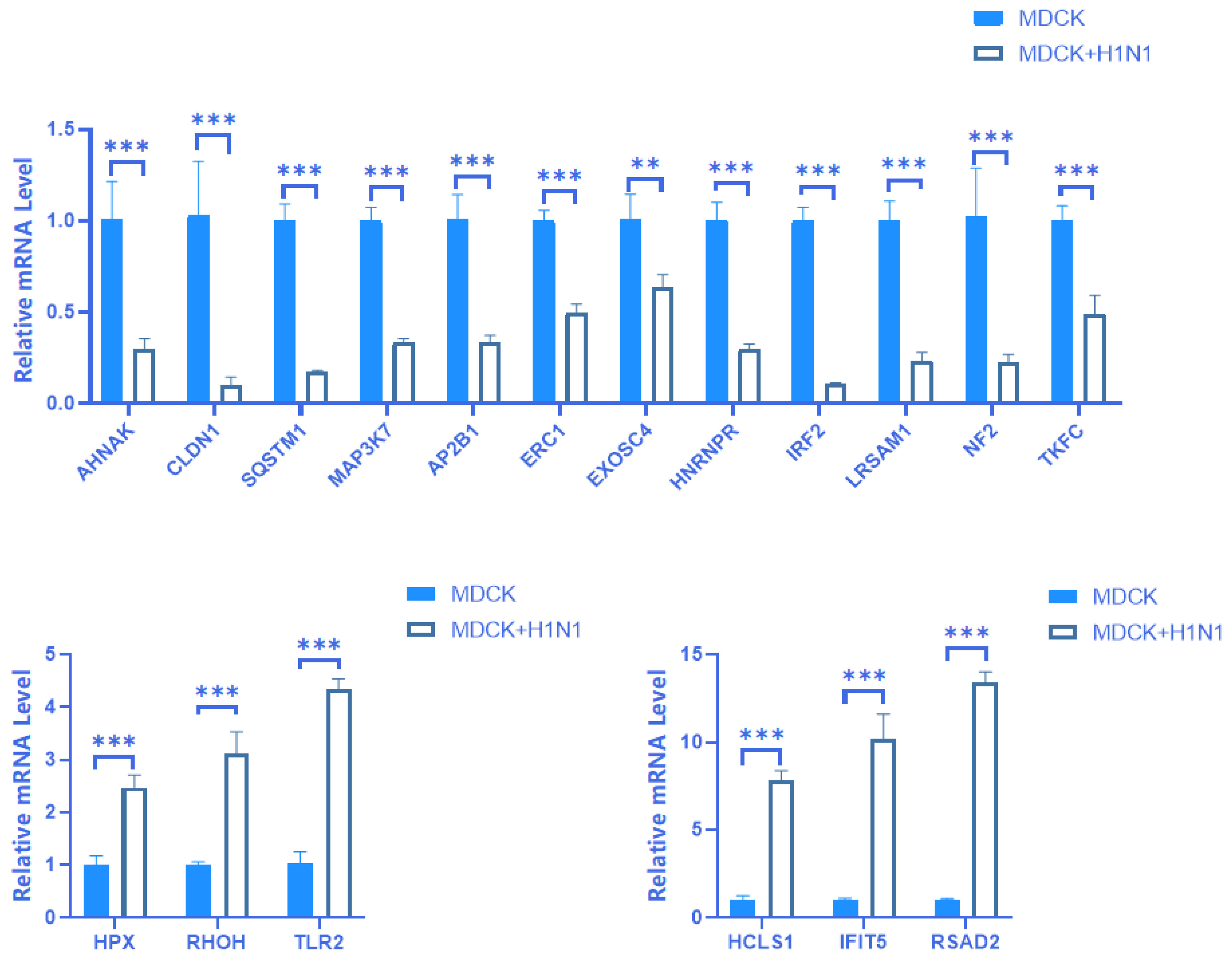
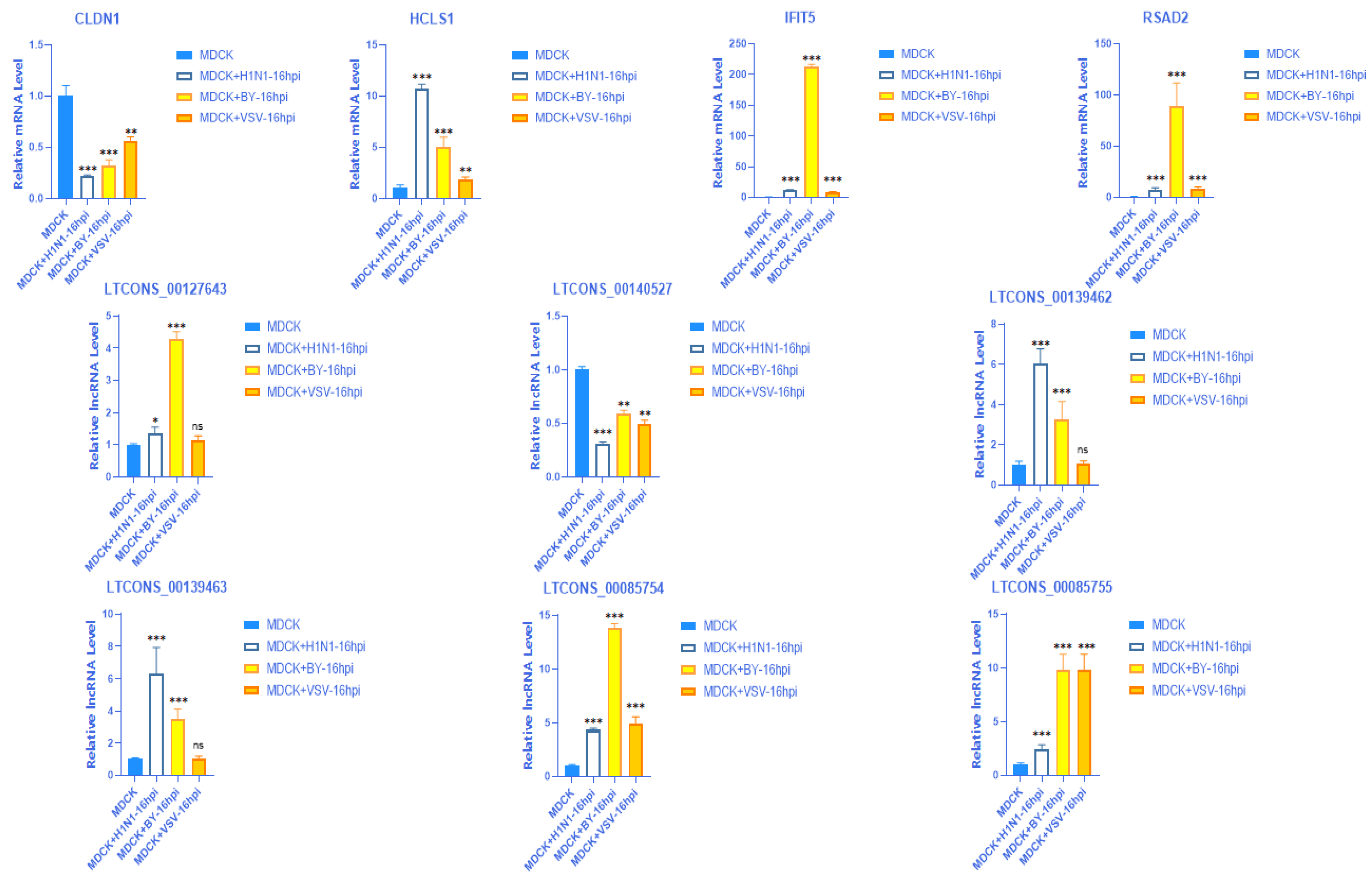
2.4. Verification of mRNA Expression of 18 Screened DEGs
2.5. Prediction and Expression Verification of lncRNA
2.6. Verification of Protein Expression of Four Target DEGs
2.7. Analysis of the lncRNA-mRNA Interaction Network Potentially Affecting IAV Replication in MDCK Cells
2.8. Validation of the Expression of 4 mRNAs with Their Matching Six lncRNAs at Different Time Points
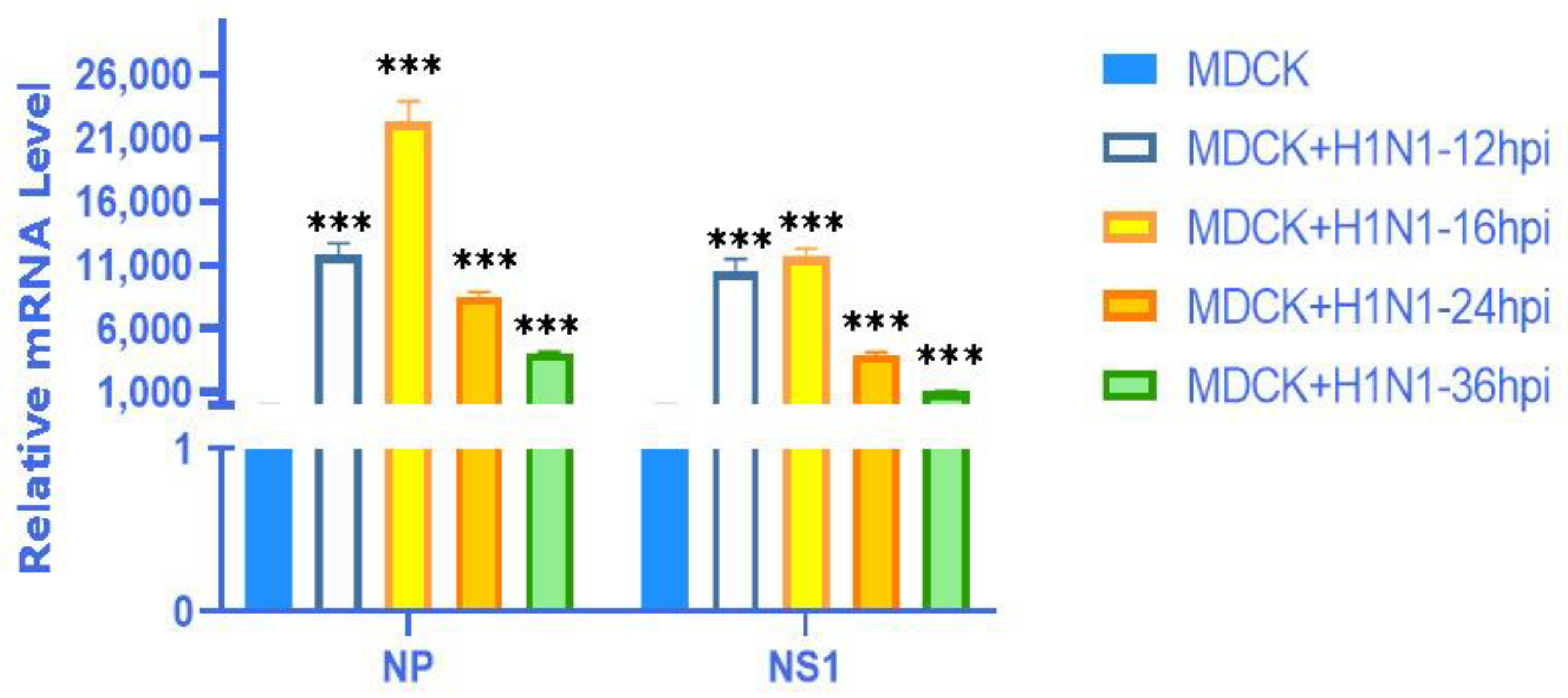
2.9. Validation of the Expression of 4 mRNAs with Their Matching 6 lncRNAs in Different Virus Strains
3. Discussion
4. Methods
4.1. Cell Culture and Virus Infection
4.2. RNA Isolation, Reverse Transcription, and RT-qPCR
4.3. Protein Expression Analysis Using Western Blot
4.4. RNA Sequencing and Data Quality Analysis
4.5. RNA-Seq Differential Gene Expression Analysis
4.6. lncRNA Target Gene Prediction
4.7. Gene Ontology and KEGG Enrichment Analysis of the RNA-Seq Data
4.8. Coding Ability Prediction
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| IAV | Influenza A virus |
| lncRNA | Long non-coding RNA |
| RNA-seq | RNA-sequencing |
| hpi | Hours post-infection |
| DE | Differentially expressed |
| DEG | Differentially expressed genes |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GO | Gene Ontology |
| MOI | Multiplicity of infection |
| qRT-PCR | Quantitative Real-time PCR |
| TLR2 | Toll-like receptor 2 |
| RSAD2 | radical S-adenosyl methionine domain containing 2 |
| IFIT5 | interferon-induced protein with tetratricopeptide repeats 5 |
| HCLS1 | hematopoietic cell-specific Lyn substrate 1 |
| CLDN1 | claudin 1 |
| HPX | hemopexin |
| RHOH | ras homolog family member H |
| AHNAK | AHNAK nucleoprotein |
| SQSTM1 | sequestosome 1 |
| P62 | heat shock 90-like protein |
| MAP3K7 | mitogen-activated protein kinase kinase kinase 7 |
| Tak1 | TGF-beta activated kinase 1 |
| AP2B1 | adaptor-related protein complex 2 subunit beta 1 |
| ERC1 | ELKS/RAB6-interacting/CAST family member 1 |
| EXOSC4 | exosome component 4 |
| HNRNPR | heterogeneous nuclear ribonucleoprotein R |
