Abstract
In December 2019, the COVID-19 disease started in Wuhan, China. The WHO declared a pandemic on 12 March 2020, and the disease started in Myanmar on 23 March 2020. In December 2020, different variants were brought worldwide, threatening global health. To counter those threats, Myanmar started the COVID-19 variant surveillance program in late 2020. Whole genome sequencing was done six times between January 2021 and March 2022. Among them, 83 samples with a PCR threshold cycle of less than 25 were chosen. Then, we used MiSeq FGx for sequencing and Illumina DRAGEN COVIDSeq pipeline, command line interface, GISAID, and MEGA version 7 for data analysis. In January 2021, no variant was detected. The second run, during the rise of cases in June 2021, showed Alpha, Delta, and Kappa variants. The third and the fourth runs in August and December showed only a Delta variant. Omicron and Delta variants were detected during the fifth run in January 2022. The sixth run in March 2022 showed only Omicron BA.2. Amino acid mutation at the receptor binding domain of Spike glycoprotein started since the second run coupling with high transmission, recurrence, and vaccine escape. We also found the mutation at the primer targets used in current RT-PCR platforms, but there was no mutation at the existing antiviral drug targets. The occurrence of multiple variants and mutations claimed vigilance at ports of entry and preparedness for effective control measures. Genomic surveillance with the observation of evolutionary data is required to predict imminent threats of the current disease and diagnose emerging infectious diseases.
1. Introduction
In December 2019, COVID-19 disease was first identified in Wuhan, China [1,2]. The WHO declared the pandemic on 11 March 2020. COVID-19 is caused by a Human Coronavirus, SARS-CoV-2. Human coronavirus (HCoV) is a member of the coronavirinae subfamily of the coronaviridae family [3].
In Myanmar, the first wave started with two cases on 23 March 2020. There were only 374 cases and 6 deaths during the first wave, which ended on 16 July [4]. The second wave started on 16 August in Rakhine State, followed by an outbreak in Yangon occurring among 32,351 cases with 765 fatalities. The first wave was comparatively moderate compared to regional peers, but there was a dramatic rise in the death toll during the second wave. Early June 2021 brought the third wave with a massive number of cases. Cases reached 144,157 with a death toll of 3334, which outnumbered the capacity of quarantine centers and hospitals. At the end of the year, the total confirmed cases reached 530,834, with 19,268 deaths. An Omicron variant was first identified on 7 January 2022, which has caused a raised number of cases reaching 613,577 people with 19,434 deaths as of 29 June 2022 [4].
SARS-CoV-2 is a fast-evolving virus because of rapid and massive genetic variations. The WHO classified viruses with Alpha, Beta, Gamma, Delta, and Omicron as the variants of concern (VOC) and Zeta, Eta, and Kappa as the variants of interest (VOI) [5]. Since December 2020, variants have occurred worldwide, which favors the virus to be fitter for transmission. The high transmission rate caused more morbidity and mortality, endangering existing healthcare systems. All these things affected Myanmar; the country started the COVID-19 variant surveillance program in late 2020, and the Defence Services Medical Research Centre started whole genome sequencing in December 2020.
This study used a WGS technique on the clinical samples collected during the waves above. We described the features of the viral genome sequences from these periods, including VOCs, VOIs, and genetic variation at the spike glycoprotein coding region and primer binding sites, and drew the dynamic of viral spread.
2. Materials and Methods
We took 159 SARS-CoV-2 PCR-positive samples from clustered cases in cities and border areas during the second, third, and Omicron waves. We used QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany) for RNA extraction, BioFlux SARS-CoV-2 Nucleic Acid Detection Kit (Bioer, Hangzhou, China), and Applied Biosystems StepOnePlus Real-Time PCR System to check Ct values. We chose samples with a Ct value below 25 for sequencing and finally obtained 101 samples.
We amplified the SARS-CoV-2 whole genome according to the Illumina COVIDSeq RUO Assay (Illumina, San Diego, CA, USA) protocol [6]. Briefly, we synthesized cDNA with the included COVIDseq First Strand cDNA synthesis kit, amplified the whole viral genome selectively with Primer Pool 1 and 2, fragmented and tagged with Bead-linked transposome. Following this, we added Illumina Nextera DNA CD indexes to the samples for identification before we pooled, purified, quantified, and normalized them. Finally, the MiSeq FGx sequencing system amplified them to form clusters and sequenced them using sequencing with synthesis (SBS) chemistry.