| IRF2 | interferon regulatory factor 2 |
| LRSAM1 | leucine-rich repeat and sterile alpha motif containing 1 |
| NF2 | NF2, moesin-ezrin-radixin-like (MERLIN) tumor suppressor |
| TKFC | triokinase and FMN cyclase |
| BY | B/Phuket/3073/2013-like virus B (Yamagata/16/88 lineage) |
| VSV | Vesicular stomatitis virus |
References
- Iuliano, A.D.; Roguski, K.M.; Chang, H.H.; Muscatello, D.J.; Palekar, R.; Tempia, S.; Cohen, C.; Gran, J.M.; Schanzer, D.; Cowling, B.J.; et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet 2018, 391, 1285–1300. [Google Scholar] [CrossRef] [PubMed]
- Nuwarda, R.F.; Alharbi, A.A.; Kayser, V. An Overview of Influenza Viruses and Vaccines. Vaccines 2021, 9, 1032. [Google Scholar] [CrossRef]
- Gallo-Ramírez, L.E.; Nikolay, A.; Genzel, Y.; Reichl, U. Bioreactor concepts for cell culture-based viral vaccine production. Expert Rev. Vaccines 2015, 14, 1181–1195. [Google Scholar] [CrossRef] [PubMed]
- Bissinger, T.; Wu, Y.; Marichal-Gallardo, P.; Riedel, D.; Liu, X.; Genzel, Y.; Tan, W.S.; Reichl, U. Towards integrated production of an influenza A vaccine candidate with MDCK suspension cells. Biotechnol. Bioeng. 2021, 118, 3996–4013. [Google Scholar] [CrossRef] [PubMed]
- Küchler, J.; Püttker, S.; Lahmann, P.; Genzel, Y.; Kupke, S.; Benndorf, D.; Reichl, U. Absolute quantification of viral proteins during single-round replication of MDCK suspension cells. J. Proteom. 2022, 259, 104544. [Google Scholar] [CrossRef]
- Ye, Q.; Phan, T.; Hu, W.S.; Liu, X.; Fan, L.; Tan, W.S.; Zhao, L. Transcriptomic Characterization Reveals Attributes of High Influenza Virus Productivity in MDCK Cells. Viruses 2021, 13, 2200. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Z.; Cao, Y. Host-Virus Interaction: How Host Cells Defend against Influenza A Virus Infection. Viruses 2020, 12, 376. [Google Scholar] [CrossRef]
- Beachboard, D.C.; Horner, S.M. Innate immune evasion strategies of DNA and RNA viruses. Curr. Opin. Microbiol. 2016, 32, 113–119. [Google Scholar] [CrossRef]
- Hwang, H.S.; Chang, M.; Kim, Y.A. Influenza-Host Interplay and Strategies for Universal Vaccine Development. Vaccines 2020, 8, 548. [Google Scholar] [CrossRef]
- Campbell, L.K.; Magor, K.E. Pattern Recognition Receptor Signaling and Innate Responses to Influenza A Viruses in the Mallard Duck, Compared to Humans and Chickens. Front. Cell. Infect. Microbiol. 2020, 10, 209. [Google Scholar] [CrossRef]
- Deliyannis, G.; Wong, C.Y.; McQuilten, H.A.; Bachem, A.; Clarke, M.; Jia, X.; Horrocks, K.; Zeng, W.; Girkin, J.; Scott, N.E.; et al. TLR2-mediated activation of innate responses in the upper airways confers antiviral protection of the lungs. JCI Insight 2021, 6, e140267. [Google Scholar] [CrossRef]
- Planès, R.; Bert, J.B.; Tairi, S.; BenMohamed, L.; Bahraoui, E. SARS-CoV-2 Envelope (E) Protein Binds and Activates TLR2 Pathway: A Novel Molecular Target for COVID-19 Interventions. Viruses 2022, 14, 999. [Google Scholar] [CrossRef] [PubMed]
- Maguire, P.; Loughran, S.; Harvey, R.; Johnson, P. A TLR5 mono-agonist restores inhibited immune responses to Streptococcus pneumoniae during influenza virus infection in human monocytes. PLoS ONE 2021, 16, e0258261. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qu, B.; He, G.; Cardona, C.; Song, Y.; Xing, Z. Critical Role of HAX-1 in Promoting Avian Influenza Virus Replication in Lung Epithelial Cells. Mediat. Inflamm. 2018, 2018, 3586132. [Google Scholar] [CrossRef]
- Hsu, W.; Shih, J.; Shih, J.; Du, J.; Teng, S.; Huang, L.; Wang, W. Cellular protein HAX1 interacts with the influenza A virus PA polymerase subunit and impedes its nuclear translocation. J. Virol. 2013, 87, 110–123. [Google Scholar] [CrossRef]
- Wang, G.; Jiang, L.; Wang, J.; Zhang, J.; Kong, F.; Li, Q.; Yan, Y.; Huang, S.; Zhao, Y.; Liang, L.; et al. The G Protein-Coupled Receptor FFAR2 Promotes Internalization during Influenza A Virus Entry. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Yang, S.; Wang, L.; Pan, X.; Liang, Y.; Zhang, Y.; Li, J.; Zhou, B. 5-Methoxyflavone-induced AMPKα activation inhibits NF-κB and P38 MAPK signaling to attenuate influenza A virus-mediated inflammation and lung injury in vitro and in vivo. Cell. Mol. Biol. Lett. 2022, 27, 82. [Google Scholar] [CrossRef]