We used Illumina DRAGEN COVIDSeq Test Pipeline to trim and assemble FASTQ files to get the FASTA format together with variant and lineage reports. The results were validated again by generating FASTA files with SC-2pipe (BioEasy Sdn Bhd, Selangor, Malaysia) in the local workstation and deposited at GISAID and NCBI GenBank. We collected 189 complete genome sequences from GISAID, which had 99.8% sequence homology with our samples. Subsequently, we removed duplicate and incomplete sequences and finally obtained 78 sequences. We performed sequence alignment using Molecular Evolutionary Genetics Analysis (MEGA) software version 7 with Multiple Sequence Comparisons with Log-Expectation (MUSCLE) algorithm [7], neighbor-joining method, and the Kimura 2 model with 1000 bootstraps for phylogenetic analysis.
In order to assess the effect on viral transmission and immune evasion, we observed the Spike glycoprotein coding region if there were identified nucleotide and amino acid mutations. Mutations at ORF1ab, Nucleocapsid (N), and Envelope (E) encoding regions were also assessed, especially at primer binding sites used in PCR assays. We also collected phenotypic data, including disease severity, vaccination status, and traveling history, to correlate with viral genotypes.
3. Results
We chose 83 sequences ranging in length from 28,205 to 29,830 nucleotides, and each contains over 99.75% of the genome (Table 1), and 77 homologous sequences from 15 countries. We drew a phylogenetic tree (Figure 1), and our sequences were distributed independently and were similar to strains from Europe and Asia. Our sequences were similar to those from Malaysia during the first run; India, France, South Korea, China, and Romania during the second run; Thailand, India, South Korea, USA, and Kosovo during the third run; Thailand, Malaysia, India, Austria, Germany, France, Brazil, and England during the fourth run; Malaysia, Philippines, South Korea, India, England, and Ecuador during the fifth run; and Thailand, India, England, China, USA, and Australia during the sixth run.

Table 1.
Genotyping of Myanmar SARS-CoV-2 sequences with demographic data.
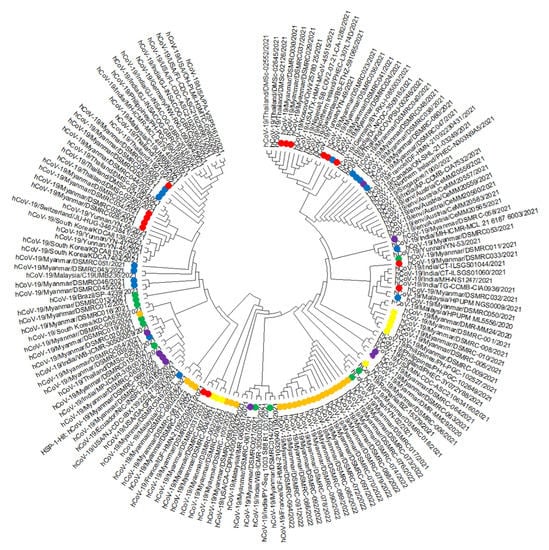
Figure 1.
Phylogenetic tree comparing Myanmar SARS-CoV-2 sequences with 78 previously published sequences. Label: Yellow = First Run, Green = Second Run, Red = Third Run, Blue = Fourth Run, Purple = Fifth Run, and Orange = Sixth Run; We performed sequence alignment using MEGA version 7 with the MUSCLE algorithm, neighbor-joining analysis method, and the Kimura 2-parameter model with 1000 bootstrap replications for phylogenetic analysis.
All our samples showed D614G mutation since the first run with additional Q677H mutation at five out of nine samples. No variant of concern (VOC) or variant of interest (VOI) existed. After six months, we did the second run and found seven VOCs (two Alpha and five Delta) and four VOIs (Kappa). Alpha variants showed a N501Y mutation at RBD, while the Delta showed L452R and T478K mutations at RBD with P681H mutation at the furin binding site (FBS). Similar to Delta variants, Kappa showed L452R and P681H mutations, but there was E484Q instead of T478K. The third and fourth runs had 15 and 16 complete sequences, showing only the Delta variant with the same mutation. Four Delta remained present during the fifth run, together with five Omicron variants. The sixth run in March 2022, with 23 samples, showed only Omicron, all having multiple mutations at RBD and FBS.