- Crequer, A.; Troeger, A.; Patin, E.; Ma, C.; Picard, C.; Pedergnana, V.; Fieschi, C.; Lim, A.; Abhyankar, A.; Gineau, L.; et al. Human RHOH deficiency causes T cell defects and susceptibility to EV-HPV infections. J. Clin. Investig. 2012, 122, 3239–3247. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Xu, C.; Wu, B.; Tang, H.; Zhao, P.; Qi, Z. SNORD126 Promotes Hepatitis C Virus Infection by Upregulating Claudin-1 via Activation of PI3K-AKT Signaling Pathway. Front. Microbiol. 2020, 11, 565590. [Google Scholar] [CrossRef]
- Mohapatra, S.; Pioppini, C.; Ozpolat, B.; Calin, G.A. Non-coding RNAs regulation of macrophage polarization in cancer. Mol. Cancer 2021, 20, 24. [Google Scholar] [CrossRef]
- Liao, Y.; Guo, S.; Liu, G.; Qiu, Z.; Wang, J.; Yang, D.; Tian, X.; Qiao, Z.; Ma, Z.; Liu, Z. Host Non-Coding RNA Regulates Influenza A Virus Replication. Viruses 2021, 14, 51. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, S.; Yang, Z.; Lin, H.; Zhu, J.; Liu, L.; Wang, W.; Liu, S.; Liu, W.; Ma, Y.; et al. Self-Recognition of an Inducible Host lncRNA by RIG-I Feedback Restricts Innate Immune Response. Cell 2018, 173, 906–919.e13. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guo, G.; Lu, M.; Chai, W.; Li, Y.; Tong, X.; Li, J.; Jia, X.; Liu, W.; Qi, D.; et al. Long Noncoding RNA Lnc-MxA Inhibits Beta Interferon Transcription by Forming RNA-DNA Triplexes at Its Promoter. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Kornienko, A.E.; Guenzl, P.M.; Barlow, D.P.; Pauler, F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013, 11, 59. [Google Scholar] [CrossRef] [PubMed]
- Barnabei, L.; Laplantine, E.; Mbongo, W.; Rieux-Laucat, F.; Weil, R. NF-κB: At the Borders of Autoimmunity and Inflammation. Front. Immunol. 2021, 12, 716469. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; He, S.; Minassian, A.; Li, J.; Feng, P. Recent advances on viral manipulation of NF-κB signaling pathway. Curr. Opin. Virol. 2015, 15, 103–111. [Google Scholar] [CrossRef]
- Acosta, P.L.; Byrne, A.B.; Hijano, D.R.; Talarico, L.B. Human Type I Interferon Antiviral Effects in Respiratory and Reemerging Viral Infections. J. Immunol. Res. 2020, 2020, 1372494. [Google Scholar] [CrossRef]
- van Woerden, G.M.; Senden, R.; de Konink, C.; Trezza, R.A.; Baban, A.; Bassetti, J.A.; van Bever, Y.; Bird, L.M.; van Bon, B.W.; Brooks, A.S.; et al. The MAP3K7 gene: Further delineation of clinical characteristics and genotype/phenotype correlations. Hum. Mutat. 2022, 43, 1377–1395. [Google Scholar] [CrossRef]
- Sood, N.; Verma, D.; Paria, A.; Yadav, S.; Yadav, M.; Bedekar, M.; Kumar, S.; Swaminathan, T.; Mohan, C.; Rajendran, K.; et al. Transcriptome analysis of liver elucidates key immune-related pathways in Nile tilapia Oreochromis niloticus following infection with tilapia lake virus. Fish Shellfish Immunol. 2021, 111, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef]
- Shen, Z.; Wei, L.; Yu, Z.B.; Yao, Z.Y.; Cheng, J.; Wang, Y.T.; Song, X.T.; Li, M. The Roles of TRIMs in Antiviral Innate Immune Signaling. Front. Cell. Infect. Microbiol. 2021, 11, 628275. [Google Scholar] [CrossRef]
- Zhao, L.; Xia, M.; Wang, K.; Lai, C.; Fan, H.; Gu, H.; Yang, P.; Wang, X. A Long Non-coding RNA IVRPIE Promotes Host Antiviral Immune Responses Through Regulating Interferon β1 and ISG Expression. Front. Microbiol. 2020, 11, 260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Pitchiaya, S.; Cieślik, M.; Niknafs, Y.S.; Tien, J.C.; Hosono, Y.; Iyer, M.K.; Yazdani, S.; Subramaniam, S.; Shukla, S.K.; et al. Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat. Genet. 2018, 50, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.; Yang, Z.; Xiong, T.; Wang, T.; Yang, J.; Huang, M.; Liu, D.; Chen, R. Avian IRF1 and IRF7 Play Overlapping and Distinct Roles in Regulating IFN-Dependent and -Independent Antiviral Responses to Duck Tembusu Virus Infection. Viruses 2022, 14, 1506. [Google Scholar] [CrossRef] [PubMed]
- Deffieu, M.; Clément, C.; Dorobantu, C.; Partiot, E.; Bare, Y.; Faklaris, O.; Rivière, B.; Ayala-Nunez, N.; Baumert, T.; Rondé, P.; et al. Occludin stalls HCV particle dynamics apart from hepatocyte tight junctions, promoting virion internalization. Hepatology 2022, 76, 1164–1179. [Google Scholar] [CrossRef]