Three out of four Delta variants during the fifth run (DSMRC 054, 058, 060) showed Nucleocapsid G28881T, and DSMRC 058, 060 showed ORF1ab G15451A nucleotide substitutions. Three Omicron variants (DSMRC 063, 064, 067) showed trinucleotide mutations (G28881A, G28882A, G28883C) at the Nucleocapsid coding region and Envelope C26270T mutation. All our sequences belonged to non-variant and four variants as well as 15 strains. Generally, they were distributed equally with each other and worldwide sequences. However, the third and fourth runs with the Delta variant became more homologous.
4. Discussion
Many variants were found in this study and were similar to those found in India, in England, and worldwide. We found similar sequences from Asia and worldwide without any from Africa and South America. Since late 2021, the Delta variant dominated until the appearance of Omicron in January 2022. The Delta variant took over other global variances after its emergence in India in December 2020 [8].
Phylogenetic analysis showed that our sequences were distributed independently and clustered together with those from 21 countries. These results showed complex geographical and temporal transmission patterns and a high virus mutation rate. Our sequences became more homologous with worldwide sequences during the third and fourth runs. Clustering with those from Europe (France, England) showed rapid disease transmission in today’s world. Clustering with other Asian countries (India, Malaysia, and Thailand) showed the border crossing of the disease through land and air. Sequences had many nucleotide changes compared to the original COVID-19 sequence from Wuhan, China (NC045512.2) [9].
The D614G mutation, found in January 2021, was first detected in January 2020 in China and Germany. It outnumbered the initial coronavirus by April/May [10]. The G614 variant had improved receptor binding and transmission, becoming the dominant global strain [9]. Molecular courting also indicated that this novel mutation ascended early in the pandemic. Genetic drift caused a high rate of a specific variant without selective advantage [11]. The Q677H mutation found in some of our samples might affect the stability of the spike protein as it lay near the furin cleavage site, but there was no evidence of its effect on the pathogenicity [12]. No differences were identified in severity between patients with Q677H and without.
The N501Y mutation observed in Alpha, and L452R and T478K in Delta variant, lied at RBD. In the U.K., South Africa, and Brazil, the N501Y mutation had increased binding affinity to ACE2. L452R showed reduced neutralization from several monoclonal antibodies [13,14,15] and convalescent plasma [13]. L452R independently appeared in different lineages between December 2020 and February 2021. This substitution might result from viral adaptation due to increased immunity in the population [9]. The T478K mutation at the interface complex with human ACE2 might also affect the affinity with human cells and viral infectivity [16]. Kristian Andersen identified a notable feature: polybasic, furin binding at positions 681–688, and subsequent cleavage of the Spike protein worsened its stability. Cleavage of subunits increased binding affinity to the ACE2 receptor markedly [17]. Unfortunately, all Delta and Kappa variants developed with the P681H mutation at FBS and would cause more transmission.
Kappa variants also showed a E484Q mutation at the RBD’s receptor-binding motif (RBM). As the central functional motif, RBM directly binds the human ACE2 receptor [18]. E484N and E484K escape from several monoclonal antibodies and antibodies in convalescent plasma [15,19]. Due to the presence of both L452R and E484Q mutations, our Kappa variant would affect the antigenicity and subsequent immune protection [18,19]. Investigations using pseudoviruses with RBD mutations revealed that the neutralizing activity of plasma from vaccinated people decreased onefold to thrice against N501Y and E484K [20].
Finally, Omicron came up with multiple mutations at RBD and FBS, which would cause the virus to be more transmissible and affect the host’s immune protection. Clinical data showed rapid transmission without evidence of the disease severity and a high mortality rate. K417 is the epitope for class 1 and class 2 antibodies [21], but mutation would affect only class 1 antibody binding. Therefore, the K417 mutation found in this study would have less effect on the polyclonal antibody response, which is responsible for class 2 antibody binding. They were more vulnerable to substitutions including E484K [22]. K417N and K417T had lower ACE2-binding affinity in addition to their antigenic impact [23].