- Kurokawa, C.; Iankov, I.D.; Galanis, E. A key anti-viral protein, RSAD2/VIPERIN, restricts the release of measles virus from infected cells. Virus Res. 2019, 263, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hinson, E.R.; Cresswell, P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2007, 2, 96–105. [Google Scholar] [CrossRef]
- Wu, Y.; Song, X.; Cui, D.; Zhang, T. IFIT3 and IFIT5 Play Potential Roles in Innate Immune Response of Porcine Pulmonary Microvascular Endothelial Cells to Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus. Viruses 2022, 14, 1919. [Google Scholar] [CrossRef]
- Wang, S.; Wan, L.; Ren, H.; Xie, Z.; Xie, L.; Huang, J.; Deng, X.; Xie, Z.; Luo, S.; Li, M.; et al. Screening of interferon-stimulated genes against avian reovirus infection and mechanistic exploration of the antiviral activity of IFIT5. Front. Microbiol. 2022, 13, 998505. [Google Scholar] [CrossRef]
- Mazel-Sanchez, B.; Boal-Carvalho, I.; Silva, F.; Dijkman, R.; Schmolke, M. H5N1 Influenza A Virus PB1-F2 Relieves HAX-1-Mediated Restriction of Avian Virus Polymerase PA in Human Lung Cells. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Yumine, N.; Matsumoto, Y.; Ohta, K.; Fukasawa, M.; Nishio, M. Claudin-1 inhibits human parainfluenza virus type 2 dissemination. Virology 2019, 531, 93–99. [Google Scholar] [CrossRef]
- Mailly, L.; Baumert, T. Hepatitis C virus infection and tight junction proteins: The ties that bind. Biochimica et biophysica acta. Biomembranes 2020, 1862, 183296. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gan, L.; Sun, P.; Wang, J.; Li, D.; Cao, Y.; Fu, Y.; Li, P.; Bai, X.; Li, K.; et al. The long non-coding RNA LNC_000397 negatively regulates PRRSV replication through induction of interferon-stimulated genes. Virol. J. 2022, 19, 40. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef]
- Knauss, J.L.; Sun, T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience 2013, 235, 200–214. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
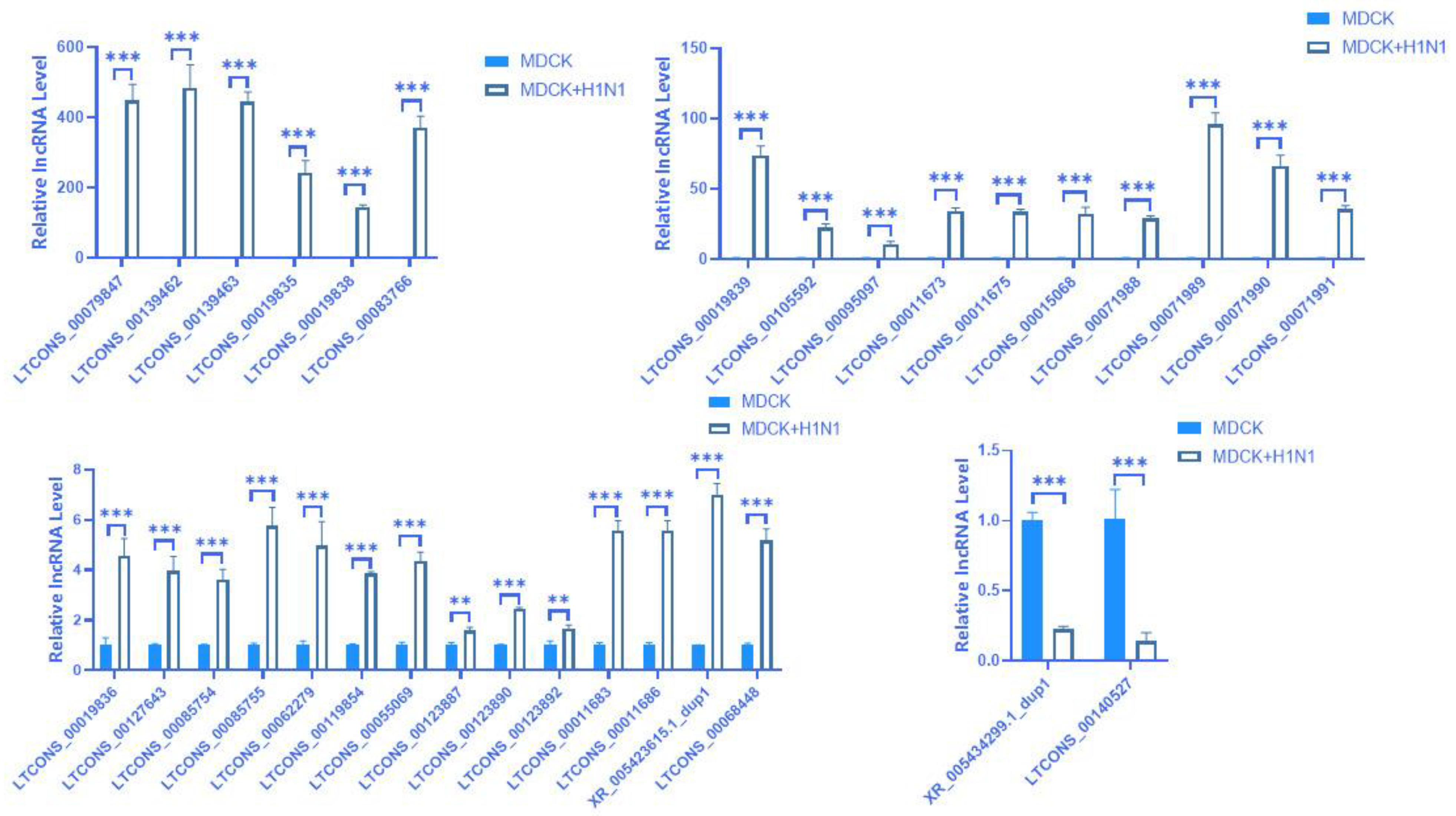
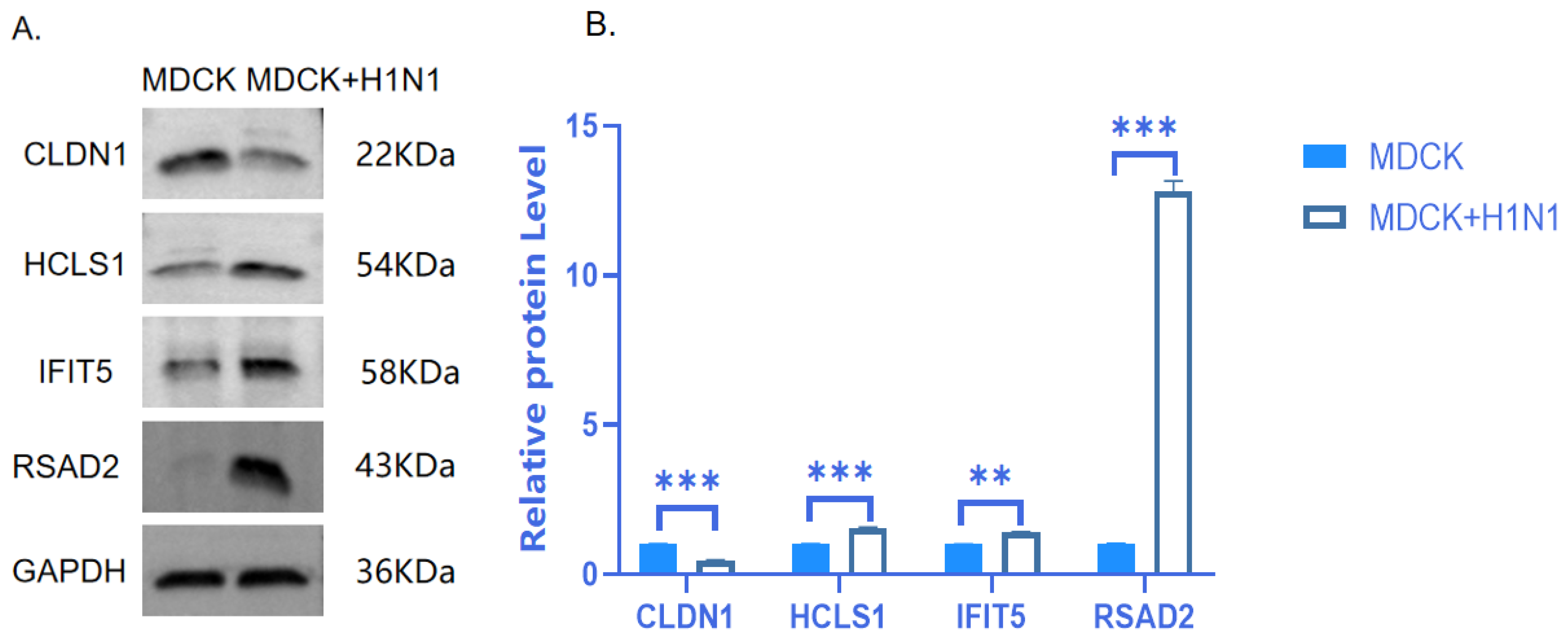
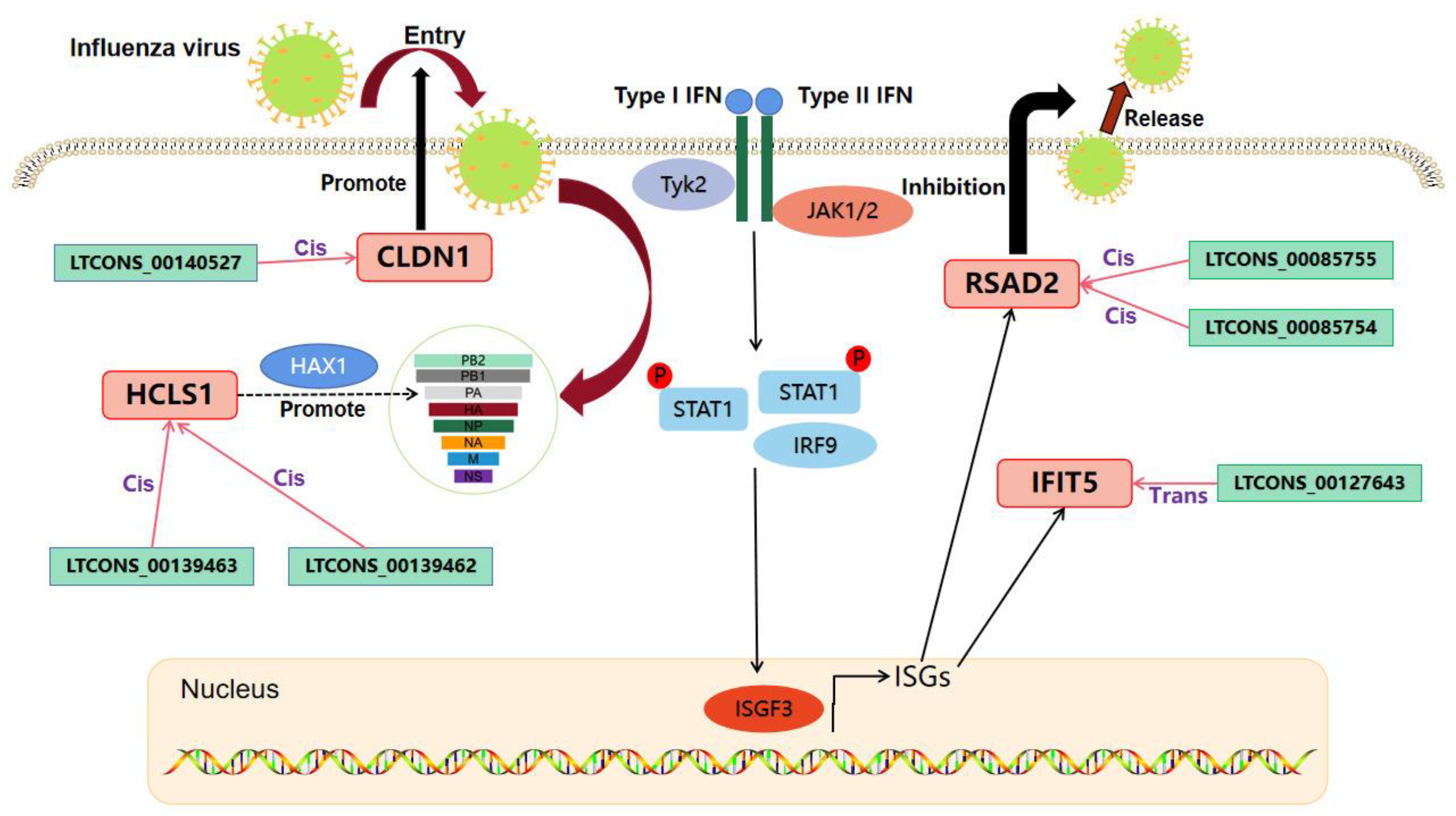
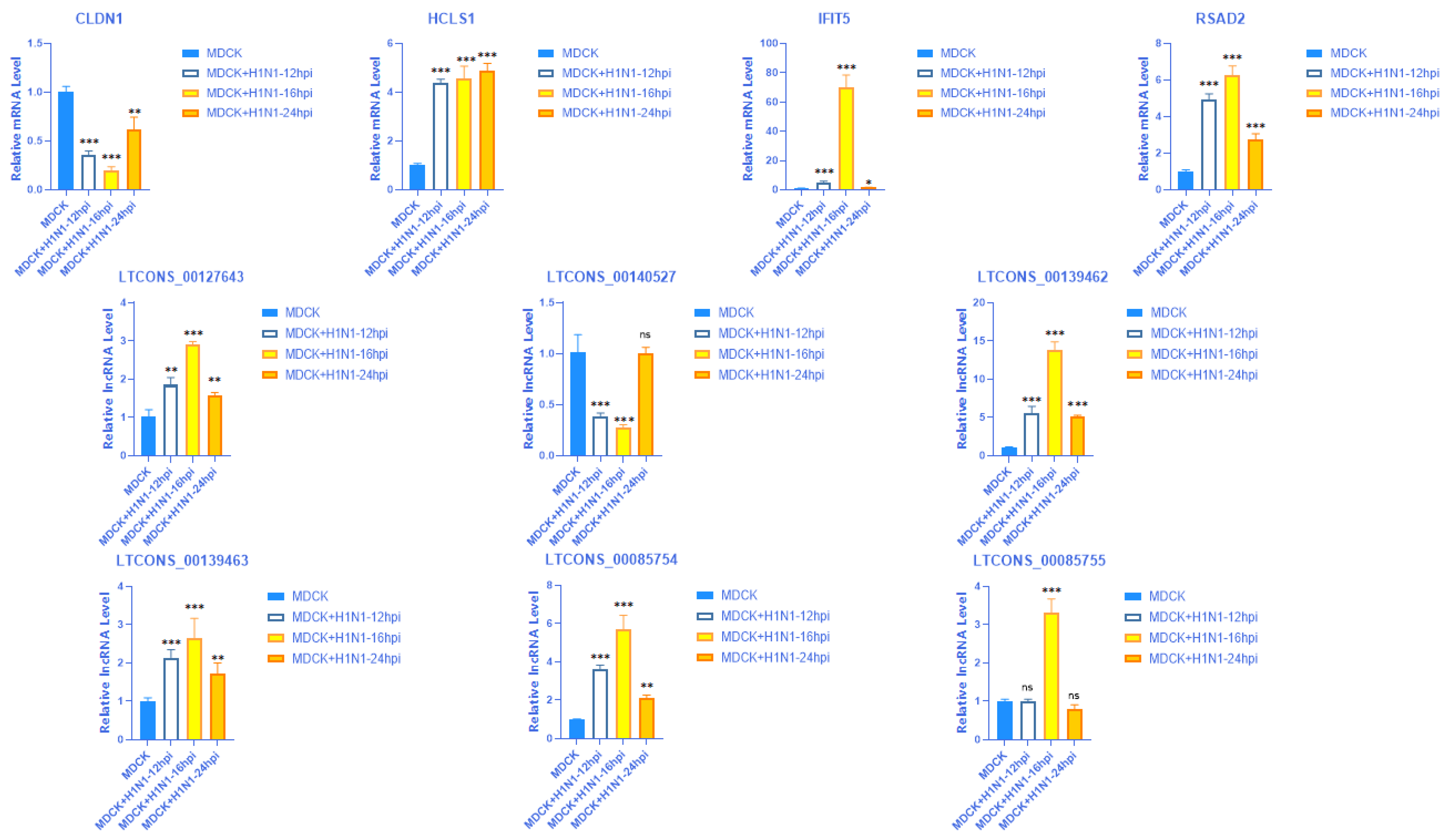
| lncRNA | log2(lncRNA) Ratio (MDCK + H1N1/MDCK) | p-Value (lncRNA) | lncRNA_Class | Target Gene | Gene ID | log2(mRNA) Ratio (MDCK + H1N1/MDCK) | p-Value (mRNA) |
|---|---|---|---|---|---|---|---|
| LTCONS_00079847 | 8.49 | 7.12 × 10−53 | cis_mRNA_up10k | TLR2 | XM_038687174.1_dup1 | 8.04 | 2.45 × 10−21 |
| LTCONS_00139462 | 11.49 | 2.62 × 10−134 | cis_mRNA_dw20k | HCLS1 | XM_038445109.1_dup1 | 7.33 | 1.79 × 10−129 |
| LTCONS_00139463 | 8.12 | 8.97 × 10−166 | cis_mRNA_dw20k | ||||
| LTCONS_00019835 | 8.58 | 1.11 × 10−28 | cis_mRNA_dw20k | RHOH | XM_038661947.1_dup1 | 6.56 | 1.91 × 10−111 |
| LTCONS_00019836 | 7.13 | 6.97 × 10−13 | cis_mRNA_dw20k | ||||
| LTCONS_00019838 | 6.83 | 1.09 × 10−168 | cis_mRNA_up10k | ||||
| LTCONS_00019839 | 6.62 | 1.04 × 10−121 | cis_mRNA_up10k | ||||
| LTCONS_00127643 | 2.43 | 1.83 × 10−90 | tran | IFIT5 | XM_038662420.1_dup1 | 5.95 | 1.49 × 10−22 |
| LTCONS_00105592 | 4.48 | 1.41 × 10−26 | cis_mRNA_up10k | HPX | XM_038430363.1_dup1 | 5.84 | 7.47 × 10−31 |
| LTCONS_00085754 | 1.58 | 1.17 × 10−6 | cis_mRNA_up10k | RSAD2 | XM_038690652.1_dup1 | 4.99 | 0 |
| LTCONS_00085755 | 3.74 | 1.21 × 10−76 | cis_mRNA_up10k | ||||
| LTCONS_00062279 | 3.04 | 0 | cis_mRNA_dw20k | SQSTM1 (p62) | XM_038681126.1_dup1 | −2.23 | 3.95 × 10−114 |
| XR_005434299.1_dup1 | −1.31 | 1.30 × 10−204 | cis_mRNA_overlap | ||||
| LTCONS_00119854 | 3.34 | 1.42 × 10−25 | cis_mRNA_dw20k | NF2 | XM_038437180.1_dup1 | −2.25 | 0 |
| LTCONS_00055069 | 3.81 | 1.70 × 10−103 | cis_mRNA_up10k | AP2B1 | XM_038677309.1_dup1 | −2.74 | 0 |
| LTCONS_00140527 | −2.69 | 1.43 × 10−9 | cis_mRNA_overlap | CLDN1 | XM_038445869.1_dup1 | −2.77 | 0 |
| LTCONS_00083766 | 7.49 | 9.19 × 10−16 | cis_mRNA_dw20k | IRF2 | XM_038690244.1_dup1 | −2.88 | 1.63 × 10−255 |
| LTCONS_00123887 | 1.14 | 2.09 × 10−10 | cis_mRNA_overlap | ERC1 | XM_038439411.1_dup1 | −2.99 | 9.22 × 10−182 |
| LTCONS_00123890 | 1.62 | 3.86 × 10−9 | cis_mRNA_overlap | ||||
| LTCONS_00123892 | 1.39 | 5.04 × 10−8 | cis_mRNA_overlap | ||||