The development of vaccines and drugs reduced disease severity and the spread of SARS-CoV-2. However, selective pressure and viral evolution yield resistant viral variants. The first FDA-approved Paxlovid is the conserved main protease (Mpro) inhibitor [24]. Mpro is a 3-chymotrypsin-like cysteine protease (3CLpro), which digests two large polyproteins (pp1a and pp1ab) to generate proteins critical for virus replication and transcription. It is encoded as nonstructural protein 5 (Nsp5) at open reading frame 1 (ORF1) [25]. Paxlovid is a useful drug in blunting SARS-CoV-2 disease pathogenesis. Nirmatrelvir is the active ingredient in Paxlovid. Ensitrelvir is another Mpro inhibitor in clinical-stage development. In Mpro, P168S mutation is the most commonly seen naturally occurring variant, followed by a single residue deletion, ΔP168. The ΔP168 mutant protease is found to have increased resistance to nirmatrelvir and ensitrelvir, but the P168S variant has no drug-resistant effect [24]. There was no P168 mutation in our study, and other studies also showed Mpro as a highly conserved protein relative to the spike protein [25].
The spike glycoprotein is used as an antigen in vaccine development as it binds to the Angiotensin-Converting Enzyme 2 receptor and enters the host cell, which is crucial in the first step of the infection process. Immunization-induced specific antibodies against the receptor binding domain block and prevent virus invasion [26]. However, we have to monitor the spike glycoprotein mutation as its coding region is one of the most variable sites in the genome [27]. The mutations at that place have effects on vaccine efficacy [28].
Currently, we are using Daan Gene and Bioflux RT-PCR and cobas6800 platforms to diagnose COVID-19. Therefore, we checked the targets of China CDC and Charité protocols. China CDC protocols target at the Nucleocapsid coding region and the ORF1ab region. Charité protocols target at the ORF1ab region and the Envelope coding region. Some Delta variants in our study showed nucleotide substitution at the China CDC forward primer binding site at the Nucleocapsid coding region (28881-28902) and Charite forward primer binding site at ORF1b (15431-15452) only. Some Omicron variants showed substitution at the China CDC forward primer binding site at the Nucleocapsid region (28881-28902) and Charite forward primer binding site at the Envelope region (26269-26294) only. These show that our molecular platforms are still effective in diagnosing COVID-19 [29].
Distribution of our sequences across the whole phylogenetic tree constructed with sequences from 15 countries worldwide, inferring globalization and high transmissibility of disease, warned us to keep stricter vigilance of imported cases. Sequence homology with Europe and Malaysia indicated keeping track of disease transmission patterns and ports of entry from these regions. Ongoing transmission from India from the First to the Sixth Run highlighted the need not to emphasize only the eastern border but to exercise more containment strategy at the west.
Multiple variants claimed sustainable plans and preparation for diagnosis, treatment, prevention, and control measures against repeatedly emerging diseases. Genomic data during waves of world outbreaks showed the dominance of the fittest Delta strain over other strains. This domination predicted the inevitable importation of this strain, and we should use this information effectively to prepare for future control measures.
5. Conclusions
Detailed information about mutations at receptor binding sites, immune epitopes, and primer binding sites offered important intelligence data to combat the novel virus. We should maintain our efforts in genomic surveillance. Finally, the whole genome sequencing platform in this study opens our capability for diagnosing emerging infectious diseases in the upcoming days in the challenging world. Multiple variants and mutations have necessitated increased vigilance at ports of entry and the development of effective control measures. Genomic surveillance with the observation of evolutionary data is required to predict imminent threats of the current disease and diagnose emerging infectious diseases.
Author Contributions
Conceptualization, methodology, software, validation, formal analysis, investigation, data curation, writing—original draft preparation, K.Z.O.; methodology, software, formal analysis, investigation, data curation, writing—original draft preparation, Z.W.H.; investigation, N.M.A., K.K.W., S.P.H., K.Z.L., P.K.A., T.W.O. and L.Y.K.; writing—review and editing, M.T.Z. and K.M.T.; supervision, K.M.; project administration, K.K.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research and APC were funded by the Directorate of Medical Services.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of the Defence Services Medical Research Centre (protocol code 056/2020, date of approval 29 December 2020).
Informed Consent Statement
Patient consent was waived because the samples were anonymous.
Data Availability Statement
Data are available at the corresponding GISAID accession number in Table 1.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lu, H.; Stratton, C.W.; Tang, Y.-W. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. J. Med. Virol. 2020, 92, 401–402. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- WHO. Coronavirus Disease 2019 (COVID-19) Situation Report–52. 2020. Available online: https://apps.who.int/iris/handle/10665/331476 (accessed on 20 June 2022).
- MOH. Ministry of Health Myanmar COVID-19 Surveillance Dashboard. 2022. Available online: https://mohs.gov.mm/ (accessed on 29 June 2022).