| LTCONS_00095097 | 5.69 | 6.72 × 10−68 | cis_mRNA_overlap | AHNAK | XM_038425334.1_dup1 | −3.55 | 0 |
| LTCONS_00011673 | 7.08 | 3.96 × 10−57 | cis_mRNA_up10k | HNRNPR | XM_038659953.1_dup1 | −3.85 | 0 |
| LTCONS_00011675 | 5.39 | 1.32 × 10−8 | cis_mRNA_up10k | ||||
| LTCONS_00011683 | 2.29 | 2.70 × 10−193 | cis_mRNA_dw20k | ||||
| LTCONS_00011686 | 4.43 | 0 | cis_mRNA_dw20k | ||||
| LTCONS_00015068 | 4.72 | 2.76 × 10−53 | cis_mRNA_up10k | ||||
| XR_005423615.1_dup1 | 3.10 | 0 | cis_mRNA_dw20k | ||||
| LTCONS_00068448 | 2.81 | 1.53 × 10−12 | cis_mRNA_up10k | MAP3K7 (MKK7) | XM_038684179.1_dup1 | −3.93 | 0 |
| LTCONS_00071988 | 5.98 | 1.55 × 10−97 | cis_mRNA_dw20k | EXOSC4 | XM_038685312.1_dup1 | −10.33 | 2.09 × 10−168 |
| LTCONS_00071989 | 7.25 | 1.30 × 10−74 | cis_mRNA_dw20k | ||||
| LTCONS_00071990 | 4.22 | 1.56 × 10−51 | cis_mRNA_dw20k | ||||
| LTCONS_00071991 | 5.38 | 8.11 × 10−72 | cis_mRNA_dw20k |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, G.; Pei, M.; Wang, S.; Qiu, Z.; Li, X.; Ma, H.; Ma, Y.; Wang, J.; Qiao, Z.; Ma, Z.; et al. Transcriptional Analysis of lncRNA and Target Genes Induced by Influenza A Virus Infection in MDCK Cells. Vaccines 2023, 11, 1593. https://doi.org/10.3390/vaccines11101593
Liu G, Pei M, Wang S, Qiu Z, Li X, Ma H, Ma Y, Wang J, Qiao Z, Ma Z, et al. Transcriptional Analysis of lncRNA and Target Genes Induced by Influenza A Virus Infection in MDCK Cells. Vaccines. 2023; 11(10):1593. https://doi.org/10.3390/vaccines11101593
Chicago/Turabian StyleLiu, Geng, Mengyuan Pei, Siya Wang, Zhenyu Qiu, Xiaoyun Li, Hua Ma, Yumei Ma, Jiamin Wang, Zilin Qiao, Zhongren Ma, and et al. 2023. "Transcriptional Analysis of lncRNA and Target Genes Induced by Influenza A Virus Infection in MDCK Cells" Vaccines 11, no. 10: 1593. https://doi.org/10.3390/vaccines11101593
APA StyleLiu, G., Pei, M., Wang, S., Qiu, Z., Li, X., Ma, H., Ma, Y., Wang, J., Qiao, Z., Ma, Z., & Liu, Z. (2023). Transcriptional Analysis of lncRNA and Target Genes Induced by Influenza A Virus Infection in MDCK Cells. Vaccines, 11(10), 1593. https://doi.org/10.3390/vaccines11101593