- WHO. Tracking SARS-CoV-2 Variants. 2022. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 22 June 2022).
- Illumina COVIDSeq Test (RUO Version). Available online: https://www.illumina.com/products/by-type/clinical-research-products/covidseq.html (accessed on 29 September 2021).
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Cavalcante, I.J.M.; Vale, M.R. Epidemiological aspects of visceral leishmaniasis (kala-azar) in Ceará in the period 2007 to 2011. Rev. Bras. Epidemiol. 2014, 17, 911–924. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. bioRxiv 2020. [Google Scholar] [CrossRef]
- Singh, D.; Yi, S.V. On the origin and evolution of SARS-CoV-2. Exp. Mol. Med. 2021, 53, 537–547. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, W. Fast-spreading SARS-CoV-2 variants: Challenges to and new design strategies of COVID-19 vaccines. Signal Transduct. Target. Ther. 2021, 6, 226. [Google Scholar] [CrossRef]
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020, 182, 1284–1294.e9. [Google Scholar] [CrossRef]
- Greaney, A.J.; Starr, T.N.; Gilchuk, P.; Zost, S.J.; Binshtein, E.; Loes, A.N.; Hilton, S.K.; Huddleston, J.; Eguia, R.; Crawford, K.H.; et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe 2021, 29, 44–57.e9. [Google Scholar] [CrossRef]
- Liu, Z.; VanBlargan, L.A.; Bloyet, L.M.; Rothlauf, P.W.; Chen, R.E.; Stumpf, S.; Zhao, H.; Errico, J.M.; Theel, E.S.; Liebeskind, M.J.; et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 2021, 29, 477–488.e4. [Google Scholar] [CrossRef] [PubMed]
- Di Giacomo, S.; Mercatelli, D.; Rakhimov, A.; Giorgi, F.M. Preliminary report on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Spike mutation T478K. J. Med. Virol. 2021, 93, 5638–5643. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; Sanchez-David, R.Y.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021, 6, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Thomson, E.C.; Rosen, L.E.; Shepherd, J.G.; Spreafico, R.; da Silva Filipe, A.; Wojcechowskyj, J.A.; Davis, C.; Piccoli, L.; Pascall, D.J.; Dillen, J.; et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated im-munity. Cell 2021, 184, 1171–1187.e20. [Google Scholar] [CrossRef] [PubMed]
- Greaney, A.J.; Loes, A.N.; Crawford, K.H.; Starr, T.N.; Malone, K.D.; Chu, H.Y.; Bloom, J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 2021, 29, 463–476.e6. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Schmidt, F.; Weisblum, Y.; Muecksch, F.; Barnes, C.O.; Finkin, S.; Schaefer-Babajew, D.; Cipolla, M.; Gaebler, C.; Lieberman, J.A.; et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021, 592, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.M.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef]
- Andreano, E.; Piccini, G.; Licastro, D.; Casalino, L.; Johnson, N.V.; Paciello, I.; Dal Monego, S.; Pantano, E.; Manganaro, N.; Manenti, A.; et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv 2020. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef]
- Moghadasi, S.A.; Heilmann, E.; Moraes, S.N.; Kearns, F.L.; von Laer, D.; Amaro, R.E.; Harris, R.S. Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors. bioRxiv 2022. [Google Scholar] [CrossRef]
- Lee, J.T.; Yang, Q.; Gribenko, A.; Perrin, B.S., Jr.; Zhu, Y.; Cardin, R.; Liberator, P.A.; Anderson, A.S.; Hao, L. Genetic Surveillance of SARS-CoV-2 M(pro) Reveals High Sequence and Structural Conservation Prior to the Introduction of Protease Inhibitor Paxlovid. mBio 2022, 13, e0086922. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Shayestehpour, M.; Mirzaei, H. The impact of spike mutated variants of SARS-CoV2 [Alpha, Beta, Gamma, Delta, and Lambda] on the efficacy of subunit recombinant vaccines. Braz. J. Infect. Dis. 2021, 25, 101606. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, Z.; Chen, Z.; Huang, X.; Xu, M.; He, T.; Zhang, Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020, 92, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Burki, T. Understanding variants of SARS-CoV-2. Lancet 2021, 397, 462. [Google Scholar] [CrossRef]
- Khan, K.A.; Cheung, P. Presence of mismatches between diagnostic PCR assays and coronavirus SARS-CoV-2 genome. R. Soc. Open Sci. 2020, 7, 200636. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).